Abstract
Staufen1 (STAU1) and Lin28B are RNA-binding proteins that are involved in neuronal differentiation as a function of post-transcriptional regulation. STAU1 triggers post-transcriptional regulation, including mRNA export, mRNA relocation, translation and mRNA decay. Lin28B also has multiple functions in miRNA biogenesis and the regulation of translation. Here, we examined the connection between STAU1 and Lin28B and found that Lin28B regulates the abundance of STAU1 mRNA via miRNA maturation. Decreases in the expression of both STAU1 and Lin28B were observed during neuronal differentiation. Depletion of STAU1 or Lin28B inhibited neuronal differentiation, and overexpression of STAU1 or Lin28B enhanced neuronal differentiation. Interestingly, the stability of STAU1 mRNA was modulated by miR-142-3p, whose maturation was regulated by Lin28B. Thus, miR-142-3p expression increased as Lin28B expression decreased during differentiation, leading to the reduction of STAU1 expression. The transcriptome from Staufen-mediated mRNA decay (SMD) targets during differentiation was analyzed, confirming that STAU1 was a key factor in neuronal differentiation. In support of this finding, regulation of STAU1 expression in mouse neural precursor cells had the same effects on neuronal differentiation as it did in human neuroblastoma cells. These results revealed the collaboration of two RNA-binding proteins, STAU1 and Lin28B, as a regulatory mechanism in neuronal differentiation.
Similar content being viewed by others
Main
Post-transcriptional regulation processes include mRNA export, RNA stability, translation involving the regulation of cell metabolism, cell fate determination, tumorigenesis and tissue regeneration through microRNA (miRNA), nonsense-mediated mRNA decay (NMD), and Staufen-mediated mRNA decay (SMD).1, 2, 3 The RNA binding proteins Lin28A and Lin28B have roles in metabolism, development, and cell growth in multiple mammalian cell types and also affect miRNA biogenesis.4, 5 Both have conserved RNA-binding motifs, including a cold-shock domain (CSD) and a Cys-Cys-His-Cys (CCHC) domain.6, 7 Furthermore, the maturation of let-7 is negatively correlated with the expression of Lin28A/Lin28B during embryonic stem cell differentiation; that is, an increase in the level of mature let-7 family members occurs in response to a decrease in Lin28A/Lin28B.4, 8, 9, 10 The main motif of miRNA and mRNA that binds Lin28, revealed by crosslinking immunoprecipitation (CLIP)-seq, is the ‘GGAG’ sequence in the hairpin loop structure of mRNA and pre-miRNA.11, 12 Lin28A also inhibits the biogenesis of miR-9 by binding to pre-miR-9 during neuronal differentiation in the mouse embryonic carcinoma cell line P19, ultimately decreasing differentiation.13 Furthermore, overexpression of Lin28A/Lin28B in mouse P19 cells induces neurogenesis with blocks of gliogenesis without affecting proliferation.14 The binding of Lin28A/Lin28B to pre- or pri-let-7 regulates glucose metabolism via the insulin-PI3K-mTOR pathway, and Lin28A enhances tissue repair by reprogramming.9, 15
Staufen1 (STAU1) is a double-stranded RNA binding protein first identified in Drosophila oogenesis and CNS development and recognizes intra- and intermolecular base-pairing with long noncoding RNA in the 3′UTR.16, 17 Binding of STAU1 to RNA initiates neuronal morphogenesis and post-transcriptional regulation, including mRNA export, relocation, translation, and mRNA decay (known as SMD);18 SMD is also involved in developmental processes such as myogenesis and adipogenesis and is likely to be involved in angiogenesis.19, 20, 21 STAU1 is distinct in neurons because it is transported to dendrites in RNA granules.22 Knockout of STAU1 in mice hippocampal neurons reduced dendritic branching and STAU1-RNP delivery,23 and downregulation of STAU1 reduced the dendritic length of secondary to quaternary branches during differentiation.24
Our hypothesis was based on the common effects and expression patterns of STAU1 and Lin28B in neuronal differentiation. Here, we observed that the levels of STAU1 and Lin28B were diminished during SH-SY5Y cell differentiation. Overexpression and depletion of STAU1/Lin28B augmented and reduced neuronal differentiation, respectively. In addition, we showed that Lin28B inhibits the maturation of miR-142-3p, which destabilizes STAU1 mRNA. Consequently, the decrease in Lin28B levels during neuronal differentiation augmented the level of mature miR-142-3p, ultimately reducing the mRNA levels of the miR-142-3p target, STAU1. In support of this observation, overexpression and inhibition of miR-142-3p reduced and increased the level of cellular STAU1 mRNA, which inhibited and promoted neuronal differentiation, respectively. Genome-wide analysis revealed that the transcripts whose expression was upregulated upon STAU1 and UPF1 depletion during differentiation may play key roles in differentiation. Finally, overexpression of STAU1 in mouse neural precursor cells (mNPCs) caused increased neural differentiation. Taken together, our findings suggest that modulation of STAU1 expression or miR-142-3p function could be used to regulate neuronal differentiation.
Results
STAU1 regulates the differentiation of SH-SY5Y cells
SH-SY5Y cells have been exclusively used to study mechanisms of neuronal differentiation due to the convenient induction of their differentiation by retinoic acid. Immunofluorescence staining results indicated that neuronal morphology gradually developed, exhibiting expanded dendrites and an increased level of synaptophysin (SYP), a presynaptic neuronal marker (Figure 1a). The differentiation was confirmed by using western blotting and quantitative reverse transcription PCR (RT-qPCR), which showed that the levels of SYP and nestin (a marker of undifferentiated cells) increased and decreased, respectively, during differentiation (Figures 1b and c). Intriguingly, protein and mRNA expression of STAU1 decreased to approximately 40% and 61% of the value of the day 0 of differentiation, respectively, during differentiation (Figures 1b and c). The effects of STAU1 expression on neuronal differentiation were determined by transiently transfecting SH-SY5Y cells with siSTAU1 or siControl and inducing the cells to differentiate. Interestingly, depletion of STAU1 reduced the expression of SYP protein and mRNA at day 7 of differentiation to approximately 40% and 60% of the value of the control siRNA transfection, respectively (Figures 1d and e). To ensure that STAU1-depletion had the same effects in differentiating cells, we downregulated the expression of STAU1 in differentiating cells (Supplementary Figures S1a and b). Consistently, depletion of STAU1 in differentiating cells also decreased the expression of SYP, indicating that STAU1 expression plays a key role in differentiation. In SMD, STAU1 binds to structured RNA with UPF1, triggering mRNA decay. If the expression of UPF1 is slightly decreased during differentiation and depletion of UPF1 impairs neuronal differentiation as STAU1 does, SMD may have a role in differentiation. To determine whether UPF1 affected differentiation, we depleted UPF1 in SH-SY5Y cells, which were then differentiated (Supplementary Figure S1c). Western blotting demonstrated that the level of UPF1 decreased during differentiation and that downregulating UPF1 abrogated neuronal differentiation as observed in STAU1-depletion, indicating that SMD may be a key regulator in differentiation. The effects of STAU1-depletion on differentiation were ascertained by overexpressing C-terminal HA-tagged STAU155 (STAU155-HA3) in SH-SY5Y cells and then inducing differentiation (Figures 1f and g). Western blotting and RT-qPCR results demonstrated that overexpression of STAU1 during differentiation increased the expression of SYP compared to expression in mock transfection. Our findings were verified by another approach, immunofluorescence detection (Figure 1h). SYP expression was detected in STAU1-depleted cells and STAU155-HA3-overexpressing cells after 7 days of differentiation, indicating that depleting STAU1 resulted in lower expression of SYP and overexpression of STAU1 led to higher expression of SYP than in mock transfected cells (Figure 1h). To determine the effects of STAU1 on neuronal differentiation, we measured dendritic lengths during differentiation (Figure 1i) and in the presence or absence of siSTAU1 or overexpressed STAU155-HA3 at day 7 of differentiation (Figure 1j). Dendritic lengths increased during differentiation, and depletion or overexpression of STAU1 decreased or increased dendritic lengths, respectively. The simple interpretation of all of these results was that SMD activity might naturally decrease in response to the decreased amounts of STAU1 and UPF1 during differentiation and that inhibition of SMD repressed neuronal differentiation (see more implications in the Discussion section). The evidence described above indicates that STAU1 expression affected neuronal differentiation.
STAU1 expression affects neuronal differentiation in SH-SY5Y cells. (a) Differentiation of SH-SY5Y cells was induced by adding 50 μM retinoic acid for the indicated number of days. The nucleus (DAPI, blue) and the expression of the presynaptic marker, synaptophysin (SYP, green), were visualized by immunofluorescence microscopy. (b,c) As (a), except that protein and gene expressions were determined by western blot (b) and RT-qPCR (c). (d and e) SH-SY5Y cells that were transiently transfected with STAU1-specific siRNA or control siRNA were differentiated by the addition of retinoic acid. The number of the column is the mean value compared to that of day 0 of differentiation. (f,g) SH-SY5Y cells were transiently transfected with C-terminally HA-tagged STAU155 (STAU155-HA3) or an empty vector (–) as a control, and then differentiation was induced. The expression of STAU1 and SYP during the differentiation period was detected by western blot (d,f) and RT-qPCR (e and g). (h) Immunofluorescence was used to visualize SYP and DAPI at day 7 of differentiation with or without depletion of STAU1 (siSTAU1) or overexpression of STAU1 (STAU155-HA3). (i) The dendritic lengths in (a) at the indicated days were measured during differentiation (n=50). (j) Similar to (i), except that the dendritic length was measured at day 7 of differentiation in STAU1-depleted or STAU155-HA3-overexpressing SH-SY5Y cells in (h) (n=60). The protein level was normalized to the level of calnexin, and the relative levels of proteins at day 0 of differentiation were set to 1.0. The mRNA level was normalized to the level of U6 snRNA, and the relative levels of mRNAs at day 0 of differentiation were set to 1.0. Insets in (a) and (h) represent the higher magnification of the indicated field. **P<0.01
miR-142-3p reduces the level of STAU1 mRNA, inhibiting neuronal differentiation
STAU1 expression was reduced according to initiation of differentiation. To identify miRNAs that target STAU1 during differentiation, we performed an in silico screen using TargetScan (www.targetscan.org). We identified miR-142 (both miR-142-5p and miR-142-3p) as putative regulators of STAU1. To determine whether miR-142-5p or miR-142-3p regulated STAU1, we transfected HeLa cells, which do not express constitutively express miR-142 (Figure 2a),25 with miR-142-5p or miR-142-3p mimics and compared the levels of STAU1 protein and mRNA with the values in control miRNA-transfected cells (Figures 2b and c). Western blotting and RT-qPCR results demonstrated that both miR-142-5p and miR-142-3p decreased STAU1 protein and mRNA levels. Because the expression of miR-142-3p increased during differentiation, but that of miR-142-5p did not, we hypothesized that miR-142-3p could be a key factor in reducing the abundance of STAU1 mRNA (see below). To determine whether miR-142-3p targeted STAU1, we inserted the putative target sequence (WT) or the mutated sequence (Mut, lacking the miR-142-3p-binding site), along with the surrounding STAU1 3′UTR, into a bicistronic reporter vector (Figure 2d). RT-qPCR was performed using HeLa cells that were cotransfected with a miR-142-3p mimic and STAU1-3′UTR-WT or STAU1-3′UTR-Mut. The presence of miR-142-3p reduced the level of the transcripts containing STAU1-3′UTR-WT by approximately two-fold but did not have any effect on STAU1-3′UTR-Mut (Figure 2e). Notably, the miR-142-3p binding sites in the 3′UTR of STAU1 are conserved among mammals (Supplementary Figure S2a). These results indicated that STAU1 was a target of miR-142-3p.
miR-142-3p targets STAU1. (a) Endogenous miRNAs in HeLa cells were quantified by semi-quantitative RT-PCR (RT-sqPCR), as described in the Materials and Methods section, to compare the levels of human mature miRNAs. (b,c) HeLa cells that did not express miR-142-3p were transiently transfected with miR-142-3p mimic, miR-142-5p mimic, or the control miRNA mimic. Western blot (b) and RT-qPCR (c) were performed to detect and measure the levels of endogenous STAU1 protein and mRNA, respectively. The level of STAU1 mRNA was normalized to the level of GAPDH mRNA. (d) Map of a representative bicistronic Firefly/Renilla luciferase (FLuc/RLuc) plasmid containing STAU1 mRNA 3′UTRs that had putative miR-142-3p binding sites (WT) or mutated binding sites (Mut). (e) An miR-142-3p mimic or control miRNA mimic was cotransfected with STAU1-3′UTR-WT (left panel) or STAU1-3′UTR-Mut (right panel) into HeLa cells. RT-qPCR was performed to quantify the relative amount of FLuc mRNA normalized to the level of RLuc mRNA. The relative level of mRNAs in control miRNA-transfected cells was set to 1.0. **P<0.01; n.s., not significant
All of these findings prompted us to test whether miR-142-3p was upregulated to decrease the level of STAU1 expression during neuronal differentiation. SH-SY5Y cells were differentiated, and the changes in cellular let-7a, miR-142-5p and miR-142-3p were quantified by RT-qPCR (Figure 3a). The level of let-7a, which is well known to be upregulated by embryonic and neuronal stem cell differentiation,9, 10, 15 increased during differentiation. Similarly, miR-142-3p was upregulated approximately 2.3-fold, suggesting that the increase in miR-142-3p expression might have downregulated STAU1 mRNA. Indeed, transfection with an miR-142-3p mimic decreased the level of STAU1 protein and mRNA and decreased SYP expression at day 7 of differentiation (Figures 3b and c). To confirm whether miR-142-3p affects differentiation, we transfected an miR-142-3p inhibitor into SH-SY5Y cells at day 5 of differentiation, which was followed by more differentiation because the level of miR-142-3p increased as differentiation processed (Figures 3d and e). Western blotting and RT-qPCR results demonstrated that the inhibitor augmented the levels of STAU1 protein and mRNA. The levels of SYP protein and mRNA at day 7 of differentiation in inhibitor transfected cells were also increased more than the values in control RNA transfected cells, as expected. All these observations suggested that the decrease in STAU1 expression that was caused by the increase in miR-142-3p reduced neuronal differentiation.
miR-142-3p regulates neuronal differentiation, presumably via reducing STAU1 expression in SH-SY5Y cells. (a) Mature miRNAs during differentiation in SH-SY5Y cells were quantified by using RT-qPCR. The relative levels of RNAs at day 0 of differentiation were set to 1.0. (b,c) SH-SY5Y cells that were transfected with an miR-142-3p mimic or a control miRNA mimic were differentiated. (d,e) SH-SY5Y cells at day 5 of differentiation that were transfected with an miR-142-3p inhibitor or a control miRNA inhibitor were differentiated for 2 days. Cell lysates were employed in western blots (b,d) and RT-qPCRs (c,e). The relative levels of miRNAs and mRNAs were normalized to the level of U6 snRNA. *P<0.05, **P<0.01
Lin28B regulates mature miR-142-3p stability
Lin28 is one of the proteins that is most studied for its decreased expression during differentiation. Lin28 binds to the ‘GGAG’ motif of pre-miRNA, leading to degradation of miRNAs.11, 13 Intriguingly, pre-miR-142, containing a putative ‘GGAG’ motif in its pre-miRNA terminal loop, is conserved in human, rat, and mouse. Moreover, the STAU1 3′UTR harbors a conserved miR-142-3p binding site (Figure 4a). Notably, SH-SY5Y cells constitutively express Lin28B and the level of Lin28B was reduced during differentiation (Figure 4b). It is thus plausible that Lin28B is capable of regulating the fate of miR-142-3p maturation, leading to an increase in miR-142-3p and a decrease in STAU1. As a first step towards evaluating whether Lin28B regulates maturation of miR-142-3p, we employed HeLa cells that did not constitutively express Lin28B and miR-142. FLAG-Lin28B-WT, FLAG-Lin28B-mCCHC (two mutations in the CCHC domains), or an empty vector were cotransfected with pre-miR-142-WT or pre-miR-142-Mut into HeLa cells; pre-miR142-Mut lost its ‘GGAG’ motif and a bulge in the stem, as depicted in Supplementary Figure S2b. Western blot verified that comparable levels of the FLAG-Lin28B variants were expressed (Figure 4c). Expression of FLAG-Lin28B-WT slightly reduced the levels of let-7a and miR-142-3p, as expected (Figure 4d). In contrast, the expression of FLAG-Lin28B-mCCHC did not significantly change the levels of miRNA. Protein-pre-miRNA binding was examined to ascertain whether Lin28B binds to pre-miR-142 (Figure 4e). An anti-FLAG antibody was used to immunopurify FLAG-Lin28B variants from lysates of HeLa cells that had been transiently cotransfected with FLAG-Lin28B-WT, FLAG-Lin28B-mCCHC, or an empty vector with or without pre-miR142-WT or pre-miR142-Mut. Comparable levels of immunoprecipitated FLAG-Lin28B-WT and FLAG-Lin28B-mCCHC were observed by western blot (Figure 4e, left panel). RT-qPCR results demonstrated that FLAG-Lin28B-WT formed mRNP 1.5-fold more effectively with pre-miR-142-WT than with pre-miR-142-Mut (Figure 4e, right panel). However, FLAG-Lin28B-mCCHC did not efficiently interact with either pre-miR-142-WT or pre-miR-142-Mut. All these results suggested that Lin28B could regulate miR-142-3p maturation and form mRNP with pre-miR-142.
Lin28B regulates the level of STAU1 via miR-142-3p maturation. (a) A diagram of the RNA structures of conserved human, rat, and mouse pre-miR-142 predicted by mFold using Gibbs free energy (ΔG). Red bold sequences indicate the putative Lin28B binding sites (GGAG), and bold black sequences are the mature miR-142-3p (left panel). The orange box indicates the conserved miR-142-3p binding sites in the human, rat, and mouse STAU1 3’UTRs (right panel). (b) Western blots indicate proteins in various cell lines. SH-SY5Y cells were differentiated for 7 days. (c and d) HeLa cells that were transiently transfected with the FLAG-Lin28B-WT (WT), the FLAG-Lin28B CCHC-mutant (mCCHC), or an empty vector (–) were cotransfected with pre-miR-142-WT or pre-miR-142-Mut. Western blot (c) and RT-qPCR (d) were used to detect the expression of FLAG-Lin28B and to quantify the level of the indicated miRNAs. The levels of miR-142-3p and let-7a were normalized to the levels of exogenous pre-miR-142 and U6 snRNA, respectively. (e) Similar to (c) and (d) except that FLAG-Lin28B-WT or -mCCHC was immunoprecipitated with anti-FLAG antibody-conjugated beads. Transfection of pFLAG (empty vector) served as a negative control (–). Western blot was performed to test whether a comparable amount of immunoprecipitated FLAG-Lin28B was present. Coimmunoprecipitated pre-miR-142 was spiked with E. coli RNA after IP. RT-qPCR was employed to quantify the relative levels of pre-miR-142 and pre-let-7a, and its results were normalized to the level of E. coli LACZ mRNA to account for variations in RNA recovery. (f,g) Depletion of Lin28B by siRNA-mediated downregulation was performed in SH-SY5Y cells. Western blot (f) and RT-qPCR (g) were employed to evaluate downregulation of Lin28B and the relative levels of mature miRNAs, pri-miRNAs, and miRNA-targeted mRNAs. (h) SH-SY5Y cells that were transfected with siControl or siLin28B were treated with 40 μg/ml 5,6-dichloro-1-β-D-ribofuranosylbenzimidazole (DRB) for the indicated duration. RT-qPCR was performed to obtain relative levels of STAU1 mRNA (normalized to the level of GAPDH mRNA). Mean values and standard errors were calculated from at least six independently performed transfections. t1/2, half-life. (i) SH-SY5Y cells that were transfected with FLAG-Lin28B were differentiated using retinoic acid. A western blot was performed to observe the indicated proteins. (j) Similar to (i) except that the endogenous Lin28B was depleted instead of FLAG-Lin28B being overexpressed. The number of the column is a mean value relative to results from mock or Lin28B-WT transfections. *P<0.05, **P<0.01; n.s., not significant
To determine whether Lin28B reduced the levels of mature miR-142-3p, Lin28B was depleted by siRNA-mediated downregulation without differentiation. Western blot results demonstrated that the expression of Lin28B was efficiently downregulated (Figure 4f). While Lin28B levels were depleted in SH-SY5Y cells, the levels of mature miRNA, pri-miRNA, and miRNA targets were quantified by RT-qPCR (Figure 4g). Depletion of Lin28B increased the level of mature miR-142-3p, while the level of let-7a was also slightly derepressed (Figure 4g). Notably, the pri-miRNA level was not significantly changed by downregulation of Lin28B. However, as a result of the increase in let-7a and miR-142-3p, the levels of HMGA2 and STAU1 mRNA were reduced to 80% of the values in the control (Figure 4g), indicating that the expression of Lin28B could regulate the level of mature miR-142-3p, ultimately leading to the modulation of STAU1 mRNA stability. To test whether STAU1 was a target of Lin28B, we depleted Lin28B and analyzed the half-life of endogenous STAU1 mRNA by blocking transcription and using RT-qPCR; the half-life of STAU1 mRNA was reduced by approximately 50% (Figure 4h).
Lin28B is a key regulator in STAU1 mRNA stability because it regulates miR-142-3p abundance; we thus investigated whether a change in Lin28B expression affected neuronal differentiation. Lin28B was overexpressed (Figure 4i) or depleted (Figure 4j), and induction of differentiation followed. Overexpression of FLAG-Lin28B exclusively enhanced the expression of SYP, suggesting that Lin28B played a key role in the differentiation of SH-SY5Y cells, as it also does in ES and neuronal precursor cells (Figure 4i). In contrast, depletion of Lin28B significantly reduced the level of SYP as differentiation proceeded (Figure 4j). All these results indicated that Lin28B positively regulated STAU1 expression during differentiation.
Because SH-SY5Y cells can grow once differentiation starts, cell growth after differentiation induction may affect the differentiation process. In addition, overexpression of STAU1 was recently reported to impair cell proliferation in HCT116 cells.26 To examine the effects of STAU1 and Lin28B expression on proliferation and, consequently, differentiation, we either overexpressed or depleted STAU1 or Lin28B and assessed cell growth by cell counting (Supplementary Figure S3). Western blot results demonstrated that overexpression and depletion of STAU1 and Lin28B were maintained until day 5 of proliferation (left panels in Supplementary Figures S3a–d); however, the level of overexpressed STAU155-HA3 at day 5 was similar to the level in mock transfected cells. Cell counting results indicated that the overexpression of Lin28B or depletion of STAU1 or Lin28B had no effect on cell growth. However, similar to a previous report,26 overexpression of STAU155-HA3 repressed cell growth at day 3 of proliferation, but did not alter cell growth at day 5 of proliferation, presumably because overexpression was diminished from the day-3 proliferation. Cell growth tests indicated that overexpression of STAU1 may affect cell metabolism, which would explain the reason that the overexpression of STAU155-HA3 increased the level of SYP by only 1.5-fold, in contrast with the results from STAU1-depletion (Figures 1d–g).
SMD elicits widespread upregulation of transcripts in SH-SY5Y cells
STAU1 is a key factor in SMD via binding to RNA with UPF1. Several lines of evidence suggested that SMD could regulate neuronal differentiation: (1) The expression of both endogenous STAU1 and UPF1 was reduced during differentiation, and (2) Downregulation of STAU1 or UPF1 decreased neuronal differentiation. All of these findings prompted us to determine which transcripts were regulated by SMD. Thus, mRNA-seq experiments were performed using lysates of UPF1- or STAU1-depleted SH-SY5Y cells. The specific downregulation of UPF1 or STAU1 was confirmed by western blot (Figure 5a). Comparative analysis using mRNA seq results revealed that 1253 and 1301 transcripts were upregulated at least twofold upon STAU1- and UPF1-depletion, respectively, and 480 transcripts were commonly upregulated (Figure 5b and Supplementary Table S1). We determined potential SMD targets by elucidating enrichment by using Gene Ontology (GO) analysis (Figure 5c). The GO processes that were commonly upregulated upon UPF1 and STAU1 depletion were morphogenesis, cell development and regulation of transport, suggesting that SMD could be involved in neuronal differentiation (Figure 5c). Moreover, because the levels of STAU1 and UPF1 were reduced during differentiation, the levels of SMD-targeted transcripts were supposed to increase, which may augment or impair neuronal differentiation. These speculations drove us to determine which transcripts were upregulated by differentiation and depletion of UPF1 and STAU1. Therefore, mRNA-seq experiments were performed using SH-SY5Y cells that had been differentiated for 7 days, compared to experiments with undifferentiated control cells. A total of 2784 transcripts were upregulated upon induction of differentiation, and 5% of these transcripts (146) were commonly upregulated with both UPF1 and STAU1 depletion (Figure 5d and Supplementary Table S1). Indeed, GO analysis demonstrated that the gene sets involved in morphogenesis and the regulation of transport were upregulated, suggesting that SMD may play a role in neuronal differentiation (Figure 5e and Supplementary Table S2). To assess whether the transcripts that were upregulated upon UPF1 and STAU1 depletion during differentiation were SMD targets, we coimmunoprecipitated transcripts with STAU155-HA3. Western blotting results indicated that IP was specific (Figure 5f). Sixteen out of the eighteen transcripts that had been upregulated upon STAU-depletion or formed mRNP complex with STAU1 in previous studies17, 27, 28 were coimmunoprecipitated with STAU155-HA3 compared to IP of mock transfection samples. Well-known SMD targets21 c-Jun and IL-7R were coimmunoprecipitated, and the negative control transcript ZNF75D (only upregulated upon UPF1-depletion) was not significantly coimmunoprecipitated (Figure 5g). Furthermore, the transcripts related to differentiation also coimmunoprecipitated with STAU155-HA3 (Figure 5h). These observations suggested that the level of STAU1 expression could modulate neuronal differentiation.
Transcriptome analysis demonstrating that SMD regulates neuronal differentiation. (a) The expression of UPF1 or STAU1 was downregulated by siRNA-mediated silencing in SH-SY5Y cells. Western blot was employed to evaluate the downregulation of UPF1 and STAU1. (b) Comparative analysis of gene expression profiling using total RNAs purified from (a). Transcripts upregulated more than twofold were selected for further analysis. The numbers in parentheses indicate the numbers of upregulated transcripts upon downregulation of UPF1 or STAU1. (c) Gene ontology analysis was performed from commonly upregulated transcripts in (b) using the DAVID and Medline databases. (d) Comparative analysis of gene expression profiling using total RNAs from differentiated and undifferentiated SH-SY5Y cells for 7 days. Transcripts upregulated more than twofold were chosen for analysis. Venn diagrams show the number of transcripts that were commonly upregulated upon the differentiation and downregulation of UPF1 or STAU1. (e) Same as (c) except that the commonly upregulated transcripts in (d) were analyzed. (f) SH-SY5Y cells were transiently transfected with STAU155-HA3 or an empty vector (–), and immunoprecipitation was performed using anti-HA-conjugated beads. Western blotting was performed to detect immunoprecipitated STAU155-HA3. (g and h) Immunoprecipitated samples in (f) were spiked with a small amount of E. coli RNA, and RT-qPCR measurements of the level of E. coli LACZ mRNA were employed to control for variations in RNA recovery. Values from immunoprecipitation using mock transfection were set to 1.0. *P<0.05, **P<0.01; n.s., not significant
STAU1 regulates differentiation of mNPCs
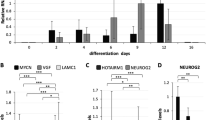
Next, mNPCs from the mouse cortex were employed to assess whether molecular regulation among Lin28B, miR-142-3p and STAU1 also controls mouse neural differentiation. Western blot and RT-qPCR results demonstrated that decreases in mouse Lin28B (mLin28B), mSTAU1, and mUPF1 and an increase in mSYP also occurred in mNPC differentiation (Figures 6a and b). Furthermore, the expression levels of mmu-let-7a and mmu-miR-142-3p miRNAs increased with differentiation, suggesting that the Lin28B:miR-142-3p:STAU1 regulation network might be used in the differentiation of mNPCs. We ascertained the function of STAU1 in mNPCs by using retroviral infection to overexpress human STAU155-HA3 in mNPCs and then inducing differentiation. The results of experiments using mNPCs were similar to those of SH-SY5Y cells. Overexpression of human STAU155-HA3 increased the level of mSYP by approximately twofold at day 12 of differentiation (Figure 6c). This increased amount of SYP was verified by immunofluorescence analysis (Figure 6d), suggesting that regulation of STAU1 enhanced mouse neural development.
STAU1 regulates differentiation of mNPCs. (a,b) mNPCs from cortex at E12.5 were differentiated for the indicated number of days. Western blot (a) and RT-qPCR (b) were performed to detect specific mouse (m) proteins (blot) and quantify the endogenous mRNAs and miRNAs (RT-qPCR). The relative levels of miRNAs and mRNAs were normalized to those of U6 snRNA and mGAPDH mRNA, respectively. The relative levels of RNAs at day 4 of differentiation were set to 1.0 in (b). **P<0.01 and *P<0.05. (c) STAU1 expression was upregulated by infection with a retrovirus expressing STAU155-HA3 and GFP or GFP alone during the differentiation of mNPCs. Western blot was performed to detect expression of the indicated proteins. (d) Same as (c) except that SYP and nuclear (DAPI) staining at day 12 of differentiation were observed under an immunofluorescence microscope. (e) The suggested model illustrates the sequential regulation of Lin28B:miR-142-3p:STAU1 mRNA during neuronal differentiation
Discussion
STAU1 has multiple roles in cellular processes, including mRNA relocation, mRNA surveillance, and translation,29, 30 by recognizing and binding to RNA secondary structures in proper neuronal function.31, 32, 33 In Caenorhabditis elegans, RNA immunoprecipitation revealed that STAU1 forms mRNP with transcripts related to cell differentiation,34 which was consistent with our observation that SMD regulated the stability of transcripts involved in neuronal differentiation (Figure 5). However, we could not exclude the possibility that the fates of transcripts were regulated by SMD or other functions of STAU1 during neuronal differentiation because STAU1 colocalized with translation-related proteins and mRNAs in dendrites,22 which could stabilize or destabilize mRNA. The expression of truncated STAU1 in mouse hippocampal neurons from homozygous mutants led to reduced dendritic trees that had less developed synapses.23 In addition, the downregulation of STAU1 in hippocampal pyramidal cells modified their dendritic spine morphology, decreasing the number of regular type dendritic spines and increasing the number of filopodium type dendritic spines35 and depletion of STAU1 reduced neuronal differentiation by diminishing the association of STAU1 with several miRNAs, leading to translational repression and a reduction of dendritic length.24 STAU2, a paralog of STAU1, also plays an important role in neurons, including being involved in mRNA localization and neuronal mRNA regulation.36, 37 Interestingly, the 3′UTR of STAU2 contains putative miR-142-3p binding sites, as STAU1 does, suggesting that miR-142-3p is also capable of modulating STAU2 expression; this possibility aligns well with our findings. Both STAU1- and STAU2-mediated mRNA decays required UPF1,38 which is also a key factor in NMD. The UPF1 level was repressed during differentiation of mouse neuronal stem cells, resulting in the stabilization of NMD targets.39 Similarly, the level of UPF1 during SH-SY5Y cell differentiation was also diminished, and depletion of UPF1 repressed neuronal differentiation (Supplementary Figure S1c).
STAU1 expression was regulated by miR-142-3p, whose expression increased with the decrease in Lin28B levels during differentiation. Lin28B also plays an important role in neurogenesis. For instance, Lin28B overexpression led to increased neurogenesis and decreased gliogenesis in mouse embryonic carcinoma cells.14 Moreover, Lin28B induced the formation of neuroblastomas with decreased let-7.40 Lin28B also repressed the maturation of the let-7 family members, which contained the same ‘GGAG’ motif in the pre-miRNA. Indeed, our findings showed that Lin28B could bind to pre-miR-142 and repress its maturation (Figure 4). The results in this study supported previous reports of the function of STAU1 in neurogenesis via several lines of evidence: (1) overexpression and downregulation of STAU1 increased and decreased neuronal differentiation, respectively (Figure 1), (2) miR-142-3p, which repressed STAU1 expression, had the opposite consequence for STAU1 regulation in neuronal differentiation (Figures 2 and 3), (3) genome-wide analysis showed that transcripts upregulated by UPF1 and STAU1 were involved in neuronal differentiation (Figure 5), and (4) modulation of STAU1 expression in mNPCs affected differentiation (Figure 6).
The agreement between our observations and those of previous reports suggests the simple network model depicted in Figure 6e. Because Lin28B inhibits miR-142-3p maturation, the level of miR-142-3p increased as the level of Lin28B decreased during differentiation. Consequently, the expression of STAU1, a target of miR-142-3p, was repressed during neuronal differentiation. However, the reason that inhibition of Lin28B or STAU1 expression or augmentation of miR-142-3p expression reduces neuronal differentiation is unknown. A plausible explanation could be that a decrease in the activity of SMD by siSTAU1 or siUPF1 increased the level of SMD targets that reduced neuronal differentiation, transport and the response of neurons to the environment, which might have reduced neuronal differentiation, that is, the function of SMD may be required for induction or maintenance of differentiation process. Our findings suggest that STAU1 controls neurogenesis via the modulation of gene expression. The effects of SMD on functional regulation and networks involved in neurogenesis should be further investigated.
Materials and methods
Cell culture and differentiation
HeLa and SH-SY5Y cells were grown in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin. Human neural precursor cells (hNPCs) were maintained in serum-free ITS medium in the presence of 20 ng/ml basic fibroblast growth factor (bFGF) and ascorbic acid (AA) on poly-L-ornithine (PLO)/fibronectin (FN)-coated culture dishes.41 To induce differentiation of SH-SY5Y cells, cells were seeded 1 day before treatment with 50 μM of retinoic acid. mNPCs that were isolated from the cortex at E12.5 were cultured in vitro for 3 days in serum-free N2 medium supplemented with the mitogens bFGF and epithelial growth factor. Differentiation of mouse NPCs was achieved by removing the mitogens.42
HeLa and SH-SY5Y cells were seeded 1 day before transfection. HeLa cells were transiently transfected with plasmid DNA or miRNA using Lipofectamine 3000 transfection reagent (Invitrogen, Waltham, MA, USA). SH-SY5Y cells were electroporated with plasmid DNA, siRNA, or an miRNA mimic using Nucleofector (Amaxa, Alpharetta, GA, USA) as described in the manufacturer’s protocol.
Plasmid construction
We constructed pmirGLO-STAU1-3′UTR WT and pmirGLO-STAU1-3′UTR Mut by digesting a pmirGLO plasmid (Promega, Madison, WI, USA) with XbaI and ligating the digested vector fragment to PCR-amplified fragments that had also been digested with XbaI. The PCR products were generated by annealing the three primers STAU1_3′UTR-5′, STAU1_3′UTR_middle-WT or STAU1_3′UTR_middle-Mutant which has the mutated sequences indicated in Figure 2d, and STAU1_3′UTR-3′ and amplifying using the two primers STAU1_3′UTR-XbaI-F and STAU1_3′UTR-XbaI-R, which are listed in Supplementary Table S3.
The C-terminally HA-tagged pEGFP-IRES3-STAU1-HA3 was constructed by digesting pEGFP-IRES with XhoI and ligating the resulting vector fragment to a PCR product that had also been digested with XhoI. The PCR product was generated using pCI-Neo-STAU1-HA3 and the two primers STAU1_XhoI-F and STAU1_XhoI-R, which are listed in Supplementary Table S3.
Whether pre-miR-142 is bound by Lin28B was determined by inserting the pre-miR-142-WT or pre-miR-142-Mut sequences into a pcDNA3.1 vector. The pcDNA3.1 vector was digested with BamHI and XhoI, and the resulting vector fragment was ligated to a Klenow product that had also been digested with BamHI and XhoI. The Klenow product was filled using two annealed primers, which were named pre-miR-142_BamHI-F-WT and pre-miR-142_XhoI-R-WT for WT and pre-miR-142_BamHI-F-Mut and pre-miR-142_XhoI-R-Mut for Mut, as listed in Supplementary Table S3.
Retrovirus production
The overexpression of C-terminally HA3-tagged STAU1 was achieved by using a STAU155-HA3-overexpressing retrovirus, which was obtained as previously described.43 Briefly, 293GPG packaging cells were transiently transfected with pEGFP-IRES3-STAU155-HA3, and after two days, medium was collected every day for 7 days by centrifuging the cells at 900 × g for 3 min. The resulting supernatant, which contained the retroviral particles, was used as an inoculum.
siRNA-mediated silencing
siRNA (IDT Technologies, Coralville, IA, USA) with the sequences listed in Supplementary Table S3 was electroporated into SH-SY5Y cells using Nucleofector(Amaxa, Alpharetta, GA, USA), based on the manufacturer’s protocol, to downregulate endogenous proteins, including STAU1, Lin28B, and UPF1. Control siRNA (siControl, IDT Technologies, Coralville, IA, USA) was used as a negative control.
Western blot
Cell lysates were separated by electrophoresis on 8–12% sodium dodecyl sulfate-polyacrylamide gels and transferred to nitrocellulose membranes (GE Healthcare Life Sciences, Amersham, Buckinghamshire, UK). Blots were probed with antibodies against the following proteins: STAU1 (Abcam, Cambridge, Cambridgeshire, UK), SYP (BD Biosciences, Baltimore, MD, USA), calnexin (Santa Cruz Biotechnology, Dallas, TX, USA), Lin28A (Cell Signaling, Danvers, MA, USA), Lin28B (Cell Signaling), mouse Lin28B (Proteintech group, Rosemont, IL, USA), UPF1 (Cell Signaling), nestin (Millipore, Darmstadt, Hessen, Germany), HA (Santa Cruz Biotechnology, Dallas, TX, USA), FLAG (GenScript, Piscataway, NJ, USA) and actin (Sigma, St. Louis, MO, USA). All antibodies except those of Lin28B are immunoreactive with human and mouse protein.
Immunostaining
To detect endogenous SYP, we fixed cells with 4% formaldehyde in 1 × PBS containing 0.2% saponin and then stained with a primary antibody against SYP and a secondary antibody labeled with Alexa Fluor 488 (for human SYP) or Cy3 (for mouse SYP) (Life Technologies, Carlsbad, CA, USA). Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI, Vector, Burlingame, CA, USA). Immunofluorescence images were observed using a fluorescence microscope (LEICA CTR5000, Mannheim, Baden-Württemberg, Germany).
RT-qPCR
To quantify the levels of mRNA and pri-miRNA, we performed RT-qPCR as previously described.44 The primers used for PCR are listed in Supplementary Table S3. The level of miRNA was measured using TaqMan PCR Master Mix (Thermo Fisher Scientific, Foster City, CA, USA). To detect the level of endogenous miRNA in HeLa cells, we added a poly(A) tail to miRNA by using poly(A) polymerase and then added an oligo-dT adaptor by using the QuantiMir kit (SBI, System Biosciences, Palo Alto, CA, USA). Semi-quantitative PCR (sqPCR) was then performed using the primers listed in Supplementary Table S3.
Immunoprecipitation
Cell lysates were obtained from HeLa cells that had been transiently transfected with pFLAG-Lin28B WT or pFLAG-Lin28B CCHC mutant6 and pcDNA-pre-miR-142-WT or pcDNA-pre-miR-142-Mut. Immunoprecipitation was performed using anti-FLAG beads (Sigma, St. Louis, MO, USA). Similarly, HA-conjugated beads (Pierce, Rockford, IL, USA) were used to immunoprecipitate STAU155-HA3. Coimmunoprecipitated RNA and protein were analyzed by using RT-qPCR and western blot, respectively.
miRNA expression
Human miR-142-5p and miR-142-3p mimics (GenePharma, Shanghai, China) or a control mimic (GenePharma) were electroporated (Amaxa) into SH-SY5Y cells and transfected (Lipofectamine 3000, Invitrogen, Waltham, MA, USA) into HeLa cells to determine whether miR-142-5p or miR-142-3p downregulates STAU1. The miR-142-3p mimic and inhibitor (Dharmacon, Lafayette, CO, USA) sequences are provided in Supplementary Table S3.
Statistical analysis
Unpaired Student’s t-tests were used to calculate the P-values. Differences with P<0.05 or 0.01, as indicated in figure legends, were considered statistically significant. Columns and error bars in figures indicate the mean values and standard errors for at least four independent experiments.
Library preparation and sequencing
Construction of the control and test RNA library was performed using the SENSE 3′ mRNA-Seq Library Prep Kit (Lexogen, Vienna, Austria) according to the manufacturer’s instructions. In brief, 500 ng of total RNA was prepared for each RNA species, an oligo-dT primer containing an Illumina-compatible sequence at its 5′ end was hybridized to the RNA, and reverse transcription was performed. After the RNA template was degraded, second-strand synthesis was initiated with a random primer containing an Illumina-compatible linker sequence at its 5′ end. The double-stranded library was purified by using magnetic beads to remove all reaction components. The library was amplified to add the complete adapter sequences that were required for cluster generation, and the finished library was purified from the PCR components. High-throughput sequencing was performed as single-end 75-sequencing using NextSeq 500 (Illumina, San Diego, CA, USA). The sequencing data have been deposited to the GEO database under accession number GSE94948.
Data analyses
The alignment file was obtained by using the Bowtie2 software tool to map the SENSE 3′ mRNA-Seq reads. The alignment file was used to assemble transcripts and estimate their abundance, and the differential expression of genes was detected by using bedtools and edgeR. The RT (Read Count) data were processed using the global normalization method in Genowiz version 4.0.5.6 (Ocimun Biosolutions, Hyderabad, Telangana, India). Gene classification was based on searches in the DAVID (http://david.abcc.ncifcrf.gov/) and Medline (http://www.ncbi.nlm.nih.gov/) databases.
Accession codes
References
Schoenberg DR, Maquat LE . Regulation of cytoplasmic mRNA decay. Nat Rev Genet 2012; 13: 246–259.
Durand S, Franks TM, Lykke-Andersen J . Hyperphosphorylation amplifies UPF1 activity to resolve stalls in nonsense-mediated mRNA decay. Nat Commun 2016; 7: 12434.
Sharp PA . The centrality of RNA. Cell 2009; 136: 577–580.
Shyh-Chang N, Daley GQ . Lin28: primal regulator of growth and metabolism in stem cells. Cell Stem Cell 2013; 12: 395–406.
Huang Y . A mirror of two faces: Lin28 as a master regulator of both miRNA and mRNA. Wiley Interdiscip Rev RNA 2012; 3: 483–494.
Heo I, Joo C, Kim YK, Ha M, Yoon MJ, Cho J et al. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell 2009; 138: 696–708.
Nam Y, Chen C, Gregory RI, Chou JJ, Sliz P . Molecular basis for interaction of let-7 microRNAs with Lin28. Cell 2011; 147: 1080–1091.
Cimadamore F, Amador-Arjona A, Chen C, Huang CT, Terskikh AV . SOX2-LIN28/let-7 pathway regulates proliferation and neurogenesis in neural precursors. Proc Natl Acad Sci USA 2013; 110: E3017–E3026.
Zhu H, Shyh-Chang N, Segre AV, Shinoda G, Shah SP, Einhorn WS et al. The Lin28/let-7 axis regulates glucose metabolism. Cell 2011; 147: 81–94.
Viswanathan SR, Daley GQ, Gregory RI . Selective blockade of microRNA processing by Lin28. Science 2008; 320: 97–100.
Cho J, Chang H, Kwon SC, Kim B, Kim Y, Choe J et al. LIN28A is a suppressor of ER-associated translation in embryonic stem cells. Cell 2012; 151: 765–777.
Wilbert ML, Huelga SC, Kapeli K, Stark TJ, Liang TY, Chen SX et al. LIN28 binds messenger RNAs at GGAGA motifs and regulates splicing factor abundance. Mol Cell 2012; 48: 195–206.
Nowak JS, Choudhury NR, de Lima Alves F, Rappsilber J, Michlewski G . Lin28a regulates neuronal differentiation and controls miR-9 production. Nat Commun 2014; 5: 3687.
Balzer E, Heine C, Jiang Q, Lee VM, Moss EG . LIN28 alters cell fate succession and acts independently of the let-7 microRNA during neurogliogenesis in vitro. Development 2010; 137: 891–900.
Shyh-Chang N, Zhu H, Yvanka de Soysa T, Shinoda G, Seligson MT, Tsanov KM et al. Lin28 enhances tissue repair by reprogramming cellular metabolism. Cell 2013; 155: 778–792.
Gong C, Maquat LE . lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3' UTRs via Alu elements. Nature 2011; 470: 284–288.
Kretz M, Siprashvili Z, Chu C, Webster DE, Zehnder A, Qu K et al. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature 2013; 493: 231–235.
Kretz M . TINCR, staufen1, and cellular differentiation. RNA Biol 2013; 10: 1597–1601.
Cho H, Kim KM, Han S, Choe J, Park SG, Choi SS et al. Staufen1-mediated mRNA decay functions in adipogenesis. Mol Cell 2012; 46: 495–506.
Gong C, Kim YK, Woeller CF, Tang Y, Maquat LE . SMD and NMD are competitive pathways that contribute to myogenesis: effects on PAX3 and myogenin mRNAs. Genes Dev 2009; 23: 54–66.
Kim MY, Park J, Lee JJ, Ha DH, Kim J, Kim CG et al. Staufen1-mediated mRNA decay induces Requiem mRNA decay through binding of Staufen1 to the Requiem 3'UTR. Nucleic Acids Res 2014; 42: 6999–7011.
Villace P, Marion RM, Ortin J . The composition of Staufen-containing RNA granules from human cells indicates their role in the regulated transport and translation of messenger RNAs. Nucleic Acids Res 2004; 32: 2411–2420.
Vessey JP, Macchi P, Stein JM, Mikl M, Hawker KN, Vogelsang P et al. A loss of function allele for murine Staufen1 leads to impairment of dendritic Staufen1-RNP delivery and dendritic spine morphogenesis. Proc Natl Acad Sci USA 2008; 105: 16374–16379.
Peredo J, Villace P, Ortin J, de Lucas S . Human Staufen1 associates to miRNAs involved in neuronal cell differentiation and is required for correct dendritic formation. PLoS One 2014; 9: e113704.
Shin C, Nam JW, Farh KK, Chiang HR, Shkumatava A, Bartel DP . Expanding the microRNA targeting code: functional sites with centered pairing. Mol Cell 2010; 38: 789–802.
Boulay K, Ghram M, Viranaicken W, Trepanier V, Mollet S, Frechina C et al. Cell cycle-dependent regulation of the RNA-binding protein Staufen1. Nucleic Acids Res 2014; 42: 7867–7883.
Furic L, Maher-Laporte M, DesGroseillers L . A genome-wide approach identifies distinct but overlapping subsets of cellular mRNAs associated with Staufen1- and Staufen2-containing ribonucleoprotein complexes. RNA 2008; 14: 324–335.
Kim YK, Furic L, Parisien M, Major F, DesGroseillers L, Maquat LE . Staufen1 regulates diverse classes of mammalian transcripts. EMBO J 2007; 26: 2670–2681.
Heraud-Farlow JE, Kiebler MA . The multifunctional Staufen proteins: conserved roles from neurogenesis to synaptic plasticity. Trends Neurosci 2014; 37: 470–479.
Park E, Maquat LE . Staufen-mediated mRNA decay. Wiley Interdiscip Rev RNA 2013; 4: 423–435.
Laver JD, Li X, Ancevicius K, Westwood JT, Smibert CA, Morris QD et al. Genome-wide analysis of Staufen-associated mRNAs identifies secondary structures that confer target specificity. Nucleic Acids Res 2013; 41: 9438–9460.
Ricci EP, Kucukural A, Cenik C, Mercier BC, Singh G, Heyer EE et al. Staufen1 senses overall transcript secondary structure to regulate translation. Nat Struct Mol Biol 2014; 21: 26–35.
Sugimoto Y, Vigilante A, Darbo E, Zirra A, Militti C, D'Ambrogio A et al. hiCLIP reveals the in vivo atlas of mRNA secondary structures recognized by Staufen 1. Nature 2015; 519: 491–494.
LeGendre JB, Campbell ZT, Kroll-Conner P, Anderson P, Kimble J, Wickens M . RNA targets and specificity of Staufen, a double-stranded RNA-binding protein in Caenorhabditis elegans. J Biol Chem 2013; 288: 2532–2545.
Lebeau G, Maher-Laporte M, Topolnik L, Laurent CE, Sossin W, Desgroseillers L et al. Staufen1 regulation of protein synthesis-dependent long-term potentiation and synaptic function in hippocampal pyramidal cells. Mol Cell Biol 2008; 28: 2896–2907.
Fritzsche R, Karra D, Bennett KL, Ang FY, Heraud-Farlow JE, Tolino M et al. Interactome of two diverse RNA granules links mRNA localization to translational repression in neurons. Cell Rep 2013; 5: 1749–1762.
Heraud-Farlow JE, Sharangdhar T, Li X, Pfeifer P, Tauber S, Orozco D et al. Staufen2 regulates neuronal target RNAs. Cell Rep 2013; 5: 1511–1518.
Park E, Gleghorn ML, Maquat LE . Staufen2 functions in Staufen1-mediated mRNA decay by binding to itself and its paralog and promoting UPF1 helicase but not ATPase activity. Proc Natl Acad Sci USA 2013; 110: 405–412.
Bruno IG, Karam R, Huang L, Bhardwaj A, Lou CH, Shum EY et al. Identification of a microRNA that activates gene expression by repressing nonsense-mediated RNA decay. Mol Cell 2011; 42: 500–510.
Molenaar JJ, Domingo-Fernandez R, Ebus ME, Lindner S, Koster J, Drabek K et al. LIN28B induces neuroblastoma and enhances MYCN levels via let-7 suppression. Nat Genet 2012; 44: 1199–1206.
Lee JE, Lim MS, Park JH, Park CH, Koh HC . PTEN promotes dopaminergic neuronal differentiation through regulation of ERK-dependent inhibition of S6K signaling in human neural stem cells. Stem Cells Transl Med 2016; 5: 1319–1329.
Oh SM, Chang MY, Song JJ, Rhee YH, Joe EH, Lee HS et al. Combined Nurr1 and Foxa2 roles in the therapy of Parkinson's disease. EMBO Mol Med 2015; 7: 510–525.
Yi SH, He XB, Rhee YH, Park CH, Takizawa T, Nakashima K et al. Foxa2 acts as a co-activator potentiating expression of the Nurr1-induced DA phenotype via epigenetic regulation. Development 2014; 141: 761–772.
Ahn S, Kim J, Hwang J . CK2-mediated TEL2 phosphorylation augments nonsense-mediated mRNA decay (NMD) by increase of SMG1 stability. Biochim Biophys Acta 2013; 1829: 1047–1055.
Acknowledgements
We would like to thank Dr. Lynne E. Maquat (University of Rochester) for the HA-STAU1 plasmid, Dr. Narry V. Kim (Seoul National University) for the pcDNA3-FLAG-Lin28B WT and pcDNA3-FLAG-Lin28B-mCCHC plasmids, and Dr. Chang-Hwan Park (Hanyang University) for the hNPCs. We especially thank Dr. Seung-Jae Lee (Seoul National University) for critical technical comments. This research was supported by the Basic Science Research Program through the National Research Foundation (NRF) of Korea, which is funded by the Ministry of Education, Science and Technology and (2015R1D1A1A01058878 to JH), by the framework of an international cooperation program managed by the NRF of Korea (NRF-2016K2A9A1A01951898 to JH) and by grant from the Medical Research Center (2017R1A5A2015395 to JH), funded by the NRF of Korea of the Ministry of Science and ICT, Republic of Korea.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by M Deshmukh
Supplementary Information accompanies this paper on Cell Death and Differentiation website
Rights and permissions
About this article
Cite this article
Oh, Y., Park, J., Kim, JI. et al. Lin28B and miR-142-3p regulate neuronal differentiation by modulating Staufen1 expression. Cell Death Differ 25, 432–443 (2018). https://doi.org/10.1038/cdd.2017.182
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cdd.2017.182