Abstract
The association between inflammation and endoplasmic reticulum (ER) stress has been described in many diseases. However, if and how chronic inflammation governs the unfolded protein response (UPR) and promotes ER homeostasis of chronic inflammatory disease remains elusive. In this study, chronic inflammation resulted in ER stress in mesenchymal stem cells in the setting of periodontitis. Long-term proinflammatory cytokines induced prolonged ER stress and decreased the osteogenic differentiation of periodontal ligament stem cells (PDLSCs). Interestingly, we showed that chronic inflammation decreases the expression of lysine acetyltransferase 6B (KAT6B, also called MORF), a histone acetyltransferase, and causes the upregulation of a key UPR sensor, PERK, which lead to the persistent activation of the UPR in PDLSCs. Furthermore, we found that the activation of UPR mediated by MORF in chronic inflammation contributes to the PERK-related deterioration of the osteogenic differentiation of PDLSCs both in vivo and in vitro. Taken together, our results suggest that chronic inflammation compromises UPR function through MORF-mediated-PERK transcription, which is a previously unrecognized mechanism that contributes to impaired ER function, prolonged ER stress and defective osteogenic differentiation of PDLSCs in periodontitis.
Similar content being viewed by others
Main
The endoplasmic reticulum (ER) primarily functions in protein biosynthesis and folding, lipid trafficking, metabolism and intracellular calcium storage. Elevated physiological demand for protein folding can cause the accumulation of misfolded or unfolded proteins in the ER lumen, a state called ER stress.1 The proper folding of proteins can be achieved by the unfolded protein response (UPR), which mitigates such ER stress and mediates cellular adaptation by an integrated signal transduction pathway through three canonical stress sensors located at the ER membrane: protein kinase RNA-like ER kinase (PERK), inositol-requiring protein 1 (IRE1) and activating transcription factor 6 (ATF6).1, 2 Chronic inflammation is associated with UPR activation, while the chronic inflammatory microenvironment can give rise to a certain number of pathological conditions, including rheumatoid arthritis, diabetes, neurodegenerative and neuromuscular inflammatory diseases, and periodontitis.3, 4, 5, 6 However, how chronic inflammation causes diseases and triggers ER stress has not been fully elucidated.
Periodontitis is a global widespread human chronic inflammatory disease.6 Accumulating evidence has demonstrated that UPR-related genes are upregulated in periodontitis, which suggests an association between periodontitis and ER stress.7, 8, 9 Periodontal ligament stem cells (PDLSCs) are a group of mesenchymal stem cells (MSCs) derived from the periodontal ligament10, 11 that show tissue specificity and impaired differentiation capability and have a close relationship with periodontitis.12, 13, 14 Controversial evidence was found in the regulation of the osteogenic differentiation of MSCs by UPR activation.15, 16, 17, 18, 19, 20 However, how ER stress results in the abnormal differentiation of PDLSCs remains largely unknown.
Histone acetyltransferases (HATs), such as MORF,21, 22 have diverse functions, such as epigenetic control,23, 24 transcriptional regulation,25, 26 DNA replication27 and cellular signaling,28, 29 due to their distinct domains. Reports have revealed that MORF-deficient mice showed defects in cerebral and bone development,30 which suggested that MORF plays a pivotal role in osteogenesis. Recent studies also showed that HATs could regulate UPR-related genes.31, 32, 33 In this study, we observed ER stress in MSCs from periodontitis patients (P-PDLSCs). To further explore the role of ER stress in periodontitis-associated chronic inflammation, prolonged ER stress in MSCs from normal patients (H-PDLSCs) was induced by long-term treatment with proinflammatory cytokines. However, short-term treatment with proinflammatory cytokines could not sustain ER stress in H-PDLSCs. As described above, our study investigated the effects of long-term inflammation on the decrease of MORF expression in PDLSCs, which transcriptionally represses the expression of the UPR sensor PERK. In addition, ER stress impairs the osteogenic differentiation of PDLSCs. Notably, the decreased osteogenic differentiation of PDLSCs induced by ER stress is also mediated by the PERK pathway but not by IRE1 or ATF6.
Results
Activation of ER stress and the UPR target genes in MSCs isolated from the chronic inflammatory microenvironment
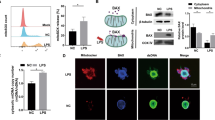
We isolated H-PDLSCs and P-PDLSCs and characterized them by flow cytometry analysis. Both H-PDLSCs and P-PDLSCs expressed the surface markers of MSCs (Supplementary Figures S1A and B). To investigate whether ER stress was induced in P-PDLSCs, we used transmission electron microscopy (TEM) analysis to observe the ER. Compared with H-PDLSCs (Figure 1a), our results showed more dilated and abundant ER in P-PDLSCs (Figure 1b). Meanwhile, we found continuous upregulation of the canonical UPR target genes PERK (also called EIF2AK3), GRP78 (HSPA5/BiP), GRP94 (HSP90B1/TRA1) and CHOP (DDIT3)34 (Figure 1c) in three consecutive passages of P-PDLSCs compared with H-PDLSCs (Passage 2–4). These results indicate that the chronic inflammatory environment induces ER stress and subsequently produces the UPR in PDLSCs.
Activation of the UPR target genes in MSCs in periodontitis-associated chronic inflammation. (a and b) Transmission electron microscopy (TEM) images of the endoplasmic reticulum (ER) in H-PDLSCs and P-PDLSCs. The overall view of cells with a low magnification (left column, scale bar, 5 μm) and a high magnification (right column, scale bar, 1 μm). The morphology of ERs is shown in the red rectangle. The results showed more dilated and abundant ERs in P-PDLSCs. (c) The expressions of UPR target genes (PERK, GRP78, GRP94, CHOP) in consecutive passages of H-PDLSCs and P-PDLSCs (Passage 2–4) were determined by real-time RT-PCR. The results were normalized to GAPDH mRNA expression. The representative results were from three independent experiments. The error bars represent the S.D. from the mean values.*P<0.05, **P<0.01, ***P<0.001
Chronic inflammation triggers the UPR and impairs the osteogenic differentiation of H-PDLSCs
To determine whether long-term inflammatory stimulation induces UPR in PDLSCs, we used interleukin-1 β (IL-1β) and tumor necrosis factor α (TNF-α), which are highly expressed in the periodontal microenvironment in patients,14, 35, 36 to treat H-PDLSCs and observe the UPR response in the cells. After continuous exposure of H-PDLSCs using IL-1β (5 ng/ml; Figure 2a) and TNF-α (10 ng/ml; Figure 2b) treatment separately for 7 days, we observed the activation of UPR by real-time RT-PCR. Notably, we found that high levels of the UPR did not decrease in three consecutive passages (Passage 2–4) with the removal of the stimulus.
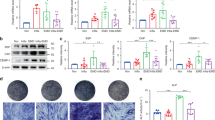
Chronic inflammation triggers the UPR and impairs the osteogenic differentiation of H-PDLSCs. (a and b) The expressions of UPR target genes (PERK, GRP78, GRP94, CHOP) were measured in H-PDLSCs cultured with IL-1β and TNF-α, respectively, for 7 days (Passage 2), and in consecutive passages of H-PDLSCs (Passage 3 and 4) with the removal of the stimulus. The results were normalized to GAPDH mRNA expression. (c and d) Western blot analysis showed the protein levels of Runx2 and OCN in H-PDLSCs treated with IL-1β (5 ng/ml) and TNF-α (10 ng/ml) after osteogenic induction for 14 days. β-Actin was used as an internal control. (e and f) Alizarin red staining of H-PDLSCs incubated with IL-1β and TNF-α was performed and quantified after 28 days of osteogenic induction. Scale bar, 50 μm, n=3. The representative results were from three independent experiments. The error bars represent the S.D. from the mean values. *P<0.05, **P<0.01, ***P<0.001
Our previous study showed the defective osteogenic differentiation of P-PDLSCs.12, 13 To determine whether the inflammatory microenvironment results in the osteogenic impairment of PDLSCs, we tested the osteogenesis of H-PDLSCs treated with IL-1β and TNF-α separately. Western blot analysis and real-time RT-PCR showed that the osteogenic marker proteins and genes of runt-related transcription factor 2 (Runx2) and osteocalcin (OCN) of H-PDLSCs were decreased with IL-1β (Figure 2c and Supplementary Figures S2A and B) and TNF-α (Figure 2d and Supplementary Figures S2C and D) treatment, respectively. Alizarin red staining demonstrated that IL-1β- and TNF-α-treated cells formed fewer mineralization nodules (Figures 2e and f). The apoptosis of P-PDLSCs and H-PDLSCs showed no significant difference at 6 h (Supplementary Figures S3A and B). After either TNF-α (10 ng/ml) or IL-1β (5 ng/ml) treatment, respectively, the apoptosis of H-PDLSCs was not increased compared with H-PDLSCs without treatment at 7 days (Supplementary Figures S3E–G). All these studies showed that chronic inflammation triggers the UPR and contributes to the deteriorative osteogenic differentiation of H-PDLSCs.
Periodontitis-associated chronic inflammation triggers UPR activation with MORF mediation
To evaluate whether inflammatory cytokines can induce the UPR after short-term stimulation, we conducted a series of time-course experiments with IL-1β- and TNF-α-treated H-PDLSCs. However, after 0–24 h of stimulation with IL-1β (Figure 3a) and TNF-α (Figure 3b), we found that the expressions of the UPR target genes were increased at the earlier stage but were returned to the original level at the later stage (within 24 h). These results indicate that long-term stimulation by inflammation may cause transcriptional changes in PDLSCs, which results in UPR activation.
Periodontitis-associated chronic inflammation triggers UPR activation with MORF mediation. (a and b) The expressions of UPR target genes (PERK, GRP78, GRP94, CHOP) were measured in H-PDLSCs treated with IL-1β and TNF-α for 0, 3, 6, 9, 12 and 24 h. (c) Western blot showed that MORF expression was decreased after transfection with MORF siRNA. (d) The expressions of UPR genes in H-PDLSCs and H-PDLSCs transfected with MORF siRNA were measured by real-time RT-PCR. The expressions of PERK, ATF4 and CHOP were elevated after the knockdown of MORF in H-PDLSCs. The results were normalized to GAPDH mRNA expression. (e) Transmission electron microscopy (TEM) images of the endoplasmic reticulum (ER) in H-PDLSCs and H-PDLSCs transfected with MORF siRNA. The overall view of cells with a low magnification (left column, scale bar, 5 μm) and a high magnification (right column, scale bar, 1 μm). The morphology of the ERs is shown in the red rectangle. The results showed more dilated and abundant ERs in H-PDLSCs transfected with MORF siRNA. (f) Western blot showed that MORF expression was increased after transfection with MORF overexpression vector. β-Actin was used as an internal control. (g) The expressions of UPR genes in H-PDLSCs and H-PDLSCs transfected with the MORF overexpression vector were measured by real-time RT-PCR. The expressions of PERK, ATF4 and CHOP were downregulated after the overexpression of MORF in H-PDLSCs. The results were normalized to GAPDH mRNA expression. The representative results were from three independent experiments. The error bars represent the S.D. from the mean values.*P<0.05, **P<0.01, ***P<0.001
Because previous studies showed that HATs played an important role in ER stress,31, 32, 33 we first screened the expressions of the HAT family in P-PDLSCs and H-PDLSCs. We identified that lysine acetyltransferase 2A (KAT2A), lysine acetyltransferase 3B (KAT3B), lysine acetyltransferase 6A (KAT6A) and MORF (also called KAT6B) of P-PDLSCs were reduced in the periodontitis-associated chronic inflammatory microenvironment (Supplementary Figure S4A). To further assess which HAT activates the UPR, we used siRNA to knockdown the expression of the HATs and observe the UPR response. Western blot analysis showed that the MORF expression of H-PDLSCs was decreased after transfection with MORF siRNA (Figure 3c and Supplementary Figure S5A). The findings revealed that knockdown of MORF resulted in higher expression of the UPR target genes PERK, activating transcription factor 4 (ATF4) and CHOP (Figure 3d), which elucidated the activation of the UPR pathway, PERK. However, knockdown of KAT2A (Supplementary Figure S4B), KAT3B (Supplementary Figure S4C) or KAT6A (Supplementary Figure S4D) did not activate the UPR. We also found more dilated and abundant ER in MORF siRNA-transfected H-PDLSCs by TEM (Figure 3e). To further elucidate the effect of MORF on ER stress, we overexpressed the MORF vector to transfect cells. We found that the overexpression of MORF decreased the expression of the UPR target genes PERK, ATF4 and CHOP (Figures 3f and g and Supplementary Figure S5B). These results suggest that periodontitis-associated chronic inflammation triggers UPR activation with MORF mediation.
MORF represses PERK and impairs MSC osteogenic differentiation in periodontitis-associated chronic inflammation
We further explored the role of MORF in the chronic inflammation and osteogenic differentiation of PDLSCs. Over a long period of stimulation of 7 days, the expression of MORF was decreased in P-PDLSCs (Figure 4a), H-PDLSCs cultured with IL-1β (Figure 4b) and TNF-α (Figure 4c), respectively, and in three consecutive passages with the removal of the stimulus (Passage 2–4). Western blot analysis showed that the osteogenic marker proteins Runx2 and OCN decreased after knockdown of MORF in H-PDLSCs (Figure 4d and Supplementary Figure S5C). Alizarin red staining showed that the formation of mineralized nodules was decreased after the knockdown of MORF in H-PDLSCs (Figure 4e).
MORF represses PERK and impairs MSC osteogenic differentiation in periodontitis-associated chronic inflammation. (a) A decreased expression of MORF in consecutive passages of P-PDLSCs was shown by real-time RT-PCR compared with H-PDLSCs (Passage 2–4). (b and c) Decreased expressions of MORF in H-PDLSCs treated with IL-1β and TNF-α for 7 days and in consecutive passages with the removal of the stimulus were shown compared with H-PDLSCs (Passage 2–4). The results were normalized to GAPDH mRNA expression. (d) Decreased expressions of osteogenic protein Runx2 and OCN were confirmed by western blot analysis after knockdown of MORF in H-PDLSCs. β-Actin was used as an internal control. (e) Alizarin red staining showed decreased mineralized nodules after knockdown of MORF in H-PDLSCs after 28 days of osteogenic induction. Scale bar, 50 μm, n=3. (f) H-PDLSCs, P-PDLSCs, H-PDLSCs transfected with MORF siRNA and H-PDLSCs transfected with MORF overexpression vector were subjected to chromatin immunoprecipitation (ChIP) assays with the binding of MORF and rabbit IgG to PERK promoters. The PERK primers for the locus indicated were CHIP-ed with anti-MORF or IgG control. (g) The increased expression of PERK in H-PDLSCs transfected with MORF siRNA and the decreased expression of PERK in H-PDLSCs with MORF overexpression vector were measured by western blot analysis. β-Actin was used as an internal control. The representative results were from three independent experiments. The error bars represent the S.D. from the mean values. *P<0.05, ***P<0.001
To test whether PERK is a direct target of MORF, we carried out chromatin immunoprecipitation (ChIP) analysis on H-PDLSCs, P-PDLSCs, H-PDLSCs transfected with MORF siRNA and H-PDLSCs transfected with overexpressed MORF. The PCR results demonstrated that DNA bearing the PERK-binding site in H-PDLSCs transfected with overexpressed MORF was prosperously immunoprecipitated by anti-MORF antibodies, which were different from H-PDLSCs, but not by control IgG. The DNA bearing the PERK-binding site in H-PDLSCs was also prosperously immunoprecipitated by anti-MORF antibodies (Figure 4f), which indicated that MORF might directly bind to PERK gene promoters. Moreover, western blot analysis showed the upregulated expression of PERK after MORF knockdown and the downregulated expression of PERK after MORF overexpression in H-PDLSCs (Figure 4g and Supplementary Figure S5D). These results suggest that MORF directly binds to PERK promoter, represses PERK gene expression and activates UPR response.
UPR activation leads to the decreased osteogenic differentiation of PDLSCs
Because decreased expression of MORF increases PERK expression and activates the UPR response and impairs osteogenic differentiation of H-PDLSCs during chronic periodontitis, we sought to determine whether activated UPR also leads to impaired osteogenic differentiation of PDLSCs. Then, we treated PDLSCs with two UPR inducers, tunicamycin (Tm) and thapsigargin (Tg), and an inhibitor 4-phenyl butyric acid (4-PBA), before osteogenic induction. The results of real-time RT-PCR confirmed that all three UPR pathways were drastically increased with both the Tm (Figure 5a) and Tg (Figure 5b) treatments for 6 h. However, the three UPR pathways of P-PDLSCs were decreased after 4-PBA treatment for 6 h (Figure 5c). Moreover, Tm treatment resulted in greater upregulation of UPR genes, compared with Tg treatment.
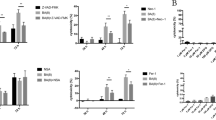
UPR activation leads to the decreased osteogenic differentiation of PDLSCs. (a and b) Increased expressions of UPR genes were found in H-PDLSCs treated with 1 μg/ml Tm and 1 μmol/l Tg for 6 h. The results were normalized to GAPDH mRNA expression. (c) Increased expressions of UPR genes were found in P-PDLSCs treated with 5 mmol/l 4-PBA for 6 h. (d and e) Decreased expressions of osteogenic protein Runx2 and OCN were shown by western blot analysis in H-PDLSCs treated with Tm and Tg after osteogenic induction. β-Actin was used as an internal control. (f and g) Alizarin red staining showed decreased numbers of mineralized nodules in H-PDLSCs treated with Tm and Tg after 28 days of osteogenic induction. Scale bar, 50 μm, n=3. (h) Increased expressions of the osteogenic proteins Runx2 and OCN were shown by western blot analysis in P-PDLSCs treated with 4-PBA after osteogenic induction. β-Actin was used as an internal control. (i) Alizarin red staining showed increased numbers of mineralized nodules in P-PDLSCs treated with 4-PBA after 28 days of osteogenic induction. Scale bar, 50 μm, n=3. The representative results were from three independent experiments. The error bars represent the S.D.from the mean values. *P<0.05, **P<0.01, ***P<0.001
Next, we used Tm, Tg and 4-PBA to treat PDLSCs separately in osteogenic induction. According to real-time RT-PCR and western blot analysis, the osteogenic marker proteins and genes of Runx2 and OCN of H-PDLSCs were decreased with both Tm (Figure 5d and Supplementary Figures S6A and B) and Tg (Figure 5e and Supplementary Figures S6C and D) treatments. Alizarin red staining showed the decreased mineralization nodules of H-PDLSCs after treatment with Tm (Figure 5f) and Tg (Figure 5g). However, the osteogenic differentiation of P-PDLSCs was rescued after 4-PBA treatment, as the osteogenic marker proteins and genes Runx2 and OCN of H-PDLSCs and the formation of mineralization nodules increased (Figures 5h and i and Supplementary Figures S6E and F). We also found that Tm (1 μg/ml) and Tg (1 μmol/l) treated PDLSCs showed no significant difference in cell apoptosis compared with the cells without treatment at 6 h (Supplementary Figures S3A, C and D). These results indicate that ER stress induced by MORF regulates the osteogenic differentiation of PDLSCs.
UPR activation regulates the osteogenic differentiation of PDLSCs through the PERK pathway
To determine whether the impaired osteogenic differentiation of PDLSCs is affected by the PERK pathway through MORF regulation, we used siRNAs to knockdown three sensors of ER stress and observed the osteogenic differentiation of P-PDLSCs. The results showed the decreased expression of PERK (Figures 6a and b and Supplementary Figures S7A and B), IRE1 (Supplementary Figures S8A and B) and ATF6 (Supplementary Figures S8C and D) by real-time RT-PCR and western blot analysis. After osteogenic induction, we found that the osteogenic marker proteins and genes of Runx2 and OCN were markedly increased in PERK siRNA-transfected P-PDLSCs compared with control siRNA-transfected P-PDLSCs (Figure 6c and Supplementary Figure S7C). The formation of mineralization nodules was also increased in PERK siRNA-transfected P-PDLSCs compared with control siRNA-transfected P-PDLSCs (Figure 6d). However, the osteogenic differentiation of P-PDLSCs was not rescued after transfection with IRE1 siRNA or ATF6 siRNA (Supplementary Figures S8E–N).
UPR activation regulates the osteogenic differentiation of PDLSCs through the PERK pathway. (a and b) The expression of PERK was downregulated in P-PDLSCs after being transfected with PREK siRNA as assayed by real-time RT-PCR and western blot. (c) The osteogenic proteins Runx2 and OCN were decreased in P-PDLSCs after knockdown of PERK as assessed by western blot. β-Actin was used as an internal control. (d) Alizarin red staining showed the increased formation of mineralized nodules in P-PDLSCs transfected with PERK siRNA after 28 days of osteogenic induction. Scale bar, 50 μm, n=3. (e) The alveolar bone loss of SD rats (n=3) was determined by micro-CT and H&E staining. Four sites for two molars (one site for each root of one tooth) were analyzed morphometrically. The results of micro-CT and HE staining showed more alveolar bone loss in both the Tm (0.1 mg/ml) and Tg (0.1 mmol/l) groups compared with the saline (10 μl) group. Injection of GSK2606414 (10 μmol/l, PERK inhibitor) rescued the bone loss caused by Tm (0.1 mg/ml). Scale bar, 200 μm, n=3. The representative results were from three independent experiments. The error bars represent the S.D. from the mean values. *P<0.05, **P<0.01, ***P<0.001
Subsequently, to verify whether UPR activation causes bone loss and PERK inhibition rescues bone formation in vivo, we used Tm and Tg to induce alveolar bone loss in rats and the PERK inhibitor GSK2606414, which was found to be a specific inhibitor of PERK,37, 38, 39, 40 to inhibit drug-induced bone loss. We injected 10 μl saline, 0.1 mg/ml Tm, 0.1 mmol/l Tg and Tm in combination with 10 μmol/l of the PERK inhibitor GSK2606414 into the maxillary palatal gingiva between the first and second upper molars of SD rats. We found that the Tm-treated group showed more bone loss than the other groups, as the highest distance from the cemento-enamel junction (CEJ) to the alveolar bone crest was observed in all the four sites (Figure 6e). However, with the administration of Tm in combination of GSK2606414, the alveolar bone loss of two molars was noticeably reduced (Figure 6e). As described above, we demonstrate that ER stress regulates the osteogenic capacity of PDLSCs through PERK both in vitro and in vivo.
Discussion
In this study, we uncovered a previously unrecognized link between UPR signaling and MSC function in periodontitis-associated chronic inflammation. For the first time, we report that prolonged ER stress activates the UPR sensor PERK and then impairs MSC differentiation in the periodontitis-associated chronic inflammatory microenvironment. Meanwhile, as summarized in Figure 7, we reveal the novel mediator role of MORF to the UPR, specifically in its expression and function.
Schematic diagram shows that the mechanisms of decreased MORF lead to prolonged ER stress in periodontitis-associated chronic inflammation. Inflammatory cytokines compromise UPR function through MORF-mediated-PERK transcription, which contributes to impaired ER function, prolonged ER stress and defective osteogenic differentiation of PDLSCs in periodontitis-associated chronic inflammation
Inflammation can activate the UPR according to different cell types. In murine fibrosarcoma L929 cells, TNF-α induced the UPR in a reactive oxygen species (ROS)-dependent fashion.41 In demyelinating disorders and central nervous inflammation, interferon-γ (IFN-γ)-induced oligodendrocyte apoptosis was associated with the PERK–eIf2α pathway.42 However, low concentrations of lipopolysaccharide (1 ng/ml) can inhibit the ATF4–CHOP pathways in mononuclear macrophages;43, 44 therefore, further studies are needed to determine the dominant factors between the inflammatory environment and ER stress. Our study first observed the most obvious changes of ER morphology by TEM, which revealed ER stress in P-PDLSCs. Next, we used inflammatory cytokines to treat PDLSCs in vitro and proved that prolonged ER stress and the UPR can be triggered in the periodontal inflammatory environment.
The mechanism by which the inflammatory microenvironment causes ER stress and triggers the UPR is unclear, but several studies suggest that proinflammatory cytokines may induce the UPR in the accumulation of ROS and release Ca2+, which causes the accumulation of unfolded or misfolded proteins and mitochondrial dysfunction to induce ER stress.41, 42 Diseases, such as type 2 diabetes, cystic fibrosis, inflammatory bowel disease and periodontitis, are reported to be associated with ER stress.7, 8, 9, 45, 46, 47 Therefore, it is crucial to determine the role of ER stress in inflammatory diseases. In our study, we used IL-1β and TNF-α, which have been proved to play crucial roles and be secreted mainly by monocytes in periodontal diseases,14, 35, 36 to treat PDLSCs and mimic a long-term inflammatory microenvironment in vitro. Moreover, IL-1β and TNF-α have also been related to the UPR-mediated acute phase response.48 Our data showed that the cytokine-induced UPR peaked in the first 12 h of treatment and then declined to pre-induction levels within 24 h. However, we found that the UPR response of PDLSCs still existed after removing the stimulation of proinflammatory cytokines and passing the cells to the next two continuing passages. These results led us to hypothesize that long-term stimulation by inflammation may cause transcriptional changes in PDLSCsand result in UPR activation.
Because previous studies showed that HATs play an important role in ER stress,31, 32, 33 we turned our attention to the HAT family. After screening the entire HAT family, we isolated MORF, which conformed to our study. MORF deficiency in chronic inflammation triggers the UPR and results in high expression of PERK and its downstream genes ATF4 and CHOP in PDLSCs. MORF binding was found to be enriched in the promoter regions of PERK by CHIP analysis. Our results support the intriguing concept that inflammation decreases the expression of MORF, which acts as a transcriptional repressor in activating UPR. However, how chronic periodontitis causes persistent decreases in the expression of MORF still needs further study. The inflammatory microenvironment may regulate the expression of MORF by epigenetic modification or through the loop, in which paracrine factors of MSCs inhibit MORF expression. In addition, MORF possesses additional modules (such as PHD fingers),49 which may mediate its binding to other transcription factors in UPR genes. This evidence illustrates the possibility that MORF may function with other transcription factors and act as a transcriptional coregulator.
Previous studies showed that UPR activation can regulate osteogenic differentiation of MSCs, but it remains controversial. For example, the ER stress inducer ATF3 can suppress bone morphogenetic protein (BMP2)-induced alkaline phosphatase (ALP) expression in MC3T3-E1 cells.18 However, in OS732 cells, fluoride-induced osteoblastic and osteoclastic differentiation was due to the activation of ALP and receptor activator for nuclear factor-κB ligand (RANKL) mediated by the PERK pathway, but had no effect on Runx2 and osteoprotegerin.19 The alcohol-induced ER stress inhibited the osteogenesis of MSCs by suppressing the Wnt/β-catenin pathway, downregulating the Osf2/Cbfa1 gene, and activating TNF-α signaling by the PERK/ATF4/CHOP pathway.20 Meanwhile, the induction of ER stress caused a loss of stemness in a PERK–eIF2α-dependent manner in intestinal stem cells.50 In our study, because the inflammatory microenvironment also induced ER stress and the UPR response, we wondered whether the ER stress response of stem cells plays a pivotal role in regulating osteogenic differentiation. We performed the osteogenic differentiation assay in vitro, which showed that chronic inflammation led to the decreased osteogenic differentiation of PDLSCs. Moreover, we used siRNA transfection in vitro to further document the role of PERK pathway in mediating the osteogenic differentiation, but not other two branches of the UPR, IRE1 and ATF6. An experimental SD-rat periodontitis model was used to confirm the role of PERK in bone formation in vivo. Since the activated UPR may induce the apoptosis of PDLSCs,51 which also contribute to the decreased bone formation. We tested the apoptosis of P-PDLSCs and found no significant difference in apoptosis between P-PDLSCs and H-PDLSCs at 6 h. The apoptosis of H-PDLSCs after either long-term TNFα or IL-1β treatment was not increased compared with H-PDLSCs without treatment at 7 days. These results suggested that chronic inflammation triggered UPR might not induce apoptosis of PDLSCs. As stated above, we drew the conclusion that UPR activation regulates the osteogenic differentiation of PDLSCs through the PERK pathway.
In summary, our study revealed the mechanism by which chronic inflammation compromises UPR function through MORF-mediated-PERK transcription, which contributes to impaired ER function, prolonged ER stress and defective osteogenic differentiation of PDLSCs.
Materials and Methods
Cell culture
Healthy human tooth samples were obtained from 15 individuals, aged 25–40 years, from teeth that being extracted for orthodontic reasons. Periodontitis teeth were collected from 15 periodontitis patients aged 25–40 years. Patients diagnosed with periodontitis were defined as those with more than one periodontal pocket (depth>6 mm), attachment loss (depth≥5 mm) and alveolar bone loss (≥2/3).6 Teeth were extracted at the Department of Periodontology, School of Stomatology, Fourth Military Medical University. Consent forms were obtained before conducting this research project. The experimental protocols were approved by the Hospital Ethics Committee (No. IRB-REV-2015005). The patients included in this study had no systemic diseases, did not smoke and took no special medications over the past year.
PDLSCs were isolated and cultured as previously described.12, 52 Briefly, to obtain single-cell suspensions, periodontal ligament tissues were separated from the middle third of the root surface and digested in 3 mg/ml collagenase I (Sigma-Aldrich Corp, St Louis, MO, USA) for 2 h at 37 °C. Cells were then plated in six-well culture dishes (Costar; Corning Inc., Corning, NY, USA) in α-minimal essential medium (α-MEM; Gibco BRL, Gaithersburg, MD, USA) with 10% fetal bovine serum (Hangzhou Sijiqing Biological Engineering Materials Co., Ltd, Zhejing, China), 0.292 mg/ml l-glutamine (Invitrogen Life Technology, Carlsbad, CA, USA), 100 units/ml penicillin (Invitrogen) and 100 mg/ml streptomycin (Invitrogen) at 37 °C in 5% CO2. Cells were cultured for 2 weeks and the medium was changed every 3 days. Then, single-cell-derived colony cultures were obtained using the limiting dilution technique to further purify the stem cells. All colonies were then pooled and expanded. We used H-PDLSCs and P-PDLSCs at passage 2–5 (P2-P5). Each experiment used the same passage.
The ER stress inducers tunicamycin (Tm, 1 μg/ml) and thapsigargin (Tg, 1 μmol/l) and inhibitor 4-phenyl butyric acid (4-PBA, 5 mmol/l) were purchased from Sigma. H-PDLSCs (P3) were cultured in α-MEM additionally supplemented with Tm and Tg for 6 h; P-PDLSCs (P3) were cultured in α-MEM additionally supplemented with 4-PBA for 6 h. The inflammatory cytokines TNF-α (10 ng/ml) and IL-1β (5 ng/ml) were from Pepro-Tech (Rocky Hill, NJ, USA). For near-term stimulation, H-PDLSCs (P2) incubated with TNF-α or IL-1β were treated for 0, 3, 6, 9, 12 or 24 h. For long-term stimulation, H-PDLSCs, P-PDLSCs and H-PDLSCs (P2) incubated with TNF-α or IL-1β were cultured for 7 days and the medium was changed every other day. Then, the stimulus was removed and the same samples were passed to passage 3 to 4 and harvested for the following tests.
Flow cytometry
PDLSCs were stained with antibodies for stem cell surface markers and analyzed by flow cytometry as described previously. Briefly, to identify the phenotypes of PDLSCs, 106 cells per ml at passage 2 were incubated with phycoerythrin (PE)-conjugated monoclonal antibodies for human CD146 (Biolegend, San Diego, CA, USA), CD34, CD90, CD14 (eBioscience, San Diego, CA, USA) and fluorescein isothiocyanate (FITC)-conjugated monoclonal antibodies for STRO-1 (BioLegend) and CD105 (eBioscience) according to the manufacturer’s instructions. The incubation procedure was carried out at 4 °C for 1 h away from light. After washing with 3% PBS, cells were subjected to flow cytometric analysis (Beckman Coulter Inc., Fullerton, CA, USA).
About 106 cells were washed, trypsinized and centrifuged. The cells were labeled for 15 min with FITC-conjugated Annexin V (7Sea Pharmatech Co., Ltd, Shanghai, China) in culture medium (final concentration 0.1–1 mg/ml), then counterstained with 2 mg propidium iodide per ml culture medium (7Sea Pharmatech Co.). At least 20 000 cells were considered using the flow cytometric analysis (Beckman Coulter Inc.).
Vector construction and cell transfection
The MORF (GenBank Accession No. 23522) cDNA was amplified by PCR using the primer pairs: forward: 5′-GGGAGACCCAAGCTGGCTAGCGGCCACCATGGTAAAACTTGCAAACCCAC-3′; reverse: 5′-GCCCTCTAGACTCGAGCGGCCGCCCCTTCTCATGTAGGAGCCATTG-3′. And then, the restriction enzymes NheI and NotI were used to clone the above fragments into the backbone vector pcDNATM6/V5-His A/C vector backbone (Addgene, Cambridge, MA, USA) using a one-step cloning kit (C112-01, Clon Express; Vazyme Biotech Co., Ltd, Nan Jing, China). The overexpression vector pcDNATM6-MORF was constructed and transfected into the PDLSCs by Lipofectamine 2000 (Invitrogen).
To inhibit chaperone expression, cells were transfected with PERK, IRE1, ATF6, MORF, KAT2A, KAT6A and KAT3B siRNA conjugated with Lipofectamine 2000 for 6 h. All the agents were purchased from Santa Cruz (Santa Cruz, Dallas, TX, USA). The cells were incubated at 37 °C in an incubator for 24–48 h before further assay.
Osteogenic differentiation in vitro
For osteogenic induction in vitro, cells were seeded on six-well culture dishes and cultured in the growth medium until the cells reached confluence. To induce osteogenic differentiation, the medium was changed to 100 μg/ml ascorbic acid, 2 mol/l β-glycerophosphate and 10 nmol/l dexamethasone (Sigma). One week after osteogenic induction, total RNA was extracted and analyzed for the presence of osteogenic genes by real-time RT-PCR. Total protein was extracted and analyzed for osteogenic markers by western blot after 14 days. After 28 days, calcium deposits formed by osteoblasts on the dishes were detected by staining with 1% Alizarin Red (Sigma). The medium was changed every 3 days, and experiments were performed in triplicate. The Alizarin red staining was performed as previously described.53 For quantification analysis, Alizarin red staining was extracted with 1% cetylpyridinium chloride (Sigma) and measured with a spectrophotometer (Epoch; BioTek, Winooski, VT, USA) at 562 nm. The results were normalized to the total protein quantity of each sample to exclude the influence of cell numbers.
Real-time RT-PCR of mRNA
RNA was extracted using TRIzol (Invitrogen) according to the manufacturer’s standard protocol. Random-primed cDNA was synthesized in a 20 μl reaction volume (Takara Bio, Shiga, Japan) for the reverse transcription of mRNA. The RT-PCR reactions were performed in a total volume of 10 μl using the SYBR Premix Ex TaqII kit (Takara) and detected on the CFX96 Real-Time System (Bio-Rad, Hercules, CA, USA). PCR was performed using standard settings: 94 °C for 3 min; 94 °C for 15 s and 60 °C for 30 s repeated for 40 cycles, then the dissociation stage. Each assay was run in triplicate for technical variations. Arbitrary mRNA concentrations were calculated using the relative standard curve method. Gene expression levels were determined using the ΔΔ−ct method normalized to the reference gene GAPDH. All primer sequences used in this study are described in Supplementary Table S1.
Western blot
The western blot analysis was performed as previously described.12 Proteins were extracted with RIPA lysis buffer with protease inhibitor cocktail (Sigma) at 4 °C. After centrifugation, the soluble protein in the extract was quantified by a G250 protein assay (Beyotime, Haimen, Jiangsu, China). Samples were loaded onto 10% Tris-glycine SDS-polyacrylamide gel (Invitrogen), transferred onto a PVDF membrane (Merck Millipore, Billerica, MA, USA) and blocked with 5% nonfat milk powder in PBST (PBS with 0.1% Tween). The membranes were incubated overnight at 4 °C with the following primary antibodies. The primary antibodies for human Runx2 (Cell Signaling, Beverly, MA, USA), OCN (Santa Cruz), PERK (Cell Signaling), MORF (Abcam, Cambridge, MA, USA), IRE1 (Cell Signaling), ATF6 (Santa Cruz) and β-actin (Cowin Biotech, Beijing, China) were used in this study. The membranes were then incubated with horseradish peroxidase (HRP)-conjugated secondary antibody (Cowin Biotech) at room temperature for 2 h. We visualized the signals by exposing the membrane to chemiluminescent HRP substrate (Thermo Scientific Inc., Rockford, IL, USA) using the Western-Light Chemiluminescent Detection System (Peiqing, Shanghai, China) according to the manufacturer’s instructions.
TEM
Cells were washed with serum-free media and fixed with 4% glutaraldehyde and 4% paraformaldehyde (Sigma, both fixatTransives were diluted in phosphate-buffered saline, pH 7.2), dehydrated in a graded ethanol series and embedded in situ in an LX-812 resin (Ladd Research Industries Inc., Williston, VT, USA). Ultrathin sections were stained with uranyl acetate (30 min) and lead citrate (10 min) and observed by a FEI Tecnai G12 Spirit BioTwin transmission electron microscope (FEI Company, Hillsboro, Oregon, USA) with an accelerating voltage of 100 kV. Digital images were captured on a Veleta CCD camera (Olympus-SIS, Münster, Germany).
ChIP
We used a CHIP assay kit (Merck Millipore; Catalog 17–371) to confirm the binding between proteins and gene promoters according to the manufacturer’s protocol. Normal rabbit IgG (Merck Millipore) was used as a negative control. We obtained the PERK promoter region sequence from Genebank (Gene ID: 9451). We chose the upstream 2000 base sequences of the CDS coding region of PERK and randomly allocated it for four sequences. We carried out ChIP analysis using the four sequences by real-time RT-PCR. The final result showed that only the second sequence of PERK promoter is effective. The primers of the promoter of PERK were: 5′-TGGTGCGATCTCAGCTCATTG-3′ (forward) and 5′-TGCCT GTAATCCCAGCCCTCT-3′ (reverse). All precipitated DNA samples were analyzed by real-time RT-PCR. The results were normalized to the input value, and all CHIP experiments were repeated at least three times.
Drug administration in experimental periodontitis of Sprague-Dawley (SD) rats
Experimental periodontitis was induced as previously described.54 All animal procedures were performed according to the guidelines of the Animal Care Committee of the Fourth Military Medical University, Xi’an, China. Twelve 8-week-old SD rats were randomly distributed into four groups: Saline; Tm (0.1 mg/ml); Tg (0.1 mmol/l); Tm (0.1 mg/ml)+GSK2606414 (10 μmol/l, PERK inhibitor, purchased from Merck Millipore) with three rats per group. Then, 10 μl drugs were injected in each group into the maxillary palatal gingiva between the first and second upper molars and repeated every other day on three separate days. All rats were anesthetized and killed by exsanguination on day 7.
Micro-CT analysis
The whole head of SD rats was removed, and the maxillary jaws were scanned and analyzed using a micro-CT system (Siemens Inveon Micro-CT, Munich, Germany). After the mandibles were scanned, rebuilt images of the bone surface were used to perform three-dimensional histomorphometric analysis with the same density. From the rebuilt images, the alveolar bone height was measured at four different sites in two molars by recording the distance from the CEJ to the alveolar bone crest. The distances were assessed as the mean distance of experiment groups and were compared with the saline group with statistical analysis.
Histological analysis
The maxillary jaws were fixed with 4% paraformaldehyde for 24 h, decalcified with 10% ethylene diamine tetraacetic acid (pH 8.0) for 4 weeks and embedded in paraffin. Embedded specimens were then sectioned into 4-mm-thick sections. For histological and histomorphometric analysis, sections were deparaffinized and stained with hematoxylin and eosin (H&E) and observed under light and polarized microscopy (BX50; Olympus Optical).
Statistical analysis
All experiments were repeated at least three times and the results were similar between repeats. The data are presented as the mean±S.D. The values between the two groups were calculated by independent two-tailed unpaired Student’s t-test, and the values between multiple comparisons were assessed with analysis of variance (ANOVA) with the Bonferroni correction. All statistical analyses were performed using Graph Pad Prism 5.0 (GraphPad Software, La Jolla, CA, USA), and P-values less than 0.05 were considered statistically significant.
Abbreviations
- ER:
-
endoplasmic reticulum
- UPR:
-
unfolded protein response
- MSCs:
-
mesenchymal stem cells
- KAT6B/MORF:
-
lysine acetyltransferase 6B
- PERK:
-
protein kinase RNA-like ER kinase
- IRE1:
-
inositol-requiring protein 1
- ATF6:
-
activating transcription factor 6
- HATs:
-
histone acetyltransferases
- P-MSCs:
-
MSCs from periodontitis patients
- H-MSCs:
-
MSCs from normal patients
- TEM:
-
transmission electron microscopy
- IL-1β:
-
interleukin-1 β
- TNF-α:
-
tumor necrosis factor α
- Runx2:
-
runt-related transcription factor 2
- OCN:
-
osteocalcin
- KAT2A:
-
lysine acetyltransferase 2A
- KAT6A:
-
lysine acetyltransferase 6A
- KAT3B:
-
lysine acetyltrans- ferase 3B
- ATF4:
-
activating transcription factor 4
- ChIP:
-
chromatin immunoprecipitation
- Tm:
-
tunicamycin
- Tg:
-
thapsigargin
- 4-PBA:
-
4-phenyl butyric acid
- CEJ:
-
the cemento-enamel junction
- ROS:
-
reactive oxygen species
- IFN-γ:
-
interferon-γ
- BMP2:
-
bone morphogenetic proteins
- ALP:
-
alkaline phosphatase
- RANKL:
-
receptor activator for nuclear factor-κ B ligand
- PE:
-
phycoerythrin
- FITC:
-
fluorescein isothiocynante
- SD rats:
-
Sprague-Dawley rats
References
Kang SW, Hegde RS . Lighting up the stressed ER. Cell 2008; 135: 787–789.
Walter P, Ron D . The unfolded protein response: from stress pathway to homeostatic regulation. Science 2011; 334: 1081–1086.
Hasnain SZ, Lourie R, Das I, Chen AC, McGuckin MA . The interplay between endoplasmic reticulum stress and inflammation. Immunol Cell Biol 2012; 90: 260–270.
Hotamisligil GS . Endoplasmic reticulum stress and the inflammatory basis of metabolic disease. Cell 2010; 140: 900–917.
Yang L, Calay ES, Fan J, Arduini A, Kunz RC, Gyfi SP et al. METABOLISM. S-Nitrosylation links obesity-associated inflammation to endoplasmic reticulum dysfunction. Science 2015; 349: 500–506.
Pihlstrom BL, Michalowicz BS, Johnson NW . Periodontal diseases. Lancet 2005; 366: 1809–1820.
Domon H, Takahashi N, Honda T, Nakajima T, Tabeta K, Abiko Y et al. Up-regulation of the endoplasmic reticulum stress-response in periodontal disease. Clin Chim Acta 2009; 401: 134–140.
Lee SI, Kang KL, Shin SI, Herr Y, Lee YM, Kim EC et al. Endoplasmic reticulum stress modulates nicotine-induced extracellular matrix degradation in human periodontal ligament cells. J Periodontal Res 2012; 47: 299–308.
Yamada H, Nakajima T, Domon H, Honda T, Yamazaki K . Endoplasmic reticulum stress response and bone loss in experimental periodontitis in mice. J Periodontal Res 2015; 50: 500–508.
Seo BM, Miura M, Gronthos S, Bartold PM, Batouli S, Brahim J et al. Investigation of multipotent postnatal stem cells from human periodontal ligament. Lancet 2004; 364: 149–155.
Wang L, Shen H, Zheng W, Tang L, Yang Z, Gao Y et al. Characterization of stem cells from alveolar periodontal ligament. Tissue Eng Part A 2011; 17: 1015–1026.
Liu Y, Liu W, Hu C, Xue Z, Wang G, Ding B et al. MiR-17 modulates osteogenic differentiation through a coherent feed-forward loop in mesenchymal stem cells isolated from periodontal ligaments of patients with periodontitis. Stem Cells 2011; 29: 1804–1816.
Li N, Liu N, Zhou J, Tang L, Ding B, Duan Y et al. Inflammatory environment induces gingival tissue-specific mesenchymal stem cells to differentiate towards a pro-fibrotic phenotype. Biol Cell 2013; 105: 261–275.
Kong X, Liu Y, Ye R, Zhu B, Zhu Y, Liu X et al. GSK3β is a checkpoint for TNF-α-mediated impaired osteogenic differentiation of mesenchymal stem cells in inflammatory microenvironments. Biochim Biophys Acta 2013; 1830: 5119–5129.
Kondo S, Hino SI, Saito A, Kanemoto S, Kawasaki N, Asada R et al. Activation of OASIS family, ER stress transducers, is dependent on its stabilization. Cell Death Differ 2012; 19: 1939–1949.
Guo FJ, Jiang R, Xiong Z, Xia F, Li M, Chen L et al. IRE1a constitutes a negative feedback loop with BMP2 and acts as a novel mediator in modulating osteogenic differentiation. Cell Death Dis 2014; 22: e1239.
Saito A, Ochiai K, Kondo S, Tsumagari K, Murakami T, Cavener DR et al. Endoplasmic reticulum stress response mediated by the PERK-eIF2(alpha)-ATF4 pathway is involved in osteoblast differentiation induced by BMP2. J Biol Chem 2011; 286: 4809–4818.
Park JK, Jang H, Hwang S, Kim EJ, Kim DE, Oh KB et al. ER stress-inducible ATF3 suppresses BMP2-induced ALP expression and activation in MC3T3-E1 cells. Biochem Biophys Res Commun 2014; 443: 333–338.
Lu P, Li X, Ruan L, Xu H, Liu Q et al. Effect of siRNA PERK on fluoride-induced osteoblastic differentiation in OS732 cells. Biol Trace Elem Res 2014; 159: 434–439.
Chen Y, Gao H, Yin Q, Chen L, Dong P, Zhang X et al. ER stress activating ATF4/CHOP-TNF-α signaling pathway contributes to alcohol-induced disruption of osteogenic lineage of multipotential mesenchymal stem cell. Cell Physiol Biochem 2013; 32: 743–754.
Champagne N, Bertos NR, Pelletier N, Wang AH, Vezmar M, Yang Y et al. Identification of a human histone acetyltransferase related to monocytic leukemia zinc finger protein. J Biol Chem 1999; 274: 28528–28536.
Champagne N, Pelletier N, Yang XJ . The monocytic leukemia zinc finger protein MOZ is a histone acetyltransferase. Oncogene 2001; 20: 404–409.
Reifsnyder C, Lowell J, Clarke A, Pillus L, Yeast SAS . silencing genes and human genes associated with AML and HIV-1 Tat interactions are homologous with acetyltransferases. Nat Genet 1996; 14: 42–49.
Akhtar A, Becker PB . Activation of transcription through histone H4 acetylation by MOF, an acetyltransferase essential for dosage compensation in Drosophila. Mol Cell 2000; 5: 367–375.
Allard S, Utley RT, Savard J, Clarke A, Grant P, Brandl CJ et al. NuA4, an essential transcription adaptor/histone H4 acetyltransferase complex containing Esa1p and the ATM-related cofactor Tra1p. EMBO J 1999; 18: 5108–5119.
Hassan AH, Neely KE, Workman JL . Histone acetyltransferase complexes stabilize swi/snf binding to promoter nucleosomes. Cell 2001; 104: 817–827.
Iizuka M, Stillman B . Histone acetyltransferase HBO1 interacts with the ORC1 subunit of the human initiator protein. J Biol Chem 1999; 274: 23027–23034.
Cao X, Südhof TC . A transcriptionally [correction of transcriptively] active complex of APP with Fe65 and histone acetyltransferase Tip60. Science 2001; 293: 115–120.
Sheridan AM, Force T, Yoon HJ, O’Leary E, Choukroun G, Taheri MR et al. PLIP, a novel splice variant of Tip60, interacts with group IV cytosolic phospholipase A(2), induces apoptosis, and potentiates prostaglandin production. Mol Cell Biol 2001; 21: 4470–4481.
Thomas T, Voss AK, Chowdhury K, Gruss P . Querkopf, a MYST family histone acetyltransferase, is required for normal cerebral cortex development. Development 2000; 127: 2537–2548.
Lew QJ, Chu KL, Lee J, Koh PL, Rajasefaran V, Teo JY et al. PCAF interacts with XBP-1S and mediates XBP-1S-dependent transcription. Nucleic Acids Res 2011; 39: 429–439.
Lew QJ, Chu KL, Chia YL, Soo B, Ho JP, Ng CH et al. GCN5 inhibits XBP-1S-mediated transcription by antagonizing PCAF action. Oncotarget 2015; 6: 271–287.
Kikuchi H, Kuribayashi F, Mimuro H, Imajoh-Ohmi S, Nakayama M, Takami Y et al. Lack of GCN5 remarkably enhances the resistance against prolonged endoplasmic reticulum stress-induced apoptosis through up-regulation of Bcl-2 gene expression. Biochem Biophys Res Commun 2015; 463: 870–875.
van Galen P, Kreso A, Mbong N, Kent DG, Fitzmaurice T, Chambers JE et al. The unfolded protein response governs integrity of the haematopoietic stem-cell pool during stress. Nature 2014; 510: 268–272.
Lester SR, Bain JL, Serio FG, Johnson RB . Relationship between the gingival sulcus depth and interleukin-1 isoform concentrations within the adjacent gingival tissue. J Periodontal Res 2009; 44: 323–329.
Yamamoto T, Kita M, Oseko F, Nakamura T, Imanishi J, Kanamura N . Cytokine production in human periodontal ligament cells stimulated with Porphyromonas gingivalis. J Periodontal Res 2006; 41: 554–559.
Axten JM, Medina JR, Feng Y, Shu A, Romeril SP, Grant SW et al. Discovery of 7-Methyl-5-(1-{[3-(trifluoromethyl)phenyl]acetyl}-2,3-dihydr-1H-indol-5-yl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine (GSK2606414), a potent and selective first-in-class inhibitor of protein kinase R (PKR)-like endoplasmic reticulum kinase (PERK). J Med Chem 2012; 55: 7193–7207.
Bi M, Naczki C, Koritzinsky M, Fels D, Blais J, Hu N et al. ER stress-regulated translation increases tolerance to extreme hypoxia and promotes tumor growth. EMBO J 2005; 24: 3470–3481.
Blais JD, Addison CL, Edge R, Falls T, Zhao H, Wary K et al. Perk-dependent translational regulation promotes tumor cell adaptation and angiogenesis in response to hypoxic stress. Mol Cell Biol 2006; 26: 9517–9532.
Kim I, Xu W, Reed JC . Cell death and endoplasmic reticulum stress: disease relevance and therapeutic opportunities. Nat Rev Drug Discov 2008; 7: 1013–1030.
Xue X, Piao JH, Nkajima A, Sakon-komazawa S, Kojima Y, Mori K et al. Tumor necrosis factor alpha (TNFalpha) induces the unfolded protein response (UPR) in a reactive oxygen species (ROS)-dependent fashion, and the UPR counteracts ROS accumulation by TNFalpha. J Biol Chem 2005; 280: 33917–33925.
Lin W, Harding HP, Ron D, Popko B . Endoplasmic reticulum stress modulates the response of myelinating ogoligodendrocytes to the immune cytokine interferon-gamma. J Cell Biol 2005; 169: 603–612.
Woo CW, Kutzler L, Kimball SR, Tabas I . Toll-like receptor activation suppresses ER stress factor CHOP and translation inhibition through activation of eIF2B. Nat Cell Biol 2012; 14: 192–200.
Woo CW, Cui D, Arellano J, Dorweiler B, Harding H, Fitzgerald KA . Adaptive suppression of the ATF4-CHOP branch of the unfolded protein response by toll-like receptor signalling. Nat Cell Biol 2009; 11: 1473–1480.
Ozcan U, Cao Q, Yilmaz E, Lee AH, Lwakoshi NN, Ozdelen E et al. Endoplasmic reticulum stress links obesity, insulin action, and type 2 diabetes. Science 2004; 306: 457–461.
Lin JH, Walter P, Yen TS . Endoplasmic reticulum stress in disease pathogenesis. Annu Rev Pathol 2008; 3: 399–425.
Kaser A, Lee AH, Franke A, Glickman JN, Zeissig S, Tilg H et al. XBP1 links ER stress to intestinal inflammation and confers genetic risk for human inflammatory bowel disease. Cell 2008; 134: 743–756.
Zhang K, Shen X, Wu J, Sakaki K, Saunders T, Rutkowski DT et al. Endoplasmic reticulum stress activates cleavage of CREBH to induce a systemic inflammatory response. Cell 2006; 124: 587–599.
Qin S, Jin L, Zhang J, Liu L, Ji P, Wu M et al. Recognition of unmodified histone H3 by the first PHD finger of bromodomain-PHD finger protein 2 provides insights into the regulation of histone acetyltransferases monocytic leukemic zinc-finger protein (MOZ) and MOZ-related factor (MORF). J Biol Chem 2011; 286: 36944–36955.
Heijmans J, van Lidth de Jeude JF, Koo BK, Rosekrans SL, Wielenga MC, van de Wetering M et al. ER stress causes rapid loss of intestinal epithelial stemness through activation of the unfolded protein response. Cell Rep 2013; 3: 1128–1139.
Xu C, Beatrice BM, John CR . Endoplasmic reticulum stress: cell life and death decisions. J Clin Invest 2005; 115: 2656–2664.
Zhang J, An Y, Gao LN, Zhang YJ, Jin Y, Chen FM . The effect of aging on the pluripotential capacity and regenerative potential of human periodontal ligament stem cells. Biomaterials 2012; 33: 6974–6986.
Chang J, Wang Z, Tang E, Fan Z, McCauley L, Franceschi R et al. Inhibition of osteoblastic bone formation by nuclear factor-kappaB. Nat Med 2009; 15: 682–689.
Li C, Li B, Dong Z, Gao L, He X, Liao L et al. Lipopolysaccharide differentially affects the osteogenic differentiation of periodontal ligament stem cells and bone marrow mesenchymal stem cells through Toll-like receptor 4 mediated nuclear factor κB pathway. Stem Cell Res Ther 2014; 5: 67.
Acknowledgements
This work was supported by the grants from the National Natural Science Foundation of China (No. 81470710 to YJ, 81470742 to QTW, 31401255 to BL, 81271137 to QTW, 81570976 to FJ and 31301140 to XNH).
Author contributions
PX, BL and YA were involved in the practical achievement of the experiments. PX, BL, YA, XNH, JS and DF collected, analyzed and interpreted the data. PX and BL drafted the manuscript. GYD, RH and FJ provided human teeth samples for the experiments. YJ and QTW designed the study, wrote the manuscript and provided administrative support. All the authors read and approved the manuscript for publication.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by JM Hardwick
Supplementary Information accompanies this paper on Cell Death and Differentiation website
Supplementary information
Rights and permissions
About this article
Cite this article
Xue, P., Li, B., An, Y. et al. Decreased MORF leads to prolonged endoplasmic reticulum stress in periodontitis-associated chronic inflammation. Cell Death Differ 23, 1862–1872 (2016). https://doi.org/10.1038/cdd.2016.74
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cdd.2016.74
This article is cited by
-
Endoplasmic reticulum stress: a novel targeted approach to repair bone defects by regulating osteogenesis and angiogenesis
Journal of Translational Medicine (2023)
-
DNA Methylation and Histone Modification in Dental-derived Mesenchymal Stem Cells
Stem Cell Reviews and Reports (2022)
-
The effect of low-level laser irradiation on the proliferation, osteogenesis, inflammatory reaction, and oxidative stress of human periodontal ligament stem cells under inflammatory conditions
Lasers in Medical Science (2022)
-
KAT6A regulates stemness of aging bone marrow-derived mesenchymal stem cells through Nrf2/ARE signaling pathway
Stem Cell Research & Therapy (2021)
-
Low-intensity pulsed ultrasound upregulates osteogenesis under inflammatory conditions in periodontal ligament stem cells through unfolded protein response
Stem Cell Research & Therapy (2020)