Abstract
Members of the p53 tumor-suppressor family are expressed as multiple isoforms. Isoforms with an N-terminal transactivation domain are transcriptionally active, while those ones lacking this domain often inhibit the transcriptional activity of other family members. In squamous cell carcinomas, the high expression level of ΔNp63α inhibits the tumor-suppressor function of TAp73β. This can in principle be due to blocking of the promoter or by direct interaction between both proteins. p63 and p73 can hetero-oligomerize through their tetramerization domains and a hetero-tetramer consisting of two p63 and two p73 molecules is thermodynamically more stable than both homo-tetramers. Here we show that cells expressing both p63 and p73 exist in mouse epidermis and hair follicle and that hetero-tetramer complexes can be detected by immunoprecipitation in differentiating keratinocytes. Through structure determination of the hetero-tetramer, we reveal why this hetero-tetramer is the thermodynamically preferred species. We have created mutants that exclusively form either hetero-tetramers or homo-tetramers, allowing to investigate the function of these p63/p73 hetero-tetramers. Using these tools, we show that inhibition of TAp73β in squamous cell carcinomas is due to promoter squelching and not direct interaction.
Similar content being viewed by others
Main
The p53 protein family has a central role in the cellular defense against tumorigenesis. Although p53 is the main tumor suppressor, p73 has additional important developmental roles1 besides its action as a tumor suppressor.2 The connection of p63 and tumor development, however, is less obvious. So far two main functions have been identified for p63 that are connected to two different isoforms of the protein. The ΔNp63α isoform that lacks the N-terminal transactivation (TA) domain is highly expressed in cells of the basal compartment of stratified epithelial tissues where it serves important functions in maintaining the proliferative potential of these cells.3, 4 The second function that is carried out by the full-length TAp63α isoform is that of a quality control factor in female oocytes.5 Mutations in the human p63 gene are the cause for six different syndromes characterized by deformations of the limbs and/or skin erosions.6, 7 In tumors, however, mutations in the p63 gene are found only infrequently, suggesting that p63 is not a typical tumor suppressor. Overexpression of the ΔNp63α isoform, however, is observed in squamous cell carcinomas, particularly in head and neck squamous cell carcinomas (HNSCCs), a group of malignancies derived from cells of the basal epithelia of the aerodigestive mucosa.8 Although it has been shown that ΔNp63α acts as a transcription factor for a certain set of genes, it inhibits the p53 and p73-induced activation of prototypical apoptosis-related genes by binding to and blocking the promoter. In addition to this indirect effect via promoter squelching, it was shown that p73 directly interacts with ΔNp63α, which may also contributes to inhibiting p73’s function as a tumor suppressor.8 In previous studies, along with others, we have shown that p63 and p73 can form mixed tetramers via their tetramerization domains (TDs).9, 10 Surprisingly, hetero-tetramers consisting of one homo p63 dimer and one homo p73 dimer are thermodynamically more stable than both homo-tetramers. This in vitro finding suggests that in cells that express both p73 as well as p63 mixed tetramers can exist. Such a direct interaction has indeed been identified in HNSCC cells where overexpression of the ΔNp63α isoform may be a selective advantage for tumor growth by inhibiting TAp73 isoforms. Both proteins also occur simultaneously in normal tissues such as the basal layer of keratinocytes.9, 11 As a first step to characterize the biological function of such hetero-tetramers, we wanted to investigate if they exist in human primary keratinocytes. In addition, we were interested to understand at the structural level exactly why the hetero-tetramer is thermodynamically more stable than the homo-tetramers and how this interaction contributes to the inhibition of TAp73β in HNSCCs. Finally, we aimed to develop a toolkit to study the function of hetero-tetramers during development.
Results
Co-expression and hetero-tetramerization of p63 and p73 in vivo
Cells in the basal layer of epithelial tissues express a high concentration of ΔNp63α.12 In the suprabasal cell layers, the expression of p63 gets gradually reduced with the increasing level of differentiation. To investigate if p63-positive cells also express p73, we stained skin sections of P5 mice (Figure 1a). Although the expression level of p63 is much higher, some cells of the basal layer as well as in the hair follicle express p73 as well. This observation is consistent with earlier reports of p73 expression in the skin11, 13, 14 as well as in multiciliated cells.15 To further investigate if cells exist that simultaneously express both p63 and p73, we used immunofluorescence (IF). This analysis showed that both in the basal layer of the skin as well as in the outer root sheath (ORS) of the hair follicle double-positive cells exist. Although in both cases several layers of p63-positive cells occur, p73 expression seems to be confined to some cells in the basal layer of the epidermis and the outer cell layer of the ORS of hair follicle (Figure 1b and Supplementary Figure S1A). Double-positive cells were also always K14 positive, whereas the K14-negative cells of the matrix of the hair follicle bulb only express p63 but not p73.
Co-expression and hetero-tetramerization of endogenous p63 and p73. (a) IHC staining of P5 mouse skin sections for p63 and p73. Sections include the epidermis and dermis, latter harboring the hair follicles. Stained epidermis and hair follicle structures are shown in detail. The black dashed line marks the transition between epidermis and dermis. Scale bar: 25 μm. (b) Representative IF staining of skin from a new-born (P5) mouse for p63 and p73. Both, in the epidermis (upper panel) and hair follicle (lower panel), p73 (green) colocalizes with p63 (red) in a subset of K14-positive cells (cyan). In the epidermis, p63 and K14-positive cells are located in the basal layer at the boundary between epidermis and dermis (white dashed lines, upper panel)). In the anagen hair follicle (white dashed lines, lower panel), p63 is expressed by the cells of the matrix (M) and the outer rood sheet (ORS). K14-positive cells from the outer layer of the ORS show colocalization of p63 and p73. The structure and expression pattern of the interfollicular epidermis (IFE) is equal to the regular epidermis. White boxes mark the magnified area. Scale bar: 25 μm. Expression level of p63, p73 and K10 during keratinocyte differentiation. Differentiation of keratinocytes was induced by supplementing the culture medium with high calcium concentrations. p63 and p73 levels were detected via western blot on consecutive days. p73 initially shows a very low basal expression. At day 2 after induction, the protein level sharply increases and is gradually lost again after day 3. p63 levels are initially on a high level, but the expression gradually decreases after day 2 (note that for the western blot in panel c a lower amount of the p63 antibody was used than for the p63 western blot shown in d). Uninduced control cells were blotted as well (‘-‘). K10 expression (red line in panel p63 symbolizing the expression profile) is detectable from day 3 on. (d) Differentiated and non-differentiated primary human keratinocyte lysate was immunoprecipitated with a p73-specific antibody and a p63-specific antibody was used for detection and vice versa. Keratinocyte differentiation was induced with high calcium concentrations (dx=day × after induction). Normal IgG was used as a control. IP, immunoprecipitation; WB, western blot
In the next step, we investigated if hetero-tetramers occur in primary human keratinocytes. Although we could detect a strong signal for p63 (ΔNp63α) in these cells, the p73 level was very weak, could, however, be increased upon keratinocyte differentiation initiated by high concentration of calcium. (Figure 1c). Interestingly, we observed a burst in p73 expression between days 2 and 4. At day 6, expression of both p63 and p73 is almost completely lost while expression of the differentiation marker K10 becomes prominent. This expression analysis is consistent with the expression pattern of p63, p73 as well as K14 and K10 in the mouse skin (Supplementary Figure S1B).
To investigate the formation of hetero-tetramers, we used co-immunoprecipitation experiments. Interaction between both proteins could be observed in both combinations, using either an anti-p63 antibody or an anti-p73 antibody for the co-immunoprecipitation experiments, demonstrating that in differentiating primary keratinocytes both proteins indeed form hetero-tetramers (Figure 1d). Differentiation of the keratinocytes was monitored by western blot analysis of the differentiation markers K14 and K10 (Supplementary Figure S1C).
Determination of the structure of the hetero-TD of p63 and p73
Further characterization of the biological function of p63/p73 hetero-tetramers is currently hampered by the fact that cells always contain a mixture of both homo-tetramers as well as the hetero-tetramer. If cells containing only the hetero-tetramer would be available, ChIPseq or RNAseq experiments could be used to characterize the hetero-tetramer-specific transcriptome and compare it with the data known from p63 and p73. Similarly, if hetero-tetramerization could be prevented the biological function of the hetero-tetramer could be analyzed with knock out models. The basis for creating such research tools would be mutational studies designed to either strengthen hetero-tetramer formation or prevent it. As a first step toward this goal, we decided to determine the NMR solution structure of the hetero-tetramer. Previous NMR investigations have revealed that a mixture of the p63 and p73 TDs not only contain the two homo-tetramers and the hetero-tetramer consisting of two homo-dimers but all possible combinations of p63 and p73 within hetero-tetramers (Supplementary Figure S2).9, 10 Such a mixture of seven different species would be far too complex for a detailed structural investigation. To be able to analyze the structure of the p632/p732 hetero-tetramer, consisting of a p63 homo-dimer and a p73 homo-dimer, we decided to selectively stabilize it by mutagenesis. We therefore mutated amino acids, which form inter-dimer salt bridges (Figure 2a) in both tetramers in such a way that they lead to an attractive interaction in case of the hetero-tetramer while destabilizing the homo-tetramers (E363K in the p73 TD and K377E in the p63 TD). To be able to analyze the results of our mutagenesis experiment, we labeled the p73 TD with 15N-lysine. In previous experiments, we have seen that the chemical shift of K372 located in the first helix is a sensitive indicator of the formation of the different hetero-tetramer species.9 Figure 2c shows a spectrum of a mixture of the two mutated domains demonstrating that only a single peak remains. This peak shows chemical shifts similar to K372 in the p632/p732 hetero-tetramer obtained with the two wild-type domains (Supplementary Figure S2). Similar results were obtained by titrating unlabeled p73 TD to 15N-lysine-labeled p63 TD (Figure 2d). These results suggested that indeed the mutations selectively stabilize the expected hetero-tetramer.
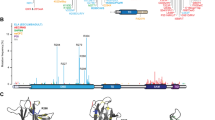
Structure of the hetero-tetramer between the TDs of p63 and p73. (a) Sequence alignment of p63 and p73 TDs. The TDs form salt bridges across the central tetrameric interface. Here we introduced charge-swapped mutants of p63 TD (K377E) and p73 TD (E363K) that are designed to exclusively form hetero-tetramers. (b) p63 TD and p73 TD form several different hetero-tetrameric species when mixed. Here we show schematically that the designed p63 TD hetero and p73 TD hetero mutants exclusively form hetero-tetramers. (c) Section of [15N, 1H]-TROSY spectra of 15N-lysine-labeled p73 TD hetero (E363K) in absence (blue) or presence of an equimolar amount of p63 TD hetero (K377E) (red), showing K372. (d) Section of [15N, 1H]-TROSY spectra of 15N-lysine-labeled p63 TD hetero (K377E) in absence (blue) or presence of an equimolar amount of p73 TD hetero (E363K) (red), showing K379. (e) The hetero-tetramer of p63/p73 (PDB 2NB1) is composed of two chains of each protein. For clarity reasons, the flexible c-termini of the p63 chains after Q409 have been omitted. p63 chains (blue) and p73 chains (red) each form an independent dimer consisting of an N-terminal antiparallel β-sheet and one subsequent helix per monomer. These helices interact with the corresponding helices from the other dimer, thus forming the central tetramerization interface. In addition, the C-terminal helix 2 leans toward the β-strand from the opposing dimer. (f) View onto the p63 β-sheet as a surface model with helices 2 of p73 shown as explicit secondary structure element. The hydrophobic outward facing amino acids of the p63 chains are depicted as colored sticks (L361 in orange, Y363 in yellow, P365 in blue). The side chain of L396 of p73 (red, shown as sticks) penetrates into a hydrophobic pocket formed mainly by P365 of p63. L361 shows a small additional stabilization, however, the main effect can be attributed to P365 (Supplementary Figure S4). NOE restraints, which could be detected in this region, are shown as green lines
We determined the structure of the TD of the p632/p732 hetero-tetramer by standard multidimensional NMR spectroscopy. A representative structure is shown in Figure 2e. The overall tertiary structure closely resembles those of both homo TDs and is, as expected, comprised of a p63 homo-dimer and a p73 homo-dimer. In principle, three different interaction regions between both dimers could be responsible for the higher stability of the p632/p732 hetero-tetramer as shown by inter chain NOEs (Supplementary Figures S3A-C). The by far largest interaction region is formed by helix 1 of each subunit, forming the central tetramerization interface. Owing to very high sequence conservation, it is unlikely that this region is responsible for the selective stabilization effect.
The second region spans the loop region between helix 1 and the first turn of helix 2 in each subunit. We tested the importance of this region by exchanging the sequence TIET in p63 to the p73 sequence (LVDS) but did not detect a significant destabilization of the p632/p732 hetero-tetramer (Supplementary Figures S3D-G).
The third region comprises helix 2 of p73 and the β-sheet of p63. In the structure of the p632/p732 hetero-tetramer, L396 of p73 is situated in a hydrophobic pocket formed by the aromatic ring of Y363 and P365 of p63 (Figure 2f). Interestingly, no corresponding interaction is detected between helix 2 of p63 and the β-stand of p73. The missing interaction of p63 helix 2 with p73 is supported by a total lack of NOE restraints during structure calculation, as well as HetNOE experiments of the complex (Supplementary Figures S3H and I).
To test whether this potential interaction is indeed relevant for the increased stability of the hetero-tetramer, a L395A/L396A mutant of the otherwise wild-type p73 TD was expressed and titrated to 15N-lysine-labeled p63 TD (Figures 3c and d). The resulting distribution of individual oligomers was compared with the one obtained with the wild-type sequences (Figures 3a and b). Furthermore, in a separate experiment the outward facing leaf of the β-strand of the p63 TD was mutated to resemble the more hydrophilic p73 TD (L361T/P365Q in Figures 3e and f) and titrated to 15N-labeled p73 TD. Mutation of L395 and L396 to alanine in p73 leads to a strong reduction in the population of the p632/p732 hetero-tetramer (Figure 3g). A similar result was obtained for the L361T/P365Q double mutation in the p63 TD. In this case, mutation of P365 seems to have the overwhelming effect (Supplementary Figure S4).
Hetero-tetramerization between the TDs of p63 and p73. All mutations described in panels c–f were designed based on the NMR solution structure of the hetero-TD. The mutations were carried out in wild-type proteins, therefore they do not carry the hetero-stabilizing mutations. (a and b) Sections of [15N,1H]-BEST-TROSY spectra of selectively lysine-labeled wild-type p63 (a) and wild-type p73 (b) showing K377 or K372, respectively. A total of eight different resonances can be detected after mixing each protein with equal amounts of unlabeled p73 or p63, respectively, and subsequent incubation for 24 h at 37 °C. Peaks corresponding to the homo-tetramers in a and b are overlaid in green and blue, respectively. Peaks corresponding to the homo-tetrameric population, as well as the hetero-tetramer of homo-dimers, are marked in each panel. The mayor single population in both cases is the hetero-tetramer of homo-dimers. (c and d) Sections of [15N,1H]-BEST-TROSY spectra of p63 and mutant p73 (L395A, L396A), prepared analogous to a and b. In contrast to a and b, the magnitude of hetero-tetramers of homo-dimers is drastically reduced. (e and f) Sections of [15N,1H]-BEST-TROSY spectra of mutant p63 (L361T, P365Q) and p73 prepared analogous to a and b. L361 and P365 are the only amino acids of the outward facing leaf of the p63 TD, which are different to p73. Both were mutated to the corresponding amino acids of p73, which also leads to a drastic reduction of the formation of hetero-tetramers of homo-dimers. (g) The intensities of all visible populations in each spectrum can be used to calculate the percentage of each possible combination of homo and hetero-tetramers in solution. For the wild-type protein (red bars), the hetero-tetramer of homo-dimers is the most stable and therefore most prominent population, whereas for both double mutants (orange and blue bars) the percentage of this population is drastically reduced, almost reaching the statistically expected values (black bars)
A different set of salt bridges selects for homo-tetramers by preventing hetero-tetramerization
As mentioned above, studying the biological functions of p632/p732 hetero-tetramers created by interaction of the different p63 isoforms with p73 isoforms would be possible by ‘knock out’ studies in which the formation of hetero-tetramers can be prevented. Based on the success of the selective stabilization of the hetero-tetramer described above, we reasoned that a similar rational design approach could be taken to create mutants that are incapable of forming hetero-tetramers but without influencing the formation of the homo-tetramers. To abolish hetero-tetramerization, we introduced charge swap mutations in the tetramerization interface of p73 that lead to attractive interactions within a p73 homo-tetramer but to charge repulsion in hetero-tetramers (E363R, K370E, E373R and R390D; see Supplementary Figure S5). Size exclusion chromatography (SEC) showed that the mutated TD forms tetramers (Figure 4a). Mixing 15N-lysine-labeled wild-type p63 TD with the unlabeled p73 mutant TD resulted in [15N, 1H]-HSQC NMR experiments with a single peak per lysine with chemical shifts characteristic of the homo-tetramer (Figure 4b, Supplementary Figures S6A and B). This result indicated that not only the p632/p732 hetero-tetramer formation was suppressed but the formation of all possible hetero-tetramer species.
Analysis of the p73 homo mutation. (a) SEC profile of p73 TD and p73 TD homo measured on a Superdex 75 10/300. (b) Sections of [15N, 1H]-TROSY spectra of 15N-lysine-labeled p73 TD homo in absence (blue) or presence (red) of an equimolar amount of p63 TD (left) or p53 OD (right) measured after incubation for one day at 37 °C. (c) SEC profiles of TAp73α (blue) and TAp73α homo (E363R, K370E, E373R, R390D) (orange) produced in RRL. (d) Crystal structure of p73 TD homo (PDB 5HOC). Side chains of E363R, K370E, E373R and R390D are shown. Data collection and refinement statistics are provided in Supplementary Table 2. (e) Structural alignment of p73 TD homo with p63 TD (PDB 4A9Z), showing a hypothetical and non-favored hetero-tetramer of p63 TD and p73 TD homo. Electrostatic repulsions between negatively charged side chains (encircled in red) or positively charged side chains (encircled in blue) explain why p73 TD homo forms exclusively homo-tetramers. (f) Close-up view of the central tetrameric interface of p73 TD homo. 2Fo-Fc density maps are shown for the side chains of E363R, K370E, E373R and R390D
To validate the mutant TD within the context of the full-length protein, we expressed ΔNp73α containing the mutations E363R, K370E, E373R and R390D in rabbit reticulocyte lysate (RRL) and analyzed the protein by SEC. The results also showed that the mutated full-length ΔNp73α eluted as a tetramer (Figure 4c). The crystal structure of the designed p73 TD homo further confirmed our strategy and demonstrated the formation of the designed salt bridges (Figures 4d–f). As expected, the p73 TD homo does not interact with both p63 TD and p53 OD (Figure 4b).
Unfortunately, the search for mutations that would prevent hetero-tetramerization without affecting homo-tetramerization was not successful in the case of p63. Although a E370K, K377E, E380R and R397E mutant showed in the NMR investigations of the isolated TD only a single conformation with chemical shifts indicative of the homo-tetramer (Supplementary Figure S6C-E), this mutant in the context of ΔNp63α expressed in RRL formed only dimers (Supplementary Figure S6F). This result showed that these mutations weaken the tetramerization leading to the formation of dimers at low concentrations. Unfortunately, the search for additional mutations that would strengthen the formation of tetramers was not successful.
Interaction of wild-type/mutant p63 with p73 in cells
To investigate the interaction of the hetero or homo mutants of p63 and p73 in a cellular environment, we performed co-immunoprecipitation experiments of transiently overexpressed wild-type or mutant ΔNp63α with wild-type TAp73β in the neuroblastoma cell line SK-N-AS. We used this specific combination as it has been shown that direct interaction between these two isoforms inhibits the tumor-suppressor activity of TAp73β in HNSCC cells.8 Introduction of the homo-tetramer mutations (E370K, K377E, E380R and R397E) in ΔNp63α led to a marked decrease in co-IP efficiency with TAp73β, indicating that hetero-tetramer formation was repressed by these mutations also in a cellular context (Figure 5). Interaction between the hetero-tetramer mutants of TAp73β (E363K) and of ΔNp63α (E377E) again showed a strong co-IP efficiency in all tested combinations.
Introduction of homo mutations in p63 or p73 abrogates interaction in cells. Wild-type ΔNp63α, ΔNp63α hetero, ΔNp63α homo and wild-type TAp73β, TAp73β hetero or TAp73β homo were co-transfected in SK-N-AS cells. Immunoprecipitation (IP) of p63 (a and b) or p73 (c and d) was performed with subsequent analysis by western blot with a p73-specific antibody or a p63-specific antibody, respectively. Western blots were quantified using ImageJ and co-IP efficiency of wild-type proteins was set to 1 b and d
Inhibition of the transcriptional activity of TAp73β by ΔNp63α is mainly based on preventing promoter binding
As a first application of this new tool, we wanted to investigate the mechanism by which the high expression level of ΔNp63α in HNSCC cells influences the transcriptional activity of TAp73β. The transcriptional activity was measured on the Bax promoter in p73-deficient SK-N-AS cells with different amounts of ΔNp63α added in ratios from 1 : 1 to 1 : 16 (p73 : p63). As expected, the addition of wild-type ΔNp63α decreased the activity of TAp73 in a concentration-dependent manner (Figure 6a). In principle, this effect may be caused by two mechanisms. On the one hand, hetero-tetramerization can lead to a decrease in activity because of the lack of the TA domains in ΔNp63α. On the other hand, p63 binds similar promoters as p73 as both proteins share a very high sequence identity in their DNA-binding domains (~85%). At high concentration, ΔNp63α could therefore displace p73 from pro-apoptotic promoters. ΔNp63α itself was not potent in inducing transcription from the Bax promoter as shown in control experiments (Figure 6). To distinguish the effects of hetero-tetramerization and promoter displacement, we performed the titration assay with different p63 and p73 mutants. Addition of the isolated p63 TD led to a strong reduction in the transcriptional activity of wild-type TAp73β (Supplementary Figure S7A). This result was expected as hetero-tetramers consisting of TAp73β and the p63 TD contain only two DBDs. It has been shown by several laboratories that dimers (and such a hetero-tetramer is a functional dimer) retains approximately 50% of the activity of the tetramer.16, 17 Using TAp73β containing the homo mutants, however, did not show a reduction in activity, proving that hetero-tetramer formation was suppressed (Supplementary Figure S7B).
Analysis of the inhibitory potential of ΔNp63α. The transcriptional activity of TAp73β or TAp73β homo is measured on the Bax promoter in transiently transfected SK-N-AS cells. TAp73β (a and c) or TAp73β homo (b and d) are titrated with increasing amounts of ΔNp63α (a and b) or a mutant of p63, which is unable to bind to DNA (ΔNp63α R279H) (c and d). Contributions of heteotetramerization and promoter squelching in the different conditions are indicated in a matrix format. Bar diagram indicate fold change of promoter induction compared with empty vector control
Next, we compared the activity of TAp73β with the activity of its homo mutant in the presence of wild-type ΔNp63α. In both cases, we measured a similar drop in activity (Figure 6b). As the p73 homo mutations effectively prevent the formation of hetero-oligomers, this drop cannot be explained with a reduced activity of the hetero-oligomers. Instead blocking the promoter-binding site by ΔNp63α is the most likely explanation. To verify this assumption, we performed the same titration experiments with a DNA-binding domain mutant of ΔNp63α (R279H). Co-expression of wild-type TAp73β with the DNA-binding mutant of ΔNp63α resulted in a significant drop in TA activity, through the formation of hetero-tetramers with only two functional DNA-binding domains (similar to the experiments with the isolated p63 TD with approximately 50% of activity expected) (Figure 6c). In contrast, titration of the p73 homo mutant with ΔNp63α (R279H) did not show any inhibitory effect (Figure 6d). These data suggested that the strongest effect of the ΔNp63α-based inhibition was due to blocking promoter binding.
Discussion
All members of the p53 protein family are expressed as many different isoforms that are created by the combination of different N-terminal promoters and C-terminal splice events.1, 12, 18 This bewildering array of different proteins becomes even more complex by the fact that all members of this family – with the exception of the C. elegans protein Cep-1 (ref. 19) – contain an oligomerization domain (OD) that is capable of forming tetramers, thus allowing the formation of a potentially astronomical number of different protein species. The high sequence identity within the ODs among the different family members increases this theoretical number even further by potentially enabling the formation of hetero-oligomers that contain different combinations of isoforms of these family members. However, not all combinations are possible and in previous experiments we have shown that, for example, the two different family members found in the tunicate species Ciona intestinalis do not form hetero-tetramers at all.20 With respect to the three mammalian family members, along with others, we have shown that p53 does not form hetero-tetramers with either p63 or p73.9, 10 However, these investigations have also demonstrated that p63 and p73 form very stable hetero-tetramers. So far, all available data suggest that the question whether two different proteins can form hetero-tetramers or not is connected to the presence of a second helix in the TD. Proteins with a second helix can form such hetero-tetramers while no stable complexes of proteins with a second helix and proteins without a second helix have been observed.
In this publication, we have systematically analyzed the reason for the high thermodynamic stability of the hetero-tetramer formed by p63 and p73 by structure determination. Of central importance for this higher stability is the interaction of helix 2 of the p73 TD with the β-sheet of the p63 TD. This interaction of hydrophobic amino acids in the C-terminus of the second helix with the hydrophobic outer leaflet of the β-sheet is only possible in the hetero combination and explains the higher thermodynamic stability compared with the homo-tetramers.
Although the structure determination explains why in in-vitro experiments a mixture of p63 and p73 TDs forms very stable hetero-tetramers, the central question is whether these hetero-tetramers exist in vivo and what is their specific function. From investigations of p53 it is known that this protein forms dimers co-translationally.21 In addition, the longest isoform of p63, TAp63α is expressed in a closed and only dimeric conformation in oocytes.16, 22 These observations suggest that all family members of the p53 family likely form dimers co-translationally and only later two dimers interact to form the fully active tetrameric state. This expression scheme makes the formation of hetero-tetramers possible, if both p63 and p73 are simultaneously expressed in the same cells. Such co-expression has been shown, for example, for keratinocytes and our co-IP experiments with differentiating primary keratinocytes indicate that hetero-oligomerization takes place in vivo. In addition, stainings of mouse epidermis and hair follicle as well as our keratinocyte differentiation experiments show that p73 is always co-expressed with p63. These data suggest that in a cell population in the basal layer of the epidermis and the ORS of hair follicle, most likely the majority of p73 exists as a complex with p63. The p63/p73 hetero complex may therefore be characteristic for a certain transition phase of keratinocyte differentiation. Direct interaction between p63 and p73 has also been reported in HNSCCs.8 This tumor type often shows a high expression level of ΔNp63α that inhibits the tumor-suppressor function of TAp73. Our analysis of the potential mechanism of this inhibition, however, suggests that most of it is probably due to promoter squelching although the formation of hetero-complexes between transactivating p73 isoforms and inactive p63 isoforms on promoter sites with a reduced number of TA domains may contribute as well.
In addition, interaction between p63 and p73 has indirectly been observed in ChIPseq analysis of the DNA occupancy of p63 and p73 in the cervical carcinoma cell line ME180. Comparison of the binding profiles of both proteins did not reveal any significant difference, suggesting that both proteins have the same binding targets.23 Furthermore, co-occupancy was detected for all investigated sites, suggesting that hetero-tetramers could be involved. This strikingly similar binding behavior is in stark contrast to the phenotypes of the p63 and p73 knock out mice. Although the p63 knock out mouse suffers from severe defects of the development of epithelial tissues, including limb truncations and skin erosion,3, 4 the p73 knock out mouse shows defects in neurogenesis, inflammation and sensory pathways.24 The results of the ChIPseq study suggests that these clear differences are not due to different transcriptional targets but may be due to the expression pattern of both proteins in different tissues. Alternatively, the transcriptional activity of both proteins may differ because of the different isoforms. It has been demonstrated that ΔNp63 and TAp63 isoforms activate different promoters. In keratinocytes only the ΔNp63α isoform is highly expressed while several different isoforms of p73 are present. The domains of these isoforms can influence the transcriptional activity by modulating the interaction with transcription factors or other binding partners and different posttranslational modifications can also cause different effects. In previous studies, we have been able to show that while the activity of tetrameric p63 seems independent of the promoters used, the activity of p73 strongly depends on the cellular and promoter context.25 Therefore, the formation of hetero-tetramers may have an important role in fine tuning the activity of p63 and p73.
An important motivation for our structural study was to create new research tools that can be used to investigate the biological function of this p632/p732 hetero-tetramer. One way to study its potential biological function would be to create knock out cell lines or model organisms. In such a knock out study, the p63 and p73 homo-tetramers would have to be fully functional while hetero-tetramerization would be suppressed. We have designed mutations in p73 that only allow the formation of p63 and p73 homo-tetramers but prevent the formation of the p632/p732 hetero-tetramer. Unfortunately, mutations in p63 also affect the dissociation constant of these homo-tetramers. Although for p73, additional changes in length of the side chains of the mutated amino acids were successful to stabilize homo-tetrameric p73, further mutations of the p63 TD were so far not successful. Consequently, knock out studies based on our homo-tetramers should mutate only p73.
Materials and Methods
Plasmids
Mammalian cell expression constructs for the p63 isoforms and mutants used in this study have been described earlier.25 All mutants were generated using the Quickchange site-directed mutagenesis protocol. For the TA assays, single copies of the BAX promoter have been cloned into pGL3 promoter vector. For E. coli expression, the TD of p63 (amino acids 358–316) and p73 (amino acids 351–398) were subcloned into a pET16b vector (Merck Millipore, Darmstadt, Germany), modified with an N-terminal His-tag and a TEV protease site. All point mutations were introduced via QuikChange (Agilent Technologies, Waldbronn, Germany).
Bacterial expression and purification
Expression was generally carried out in T7 Express cells (New England Biolabs, Frankfurt, Germany). For unlabeled expression, cells were grown at 37 °C in 2xYT medium to an optical density of 0.7 and induced with IPTG to a final concentration of 1 mM. After induction, temperature was reduced to 22 °C and the expression was carried out overnight. For labeled expression, cells were grown at 37 °C in 2xYT medium and harvested when the optical density reached 0.4. After centrifugation, the medium was discarded and the cells were resuspended in M9 minimal medium supplemented with the appropriate isotopic labeling (1 g 15NH4Cl and 4 g glucose for 15N labeling; 1 g NH4Cl and 2 g 13C-glucose for 13C labeling and 1 g 15NH4Cl and 2 g 13C-glucose for double labeling, respectively). The cells were then grown to an OD of 0.6 or at least 1 h before induction with IPTG. After induction, temperature was reduced to 22 °C and the expression was carried out overnight.
Harvested cells were resuspended in IMAC-A buffer (25 mM Tris, 200 mM NaCl, pH 7.8) supplemented with DNAse, lysozyme and protease inhibitor cocktail (self-made) for cell disruption. Cells were lysed by sonication (5x30 s with breaks of 1 min between each pulse). Cell debris was pelleted by centrifugation (SEV34 rotor 17 000 r.p.m., 30 min). The supernatant was filtered and subsequently loaded on a HisTrap column (GE Healthcare, München, Germany), previously equilibrated with IMAC-A buffer. The column was washed with IMAC-A buffer containing 30 mM imidazole until baseline was reached. The protein was eluted with IMAC-B buffer (25 mM Tris pH 7.8, 200 mM NaCl, 500 mM imidazole). Protein-containing fractions were pooled and TEV protease was added before dialysis overnight into IMAC-A buffer. The TEV protease and the cleaved His-Tag were removed by reverse IMAC purification.
Complex formation
For complex formation, the hetero mutants of both TDs were mixed in an equimolar amount, diluted into AEX-A buffer (25 mM Tris pH 8.5) and incubated for 1 h at room temperature. They were subsequently loaded onto a HiTrap Q column (GE Healthcare) and eluted with a linear gradient to 1 m NaCl. In general, three peaks were observable, corresponding to the hetero-tetramer and both homo-tetramers. Fractions containing the hetero-tetramer were pooled and concentrated before gel filtration into NMR buffer (25 mM HEPES, 50 mM NaCl, pH 7.0). Gel filtration was performed on a HiLoad Superdex 75 16/600 column (GE Healthcare).
NMR spectroscopy
All experiments were carried out at 310 K on Bruker (Rheinstetten, Germany) Avance spectrometers with proton frequencies of 500, 600, 700, 800, 900 or 950 MHz. The backbone assignments were done with 3D HNCACB and HN(CA)CO spectra. Side chains were assigned with the help of 3D [15 N,1H]-BEST-TROSY-(H)C(C)(CO)NH-TOCSY and [15 N,1H]-H(CC)(CO)NH-TOCSY spectra. For aromatic side chains, (HB)CB(CDCD)HD-HSQC, (H)CB(CG)CCH-TOCSY and [15 N,1H]-TROSY-Har(CCgbaCO)NH-TOCSY spectra were used. Distance restraints were obtained from 3D 15N-edited-NOESY-HSQC (τm 60 ms) and 3D 13C-edited-NOESY-HSQC (τm 50 ms) spectra. In addition, exclusively intermolecular restrains were obtained from [13C,15 N]-filtered-13C-edited-NOESY-HSQC (τm 80 ms) and [13C,15 N]-filtered-15N-edited-NOESY-HSQC (τm 100 ms) spectra. Hydrogen bonds inside the N-terminal β-strands were measured by long range HNCO experiments. In addition, all other backbone residues, which could be observed in HSQC experiments after 1 week of incubation in buffer containing 100% 2H2O at 37 °C were considered to take part in hydrogen bonds.
Structure determination
Structural restraints were obtained from automated NOESY peak assignment using the consensus structure bundle method26 of the CYANA 3.97 software package27 based on 10 different intra- and intermolecular NOESY spectra (Supplementary Table 1). The consensus structure calculation method was performed as 20 individual structure calculations using the standard CYANA protocol and a cutoff of 0.6 was applied for the selection of consensus peak assignments.
The completeness of the chemical shift assignments for the non-labile 1H and the backbone HN was 94.3%. An initial consensus structure calculation was used to refine input NOESY peak lists by selecting only those peaks that could be assigned in a minimum of 60% of the individual structure calculations. These refined NOESY peak lists were used as input for a second consensus structure calculation. 99% of the NOESY cross peaks were assigned (Supplementary Table 1) and the final consensus set of distance restraints was composed of 2582 unambiguous upper distance limits.
Dihedral angle restraints were obtained from chemical shift analysis of the HN, N, C’, Cα, Cβ and Hα atoms using the program TALOS+.28
Each of the 220 experimental H-bonds were defined by a upper- and lower distance limit. Upper limit values were set to 3.2 Å for heavy atom distances and to 2.2 Å for distances between heavy atoms and protons. Lower limit values were set to 2.7 Å and 1.8 Å, respectively.
The structure calculation was performed with CYANA using 200 random starting conformers, and 20 000 torsion angle dynamic steps. The resulting structures were sorted according to their target function values, and the 20 structures with the lowest target function values were selected. Restrained energy refinement was carried out using the OPALp29 program, which uses the AMBER 94 force field.30
X-ray crystallography
Crystals of p73 TD homo were grown by vapor diffusion at 4 °C in a 150 nl sitting drop by mixing 100 nl protein (20.56 mg/ml) and 50 nL mother liquor (2 m sodium chloride, 0.1 m sodium phosphate, 0.1 m potassium phosphate, 0.1 m HEPES pH 7.5) equilibrated against 20 μl reservoir containing mother liquor. A crystal was cryo-protected using mother liquor supplemented with 20% glycerol and diffracted to 1.36 Å at Diamond Light Source I04-1.
Data reduction was performed with XIA2 (ref. 31) and analysis with PHENIX.xtriage. Molecular replacement using 2WQI was performed with PHASER32 and models were refined using PHENIX.refine33 and COOT. The final model was validated using Molprobity.34 Data collection and refinement statistics are summarized in Supplementary Table S2.
Population determination
For population determination, wild-type TDs or the corresponding mutants were gel filtered into KPKCl buffer (20 mM potassium phosphate pH 7.8, 100 mM KCl, 0.5 mM EDTA). In each experiment, one protein was 15N-lysine labeled, whereas the other protein was left unlabeled. The proteins were mixed in equimolar amounts and incubated for 24 h at 37 °C before measurement. [15 N-1H]-BEST-TROSY spectra were recorded at 310 K and peak intensities were quantified with Sparky 3.114.35 If multiple peaks, representing the same population, were present in the spectra the peak values were averaged and a S.D. is given in the respective figures.
Size exclusion chromatography
Analytical SEC was performed in phosphate buffer (50 mM sodium phosphate pH 7.8, 100 mM NaCl) at 4 °C using a Superose 3.2/300 column (GE Healthcare) (injection volume 50 μl; flow rate 50 μl/min; fraction size 50 μl). SEC fractions were quantified by western blotting.
Analytical SEC of TDs was performed in KPKCl buffer at 4 °C using a Superdex 10/300 column (GE Healthcare).
Western blotting
Western blotting was performed as previously described.36 The following antibodies were used: anti-p63 (H-129, Santa Cruz Biotechnology, Heidelberg, Germany), anti-p63 (4A4), anti-p63 (D2K8X, CST, Leiden, Netherlands), anti-p73α/β (ER-15, Merck Millipore), pan-p73 (Abcam, Cambridge, UK, EP436Y), anti-GAPDH (Merck Millipore) and anti-myc (clone 4A6, Merck Millipore). Quantification of western blot signals was performed using ImageJ (National Institutes of Health, MD, USA). The specificity of the p63 and p73 antibodies used has been tested (Supplementary Figure S8).
Cell culture and co-immunoprecipitation
SK-N-AS cells (p73 deficient) were maintained in DMEM, 10% FBS (PAA, GE Healthcare), 2 mM l-glutamine (PAA, GE Healthcare) and 1x MEM nonessential amino acids (Gibco, Thermo Fisher Scientific, Dreieich, Germany). Primary pooled human neonatal keratinocytes were purchased from Life technologies and cultured in EpiLife medium (M-EPI-500-CA, Life Technologies) supplemented with growth supplements (S-001- K, Thermo Fisher Scientific). Differentiation of HEKs was induced by adjusting the medium to 1.2 mM CaCl2 for time periods indicated.
Co-immunoprecipitations were performed as described previously.25 In short, HEKs or transiently transfected cells (Effectene, Qiagen, Hilden, Germany) were harvested and lysed in 50 mM Tris pH 7.5, 150 mM NaCl containing 0.75% NP-40, 1 mM DTT, PhosStop and EDTA-free complete (Roche, Grenzach-Wyhlen, Germany) or in RIPA buffer (NEB). After centrifugation, the supernatant was incubated with 2 μg of anti-p63 (H-129, Santa Cruz Biotechnology), anti-p73 (ER-15, Merck Millipore), normal IgG (rabbit or mouse, Santa Cruz Biotechnology) or anti-p63 (D2K8X, CST) in a 1:800 dilution overnight at 4 °C. Immunocomplexes were removed from the lysate using Protein G Dynabeads (Thermo Fisher Scientific), washed four times with 50 mM Tris pH 7.5, 150 mM NaCl containing 0.1% Tween-20 and eluted with LDS-sample buffer (Thermo Fisher Scientific) for 10 min at 70 °C. Samples were analyzed by western blotting. Immunoprecipitation efficiency was calculated by normalization of the IP western blot signal to the respective input signal. Each Co-IP was performed in triplicates at least.
Immunohistochemistry (IHC) and IF
Skin from P5 WT mice was fixed in formalin, embedded in paraffin and cut in 6 μm thick sections (Morphisto GmbH, Frankfurt, Germany). For IHC and IF staining, sections were deparaffinized, rehydrated and boiled for 30 min in 100 mM citrate buffer with pH 6.0 for antigen retrieval. Skin sections were blocked with 10% normal donkey serum (Santa Cruz Biotechnology, sc-2044) in TBSt (0,1% Tween-20) for 1 h at room temperature. Sections were incubated overnight at 4 °C with one or more of the following primary antibodies: p63 (Santa Cruz Biotechnology, H-129, 1:50), p73α/β (Merck Millipore, ER-15, 1:50), pan-p73 (Abcam, EP436Y, 1:50), K14 (Santa Cruz Biotechnology, C-14, 1:100) and/or K10 (Santa Cruz Biotechnology, RKSE60, 1:100) in 1% BSA in TBSt.
For IHC, sections were incubated with biotin-conjugated secondary antibodies (Vector Labs, Peterborough, UK) for 1 h at room temperature in 1% BSA in TBSt and developed using the ABC DAB Peroxidase System (Vector Labs). Nuclei were stained in Mayer’s hematoxylin for 5 min followed by dehydration and mounting of the sections.
For IF, p73 was detected using biotin-conjugated secondary antibodies and streptavidin-HRP in combination with the Tyramide Signal Amplification (TSA) System (Thermo Fisher Scientific). p63, K10 and K14 were detected using Alexa Fluo secondary antibodies (Thermo Fisher Scientific). For nuclear staining, sections were mounted with mowiol (Carl Roth, Karlsruhe, Germany) supplemented with DAPI (Thermo Fisher Scientific). All fluorescence images were obtained using a Leica (Wetzlar, Germany) TCS SP5 confocal microscope.
TA assay
TA assays were performed as described previously.25,36 The assay was performed with SK-N-AS cells using the Dual-Glo Luciferase reporter assay (Promega, Mannheim, Germany). Cells were transfected using Effectene (Qiagen) in a 12-well plate format, harvested and subsequently assayed for Renilla and Firefly luciferase activity in 96-well plates in quadruplicates. For titration assays, the concentration of TAp73β or TAp73β homo was kept constant at 75 ng per assay. Different mutants and isoforms of p63 were titrated in ratios from 1 : 1 to 1 : 16. The total amount of DNA was kept constant with addition of empty pcDNA3 accordingly. The remaining sample volume has been used for western blot analysis. Outliers have been identified using Grubb’s test and corrected data have been averaged. A total of three independent experiments have been performed. Means were compared using Student’s t-test.
Abbreviations
- TD:
-
tetramerization domain
- OD:
-
oligomerization domain
- TA:
-
transactivation
- TI:
-
transcriptional inhibitory
- SEC:
-
size exclusion chromatography
References
Kaghad M, Bonnet H, Yang A, Creancier L, Biscan JC, Valent A et al. Monoallelically expressed gene related to p53 at 1p36, a region frequently deleted in neuroblastoma and other human cancers. Cell 1997; 90: 809–819.
Rufini A, Agostini M, Grespi F, Tomasini R, Sayan BS, Niklison-Chirou MV et al. p73 in Cancer. Genes Cancer 2011; 2: 491–502.
Yang A, Schweitzer R, Sun D, Kaghad M, Walker N, Bronson RT et al. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature 1999; 398: 714–718.
Mills AA, Zheng B, Wang XJ, Vogel H, Roop DR, Bradley A . p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature 1999; 398: 708–713.
Suh EK, Yang A, Kettenbach A, Bamberger C, Michaelis AH, Zhu Z et al. p63 protects the female germ line during meiotic arrest. Nature 2006; 444: 624–628.
Rinne T, Brunner HG, van Bokhoven H . p63-associated disorders. Cell Cycle 2007; 6: 262–268.
McGrath JA, Duijf PH, Doetsch V, Irvine AD, de Waal R, Vanmolkot KR et al. Hay-Wells syndrome is caused by heterozygous missense mutations in the SAM domain of p63. Hum Mol Genet 2001; 10: 221–229.
Rocco JW, Leong CO, Kuperwasser N, DeYoung MP, Ellisen LW . p63 mediates survival in squamous cell carcinoma by suppression of p73-dependent apoptosis. Cancer Cell 2006; 9: 45–56.
Coutandin D, Löhr F, Niesen F, Ikeya T, Weber T, Schäfer B et al. Conformational stability and activity of p73 require a second helix in the tetramerization domain. Cell Death Differ 2009; 16: 1582–1589.
Joerger AC, Rajagopalan S, Natan E, Veprintsev DB, Robinson CV, Fersht AR . Structural evolution of p53, p63, and p73: implication for heterotetramer formation. Proc Natl Acad Sci USA 2009; 106: 17705–17710.
De Laurenzi V, Rossi A, Terrinoni A, Barcaroli D, Levrero M, Costanzo A et al. p63 and p73 transactivate differentiation gene promoters in human keratinocytes. Biochem Biophys Res Commun 2000; 273: 342–346.
Yang A, Kaghad M, Wang Y, Gillett E, Fleming MD, Dotsch V et al. p63, a p53 homolog at 3q27-29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol Cell 1998; 2: 305–316.
De Laurenzi V, Costanzo A, Barcaroli D, Terrinoni A, Falco M, Annicchiarico-Petruzzelli M et al. Two new p73 splice variants, g and d, with different transcriptional activity. J Exp Med 1998; 188: 1763–1768.
Kamiya M, Takeuchi Y, Katho M, Yokoo H, Sasaki A, Nakazato Y . Expression of p73 in normal skin and proliferative skin lesions. Pathol Int 2004; 54: 890–895.
Marshall CB, Mays DJ, Beeler JS, Rosenbluth JM, Boyd KL, Santos Guasch GL et al. p73 is required for multiciliogenesis and regulates the Foxj1-associated gene network. Cell Rep 2016; 14: 2289–2300.
Deutsch GB, Zielonka EM, Coutandin D, Weber TA, Schafer B, Hannewald J et al. DNA damage in oocytes induces a switch of the quality control factor TAp63alpha from dimer to tetramer. Cell 2011; 144: 566–576.
Davison TS, Nie X, Ma W, Lin Y, Kay C, Benchimol S et al. Structure and functionality of a designed p53 dimer. J Mol Biol 2001; 307: 605–617.
Marcel V, Dichtel-Danjoy ML, Sagne C, Hafsi H, Ma D, Ortiz-Cuaran S et al. Biological functions of p53 isoforms through evolution: lessons from animal and cellular models. Cell Death Differ 2011; 18: 1815–1824.
Ou HD, Löhr F, Vogel V, Mäntele W, Dötsch V . Structural evolution of C-terminal domains in the p53 family. EMBO J 2007; 26: 3463–3473.
Heering J, Jonker HR, Lohr F, Schwalbe H, Dotsch V . Structural investigations of the p53/p73 homologs from the tunicate species Ciona intestinalis reveal the sequence requirements for the formation of a tetramerization domain. Protein Sci 2016; 25: 410–422.
Nicholls CD, McLure KG, Shields MA, Lee PW . Biogenesis of p53 involves cotranslational dimerization of monomers and posttranslational dimerization of dimers. Implications on the dominant negative effect. J Biol Chem 2002; 277: 12937–12945.
Coutandin D, Osterburg C, Srivastav RK, Sumyk M, Kehrloesser S, Gebel J et al. Quality control in oocytes by p63 is based on a spring-loaded activation mechanism on the molecular and cellular level. Elife 2016; 5: e13909.
Yang A, Zhu Z, Kettenbach A, Kapranov P, McKeon F, Gingeras TR et al. Genome-wide mapping indicates that p73 and p63 co-occupy target sites and have similar dna-binding profiles in vivo. PLoS One 2010; 5: e11572.
Yang A, Walker N, Bronson R, Kaghad M, Oosterwegel M, Bonnin J et al. p73-deficient mice have neurological, pheromonal and inflammatory defects but lack spontaneous tumours. Nature 2000; 404: 99–103.
Luh LM, Kehrloesser S, Deutsch GB, Gebel J, Coutandin D, Schafer B et al. Analysis of the oligomeric state and transactivation potential of TAp73alpha. Cell Death Differ 2013; 20: 1008–1016.
Buchner L, Guntert P . Increased reliability of nuclear magnetic resonance protein structures by consensus structure bundles. Structure 2015; 23: 425–434.
Guntert P, Buchner L . Combined automated NOE assignment and structure calculation with CYANA. J Biomol NMR 2015; 62: 453–471.
Shen Y, Delaglio F, Cornilescu G, Bax A . TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J Biomol NMR 2009; 44: 213–223.
Koradi R, Billeter M, Guntert P . Point-centered domain decomposition for parallel molecular dynamics simulation. Comput Phys Commun 2000; 124: 139–147.
Ponder JW, Case DA . Force fields for protein simulations. Adv Protein Chem 2003; 66: 27–85.
Winter G . xia2: an expert system for macromolecular crystallography data reduction. J Appl Crystallogr 2010; 43: 186–190.
McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ . Phaser crystallographic software. J Appl Crystallogr 2007; 40: 658–674.
Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 2010; 66 (Pt 2): 213–221.
Chen VB, Arendall WB 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 2010; 66 (Pt 1): 12–21.
Goddard TD, Kneller DG . SPARKY 3. University of California, San Francisco, CA, USA.
Straub WE, Weber TA, Schafer B, Candi E, Durst F, Ou HD et al. The C-terminus of p63 contains multiple regulatory elements with different functions. Cell Death Dis 2010; 1: e5.
Acknowledgements
The research was funded by the DFG (DO 545/8-1), the Center for Biomolecular Magnetic Resonance (BMRZ) and the Cluster of Excellence Frankfurt (Macromolecular Complexes). DC was supported by a Boehringer Ingelheim Fonds PhD Fellowship. The Structural Genomics Consortium is a registered charity (number 1097737) that receives funds from the Canadian Institutes for Health Research, the Canadian Foundation for Innovation, Genome Canada through the Ontario Genomics Institute, GlaxoSmithKline, Karolinska Institutet, the Knut and Alice Wallenberg Foundation, the Ontario Innovation Trust, the Ontario Ministry for Research and Innovation, Merck & Co., Inc., the Novartis Research Foundation, the Swedish Agency for Innovation Systems, the Swedish Foundation for Strategic Research, and the Wellcome Trust. P.G. is supported by a Lichtenberg professorship of the Volkswagen Foundation.
Author contributions
JG, LL, DC, PG, SK and VD designed the experiments. JG, LL, DC, CO, FL, BS, AF, MS, LB, TK, ES and SM performed the experiments and analyzed the data. JG, LL, DC and VD prepared the manuscript.
Accession numbers
The coordinates of the structures reported here have been deposited in the Protein Data Bank (accession numbers 2NB1 and 5HOC).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by G Melino
Supplementary Information accompanies this paper on Cell Death and Differentiation website
Supplementary information
Rights and permissions
About this article
Cite this article
Gebel, J., Luh, L., Coutandin, D. et al. Mechanism of TAp73 inhibition by ΔNp63 and structural basis of p63/p73 hetero-tetramerization. Cell Death Differ 23, 1930–1940 (2016). https://doi.org/10.1038/cdd.2016.83
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cdd.2016.83
This article is cited by
-
p63: a crucial player in epithelial stemness regulation
Oncogene (2023)
-
Disease-related p63 DBD mutations impair DNA binding by distinct mechanisms and varying degree
Cell Death & Disease (2023)
-
DARPins detect the formation of hetero-tetramers of p63 and p73 in epithelial tissues and in squamous cell carcinoma
Cell Death & Disease (2023)
-
Designed Ankyrin Repeat Proteins as a tool box for analyzing p63
Cell Death & Differentiation (2022)
-
Structural diversity of p63 and p73 isoforms
Cell Death & Differentiation (2022)