Abstract
FK506-binding protein 51 (FKBP51) regulates the activity of the glucocorticoid receptor (GR), and is therefore a key mediator of the biological actions of glucocorticoids. However, the understanding of the molecular mechanisms that govern its activity remains limited. Here, we uncover a novel regulatory switch for GR activity by the post-translational modification of FKBP51 with small ubiquitin-like modifier (SUMO). The major SUMO-attachment site, lysine 422, is required for FKBP51-mediated inhibition of GR activity in hippocampal neuronal cells. Importantly, impairment of SUMO conjugation to FKBP51 impacts on GR-dependent neuronal signaling and differentiation. We demonstrate that SUMO conjugation to FKBP51 is enhanced by the E3 ligase PIAS4 and by environmental stresses such as heat shock, which impact on GR-dependent transcription. SUMO conjugation to FKBP51 regulates GR hormone-binding affinity and nuclear translocation by promoting FKBP51 interaction within the GR complex. SUMOylation-deficient FKBP51 fails to interact with Hsp90 and GR thus facilitating the recruitment of the closely related protein, FKBP52, which enhances GR transcriptional activity. Moreover, we show that the modification of FKBP51 with SUMO modulates its binding to Hsp90. Our data establish SUMO conjugation as a novel regulatory mechanism in the Hsp90 cochaperone activity of FKBP51 with a functional impact on GR signaling in a neuronal context.
Similar content being viewed by others
Main
Glucocorticoids (GCs) are the main mediators of the stress response. They exert a wide range of biological actions, particularly at the central nervous system, where they regulate neuronal function, proliferation and neurogenesis.1 FK506-binding protein 51 (FKBP51) is a cochaperone that modulates GC-dependent responses by regulating the activity of their receptor, the glucocorticoid receptor (GR). FKBP51 belongs to the peptidyl prolyl isomerase superfamily and the tetratricopeptide repeat (TPR)-containing immunophilins.2 The TPR domain, which is involved in protein–protein interactions, mediates its binding to Hsp90.3, 4 Through its interaction with Hsp90, FKBP51 is recruited to the GR heterocomplex to finally inhibit GR activity.5 Therefore, FKBP51 is critical for GR in vivo function and a key mediator during stress.6 Ligand binding to the GR induces an exchange between FKBP51 and the closely related TPR protein FKBP52,7 which in turn enhances GR activity.8
Abnormal FKBP51 function has been implicated in a wide variety of diseases, in particular stress-related disorders associated with impaired GR signaling.2, 9 Interestingly, it has also been suggested that FKBP51 might be involved in changes in hippocampal plasticity and alterations in its structure, with a final impact on the response to stress.10 FKBP51 arises thus as an interesting target for the treatment of these disorders. However, the molecular mechanisms that regulate FKBP51 activity are not clearly understood.
Protein SUMOylation is a post-translational modification (PTM) that consists in the covalent attachment of small ubiquitin-like modifiers (SUMOs) to target proteins through an enzymatic process that involves an E1 activating enzyme, an E2 conjugating enzyme and E3 SUMO ligases. The consequences of SUMO conjugation vary within different substrates, and include alterations in protein stability, activity and localization.11, 12
Although many PTMs have been found to modulate GR activity,13, 14, 15 to date no PTM on FKBP51 has been investigated. In particular, GR activity is modulated by SUMO conjugation to GR itself and other proteins belonging to the GR chaperone complex,16, 17, 18, 19 raising the intriguing possibility that SUMOylation of proteins from the GR heterocomplex might be a common and important upstream regulatory mechanism.
In order to discover novel players that modulate the activity of FKBP51 and hence may become of direct clinical relevance, a detailed understanding of the molecular mechanisms underlying the role of FKBP51 in GR signaling is a prerequisite. Here, we demonstrate that the inhibition of GR activity by FKBP51 is regulated by its conjugation to SUMO, uncovering a novel regulatory mechanism in the response to GCs with a biological impact on neuronal function.
Results
FKBP51 is modified by SUMO conjugation
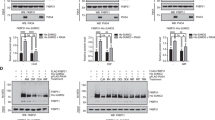
To investigate whether FKBP51 is modified by SUMO conjugation, we performed in vitro SUMOylation assays. The occurrence of slower-migrating bands in western blot assays (Figure 1a) shows that FKBP51 is indeed conjugated to SUMO in vitro. We next determined whether FKBP51 is also SUMOylated in a cellular context. As shown in Figure 1b, FKBP51 is modified by the three SUMO isoforms although SUMO2 shows the greatest potency. In addition, expression of Ubc9, the E2 conjugating enzyme, increases SUMO2 conjugation to FKBP51 (Figure 1c). Consistently, FKBP51 interacts with Ubc9 (Figure 1d). We confirmed the identity of FKBP51-SUMO2 conjugates by performing an in vitro deSUMOylation assay in the presence of the SUMO isopeptidase SENP2 (Figure 1e). Figure 1f shows the SUMOylation of endogenous FKBP51. Endogenous SUMO-modified FKBP51 is further observed when immunoprecipitating SUMO2/3 from non-transfected cells (Figure 1g). Altogether, our findings strongly support that the endogenous protein is a bona fide substrate for modification with SUMO.
FKBP51 is modified by SUMO conjugation. (a) FKBP51 was SUMOylated in vitro and analyzed by western blotting using anti-FKBP51 antibody. (b) HEK293T cells were transfected with the indicated plasmids. SUMOylated proteins were purified by Ni2+ affinity chromatography and analyzed by western blotting using anti-FKBP51 antibody. (c) HEK293T cells were transfected with the indicated plasmids and analyzed as in (b). (d) Pull-down assays were performed using HEK293T lysates expressing FLAG-FKBP51 and analyzed by western blotting using anti-FKBP51 antibody. (e) HEK293T lysates expressing FLAG-FKBP51, HA-SUMO2 and V5-Ubc9 were immunoprecipitated with anti-FKBP51 antibody, and immunoprecipitates were incubated with or without recombinant SENP2. Samples were analyzed by western blotting using anti-FKBP51 antibody. (f) HEK293T cells were transfected with the indicated plasmids and analyzed as in (b). (g) Lysates from HEK293T were immunoprecipitated with anti-SUMO2/3 antibody (ab) or non-immune antibody (NI ab) and analyzed by western blotting using the indicated antibodies. *Non-specific. (h) HT22 cells were transfected with the indicated plasmids and analyzed as in (b)
FKBP51 expression is increased both upon GC treatment and under stress in the hippocampus, where it is postulated to have a critical role in stress-related diseases.20 Therefore, we used HT22 cells, a murine hippocampal neuronal cell line,21 to study SUMOylation of FKBP51 in this context. As shown in Figure 1h, FKBP51 is SUMOylated in HT22 cells; and the expression of SENP1 decreases its conjugation to SUMO. Moreover, we analyzed protein expression of FKBP51 and SUMO2/3 in mouse hippocampus showing that hippocampal neurons expressing SUMO2/3 also express FKBP51, which suggests that FKBP51 might be a SUMO conjugation target in vivo as well (Supplementary Figure 1).
Lysine 422 is essential for SUMOylation of FKBP51
To map putative target lysines for SUMO conjugation, we performed an in silico analysis using the software SUMOsp 2.0 and SUMOplot. Lysines 138 and 422 bear the highest probability of SUMO attachment (Figure 2a). As depicted in Figure 2b, lysine 138 resides near the FK1 domain while lysine 422 is in the C-terminal tail of FKBP51. Analysis of sequence conservation reveals that both sites lie within regions of high local conservation, especially considering that surrounding sequences downstream of the second site are more divergent (Figure 2b). We mutated both lysines into non-SUMOylable but charge-conserving arginines separately (K138R and K422R mutants) and in combination (2KR), and analyzed FKBP51 SUMOylation. The 2KR mutant is used as a control. Figure 2c shows that SUMOylation is impaired when K422 is mutated while K138R mutation has no effect. SUMOylation usually occurs at lysine residues within the consensus sequence ψKxE.22, 23 In agreement, lysine 422 is the only residue of FKBP51 within such a motif. Mutating the SUMOylation site (K422) has no significant effect on Ubc9 binding (Figure 2d). However, K422R FKBP51 SUMOylation levels are considerably decreased. These results indicate that K422 is the major acceptor of SUMOylation.
FKBP51 is SUMOylated at lysine 422. (a) Putative SUMOylation sites with the highest probability. (b) Schematic representation of FKBP51: FK1, FK2 and TPR domains. The two putative SUMO-attachment sites are depicted. FKBP51 sequences are aligned centered on human K138 and K422. SUMOylation motifs are boxed. (c) HT22 cells were transfected with the indicated plasmids. SUMOylated proteins were purified by Ni2+ affinity chromatography and analyzed by western blotting using anti-FKBP51 antibody. *Non-specific. (d) Pull-down assays were performed using HEK293T lysates expressing wt, K422R or 2KR FLAG-FKBP51 and analyzed by western blotting using anti-FKBP51 antibody
FKBP51 SUMOylation modulates GR activity
To test whether FKBP51 SUMOylation impacts on GR signaling, we explored its effect on GR transcriptional activity. Stimulation of HT22 cells with the GC dexamethasone (Dex) induces GR transcriptional activity, which is inhibited when wild-type (wt) FKBP51 is expressed. Interestingly, expression of SUMOylation mutants K422R and 2KR fails to inhibit GR transcriptional activity (Figure 3a), suggesting that SUMO conjugation to FKBP51 is necessary for its inhibitory effect. Similar results were observed using FKBP51 knock-out mouse embryonic fibroblasts (MEFs), excluding any influence of endogenous FKBP51 (Figure 3b), and HEK293T cells (Supplementary Figure 2a). These results cannot be explained by alterations in expression levels or protein stability since the SUMOylation mutant is expressed to the same level as the wt protein (Figures 3a and b and Supplementary Figure 2a, western blot panels). Moreover, K422R mutation has no effect on protein half-life as shown in the presence of the protein synthesis inhibitor cycloheximide (Supplementary Figure 2b).
SUMO conjugation to FKBP51 is critical for its inhibitory effect on GR transcriptional activity and impacts on GR endogenous target gene expression and Dex inhibition of neurite outgrowth. (a and b) HT22 (a) and FKBP51−/− MEF (b) cells were transfected with TAT4-luc and GR plasmids, and with or without wt, K422R or 2KR FKBP51, and stimulated with Dex 1 nM or vehicle B for 8 h (a) and 16 h (b). Results are expressed as mean±S.E.M. (n=4). Protein levels were analyzed by western blotting. (c) HT22 cells transfected with GR plasmids with or without wt or K422R FKBP51 were stimulated with Dex 1 nM or vehicle B for 3 h, and SGK1 and GILZ mRNA levels were analyzed. Results are expressed as mean±S.E.M. (n=3). (d and e) HT22 (d) and Neuro-2a (e) cells, transfected with GR and RFP plasmids with or without wt or K422R FKBP51, were stimulated with Dex 100 nM for 24 h (d) and 10 nM for 12 h (e) or vehicle B and then differentiated. Representative images of RFP+ cells (green) are shown. Arrows indicate neurites. Results are expressed as mean±S.E.M. (n=3). *P<0.05, **P<0.01, ***P<0.001
To further analyze the functional relevance of FKBP51 SUMOylation on GR signaling, we studied the expression of two endogenous target genes, serum GC kinase 1 (SGK1) and GC-induced leucine zipper (GILZ), that regulate neuronal functional properties. We determined SGK1 and GILZ mRNA expression levels in HT22 cells expressing wt or K422R FKBP51. As shown in Figure 3c and Supplementary Figure 3, Dex stimulation induces SGK1 and GILZ expression, while wt FKBP51 impairs such increment. Consistent with our results yielded by reporter assays, the K422R mutant does not inhibit Dex-induced expression of SGK1 and GILZ, and K422R FKBP51 expressing cells show significantly higher expression of both genes under Dex treatment when compared with cells expressing wt FKBP51. These results corroborate the critical role of SUMO conjugation to FKBP51 in the regulation of GR activity.
To rule out the possibility that mutation of the K422 has a deleterious effect on protein folding, we analyzed the effect of this mutant on NF-κB pathway.24 As shown in Supplementary Figure 4, both wt and K422R FKBP51 inhibit NF-κB activity and interact with NF-κB, showing that mutation of K422 does not alter the general function of the protein.
Considering that neurite extension is one of the first cellular changes associated with neuronal differentiation and that GCs are negative regulators of neuronal differentiation,25 we evaluated the impact of FKBP51 SUMOylation on Dex-dependent regulation of neurite outgrowth in HT22 cells. Consistent with previous results,26 Dex decreases neurite outgrowth. Expression of wt FKBP51 abolishes the effect of Dex treatment while K422R mutant fails to prevent it (Figure 3d). We further validated these findings in Neuro-2a cells (Figure 3e), extensively used as a cell model of neuronal differentiation.27 These results show that FKBP51 through its SUMOylation is a key factor for Dex-mediated effects on neurite projection.
SUMO conjugation to FKBP51 and its effect on GR activity are modulated by the E3 ligase PIAS4 and heat shock
We next determined whether SUMO attachment to FKBP51 could be modulated by SUMO E3 ligases, and found that PIAS4 significantly enhances its SUMOylation (Figure 4a). We confirmed these results by purifying SUMOylated proteins from cells expressing FKBP51 and SUMO2 in the presence or absence of PIAS4 (Figure 4b). Consistently, FKBP51 interacts with PIAS4 (Figure 4c). PIAS4 overexpression also evidences SUMO conjugation to the endogenous protein (Figure 4d). To confirm that FKBP51 is directly targeted by PIAS4, we performed in vitro SUMOylation assays using recombinant FKBP51 and PIAS4 (Figure 4e), demonstrating that in vitro SUMO conjugation to FKBP51 is enhanced in the presence of the ligase. Altogether, these results show that PIAS4 modulates the occurrence of SUMO-FKBP51 conjugates.
PIAS4 and heat stress modulate SUMO conjugation to FKBP51 and its inhibitory effect on GR activity. (a) Samples from HT22 cells transfected with the indicated plasmids and PIAS4, PIAS1, Pc2 or Topors were analyzed by western blotting using the indicated antibodies. (b) HT22 cells were transfected with the indicated plasmids. SUMOylated proteins were purified by Ni2+ affinity chromatography and analyzed by western blotting using anti-FKBP51 antibody. (c) Lysates from HEK293T cells transfected with the indicated plasmids were immunoprecipitated with anti-FLAG antibody and analyzed by western blotting using the indicated antibodies. (d) HEK293T cells were transfected with the indicated plasmids and analyzed as in (b). (e) FKBP51 was SUMOylated in vitro with or without recombinant PIAS4 (250 ng) and analyzed by western blotting using the indicated antibodies. (f) HT22 cells were transfected with TAT4-luc and GR plasmids with or without wt or K422R FKBP51 and PIAS4, and stimulated with Dex 1 nM or vehicle B for 8 h. Results are expressed as mean±S.E.M. (n=3) (upper panel). Samples from HT22 cells transfected with the indicated plasmids were analyzed as in (b) (lower panel). (g) HT22 cells were transfected with TAT4-luc and GR plasmids with or without wt or K422R FKBP51, together with 20 nM scramble or PIAS4 siRNA. Cells were stimulated with Dex 1 nM or vehicle B for 8 h. Results are expressed as mean±S.E.M. (n=3) (upper panel). PIAS4 mRNA levels were analyzed by RT-qPCR (lower panel). (h) HT22 cells were transfected with the indicated plasmids. Cells subjected to heat shock − 42 ºC, HS – for 30 min or left untreated −37 ºC – were analyzed as in (b). (i) HT22 cells were transfected with MMTV-luc and GR plasmids with or without wt or K422R FKBP51. Cells were stimulated with Dex 1 nM or vehicle B, and subjected to HS for 1 h or left untreated. Cells were allowed to recover for 5 h at 37 ºC. Results are expressed as mean±S.E.M. (n=3). *P<0.05, **P<0.01, ***P<0.001
We next examined the effect of PIAS4 on GR transcriptional activity to further evaluate how modulating SUMOylation of FKBP51 impacts on GR signaling. Figure 4f (upper panel) shows that overexpression of PIAS4 potentiates wt FKBP51 inhibition of GR activity. In the presence of the K422R mutant, however, PIAS4 has no longer effect on FKBP51 action on GR activity, suggesting that potentiation of FKBP51 inhibition on GR-dependent transcription by PIAS4 is mediated by SUMOylation-competent FKBP51. Accordingly, FKBP51-SUMO conjugates are increased in the presence of PIAS4 while K422R SUMOylation levels remain considerably lower compared with wt FKBP51, even when overexpressing PIAS4 (Figure 4f, lower panel). To validate these findings, we specifically silenced PIAS4 (40%, Figure 4g, lower panel). As shown in Figure 4g, this reduction in endogenous PIAS4 expression reduces wt FKBP51 suppression of GR activity, while SUMOylation-deficient K422R is insensitive toward PIAS4 inhibition.
Stressful conditions such as heat shock are known to induce global protein SUMOylation28 and also PIAS4 expression.29 Heat stress induces FKBP51 SUMOylation (Figure 4h) and also potentiates its inhibitory effect on GR activity in a K422-dependent manner (52 versus 80% of inhibition, Figure 4i).
Altogether, these findings strongly support the role of SUMO conjugation to FKBP51 in its inhibitory effect of GR activity.
SUMO conjugation to FKBP51 modulates its activity as Hsp90-based glucocorticoid receptor cochaperone
It has been reported that FKBP51 decreases GR hormone-binding affinity30 and nuclear translocation,5 modulating GC-induced neuronal gene expression.31 To explore the mechanisms underlying the effect of FKBP51 SUMOylation on GR activity, we first performed in vitro binding assays. Figure 5a shows that wt FKBP51 decreases Dex binding to the GR while K422R and 2KR FKBP51 mutants fail to alter GR binding affinity. Next, we analyzed the impact of FKBP51 SUMOylation on GR nuclear translocation through live-imaging experiments. Expression of wt FKBP51, and not K422R mutant, interferes with hormone-induced GR translocation (Figure 5b). The possibility that the above findings were due to a difference in subcellular localization between wt and SUMOylation-deficient FKBP51 was ruled out by the fact that both proteins display the same expression pattern and are properly co-expressed with GR (Figure 5b). Moreover, while wt FKBP51 still reduces GR translocation under saturating levels of Dex,5 no effect of the K422R mutant is observed (Supplementary Figure 5), thus supporting that translocation is modulated by FKBP51 SUMOylation, independently of its effect on GR hormone-binding affinity.
FKBP51 SUMOylation is critical for its role as a GR cochaperone. (a) HEK293T cytoplasmic lysates expressing wt, K422R or 2KR FLAG-FKBP51 and HA-GR were used for in vitro ligand binding assay. Results are expressed as mean±S.E.M. (n=3). Lysates were analyzed by western blotting using the indicated antibodies. (b) HT22 cells transfected with YFP-GR plasmid with or without wt or K422R FKBP51 were stimulated with Dex 10 nM. Results are expressed as mean±S.E.M. of one representative experiment (n=3) (left panel). Representative images for each condition at t=0 s and t=3600 s are shown (right, upper panel). Cells transfected with YFP-GR plasmid with or without wt or K422R FKBP51 were analyzed by immunofluorescence using anti-FKBP51 antibody (right, bottom panel). (c) Lysates from HEK293T cells transfected with the indicated plasmids were immunoprecipitated with anti-FLAG antibody and analyzed by western blotting using the indicated antibodies. (d) Lysates from HEK293T cells transfected with the indicated plasmids were immunoprecipitated with anti-HA antibody and analyzed by western blotting using the indicated antibodies. (e) In vitro SUMOylated FKBP51 was incubated with FLAG-Hsp90 purified from HEK293T cells and immunoprecipitated with anti-FKBP51 antibody. Immunoprecipitates were analyzed by western blotting using the indicated antibodies (left panel). Pull-down assays were performed using in vitro SUMOylated FKBP51 and analyzed by western blotting using the indicated antibodies (right panel). (f) Lysates from HEK293T cells transfected with the indicated plasmids were immunoprecipitated with anti-FKBP51 antibody (ab) or non-immune antibody (NI ab) and analyzed by western blotting using the indicated antibodies. *Non-specific. #IgG
These experiments suggest that FKBP51 SUMOylation modulates GR activity by preventing both GR binding to its hormone and its translocation to the nucleus.
Interestingly, these regulatory mechanisms require prior recruitment of FKBP51 to the GR heterocomplex through its binding to Hsp90. Of note, the major SUMO acceptor site within FKBP51 sequence, K422, is located in the protein C-terminal domain, which is critical in the interaction with Hsp90.3 Therefore, in order to examine whether FKBP51 SUMOylation affects its interaction with Hsp90, we performed co-immunoprecipitation assays. As shown in Figure 5c, wt FKBP51 interacts with both Hsp90 and GR. In contrast, interaction with Hsp90 and GR is impaired by K422R mutation (Figure 5c), suggesting that FKBP51 SUMOylation is involved in its binding to Hsp90 and consequently its recruitment to the GR chaperone complex.
Recruitment of cochaperones to the GR complex by Hsp90 is known to be a competitive process.32 Upon hormone stimulation, there is a switch from FKBP51 to the closely related TPR protein, FKBP52, which positively regulates GR activity.5, 7, 8 Therefore, we analyzed whether impairment of SUMO conjugation to FKBP51 alters the cochaperone composition of this complex. Figure 5d shows that wt FKBP51 and not K422R mutant is associated to the GR, thus corroborating our previous results. Interestingly, recruitment of FKBP52 to the GR complex is enhanced in the presence of K422R mutant compared with wt FKBP51. These results suggest that impairment of SUMO conjugation to FKBP51 prevents its interaction within the GR chaperone complex allowing for the recruitment of FKBP52 instead and therefore facilitating GR nuclear translocation and activity, and support the role of SUMO conjugation to FKBP51 in its recruitment by Hsp90 to the GR complex.
To further study this mechanism, we analyzed binding of Hsp90 to in vitro SUMOylated FKBP51. As shown in Figure 5e (left panel), Hsp90 is precipitated to a greater extent by SUMOylated FKBP51 compared with the unconjugated FKBP51, pointing to the role of FKBP51 SUMOylation in promoting the interaction with Hsp90. In line with this notion, Figure 5e (right panel) shows that GST-Hsp90 preferentially binds SUMO-FKBP51 conjugates, as an increased ratio of SUMO-modified versus unmodified FKBP51 is recovered in GST-Hsp90 pull-down assays using in vitro SUMOylated FKBP51. To confirm our findings, we immunoprecipitated endogenous FKBP51 in the presence of PIAS4 and observed its interaction with Hsp90. The interaction between endogenous FKBP51 and Hsp90 is modulated in the presence of a PIAS4 shRNA (Figure 5f). Altogether, these results demonstrate the importance of SUMO conjugation to FKBP51 in its activity as an Hsp90 cochaperone and support that SUMOylation of FKBP51 promotes its interaction with Hsp90 at the endogenous level.
Discussion
In this work, we describe the functional impact on GR activity of SUMO conjugation to FKBP51. This is the first work to explore the modification of FKBP51 by a PTM and to demonstrate that FKBP51 function is tightly post-translationally regulated. We demonstrate that FKBP51 is SUMOylated at K422 and that SUMOylation of FKBP51 is important for its inhibitory effect on GR activity. Upon analyzing the biological consequences of FKBP51 SUMOylation, we report that disrupting this site-specific PTM has a strong impact on GR-dependent neuronal signaling and differentiation. We explored the molecular mechanism underlying the effect of FKBP51 SUMOylation on GR activity (Figure 6). We found that SUMO modification of FKBP51 mediates the inhibitory effect exerted by FKBP51 on GR hormone-binding affinity and GR nuclear translocation, and the interaction of FKBP51 and FKBP52 within the GR chaperone complex. The fact that FKBP51 SUMOylation is involved in its binding to Hsp90 suggests that SUMOylation of FKBP51 is necessary to allow for its association to the GR complex through its interaction with Hsp90, which in turn, is required for the effect of FKBP51 on GR activity. The critical role of FKBP51 SUMOylation in the regulation of GR activity is supported by results using the SUMO-deficient FKBP51 mutant, together with those obtained by modulating FKBP51-SUMO conjugation levels through PIAS4 expression and heat stress. Altogether, we uncover a novel regulatory mechanism of FKBP51 activity as an Hsp90 cochaperone that has an essential role in GR signaling in a hippocampal neuronal context and possibly beyond.
SUMO conjugation to FKBP51 regulates GR signaling. Modification of FKBP51 by SUMO attachment promotes its interaction with Hsp90 and its recruitment to the GR chaperone complex. Within this complex, FKBP51 decreases GR ligand affinity and nuclear translocation, which in turn, leads to inhibition of GR transcriptional activity. In this way, SUMOylated FKBP51 finally impacts on GR-dependent neuronal signaling and differentiation. While enhancement of FKBP51 SUMOylation through PIAS4 or upon cellular stress further strengthens FKBP51 inhibitory effect on GR signaling, its impairment favors the recruitment of FKBP52 to the GR heterocomplex. Contrary to FKBP51, the closely related cochaperone FKBP52 positively impacts on GR signaling, finally enhancing its transcriptional activity. S: SUMO
Upon investigating the functional consequences of SUMO attachment to FKBP51 on GR activity, we found that FKBP51 SUMOylation impacts not only on GR-dependent gene expression but also on the effect of GCs on neuronal differentiation. We demonstrate that FKBP51 SUMOylation affects the expression of GILZ and SGK1, which are upregulated by GCs in the hippocampus.33, 34 In line with these results, our data show that GCs also induce the expression of SGK1 and GILZ in HT22 hippocampal neuronal cells. Moreover, we show that only SUMOylation-competent FKBP51 is involved in their regulation.
Apart from their role in neuronal function and the stress response,35, 36 the expression of GILZ and SGK1 is also related to alterations in hippocampal structure.37 In particular, SGK1 was demonstrated to mediate the inhibitory effect of GCs on neuronal differentiation in human hippocampal progenitor cells.33 In this respect, our data show that Dex inhibition of neurite outgrowth is regulated by FKBP51 in a SUMOylation-dependent manner. We show that expression of FKBP51 prevents both GR-mediated downregulation of neurite outgrowth together with the upregulation of SGK1; further and most importantly, that these two effects are dependent on SUMO conjugation to FKBP51. FKBP51 SUMOylation might be thus a critical modulator of the SGK1-dependent effects of GCs on hippocampal neurogenesis.
Within the central nervous system, an increase in SUMO protein modification is considered to be associated with neuronal plasticity and neuroprotection against various stressful conditions.38, 39 In this regard, our data show that FKBP51 SUMOylation is increased under heat-shock stress in a hippocampal neuronal context and raises thus the interesting possibility that SUMO conjugation to FKBP51 during stress in the hippocampus might be involved in neuronal protection.
Our work shows for the first time the occurrence of endogenous SUMO-FKBP51 conjugates and identifies lysine 422, which lies within a SUMOylation consensus sequence, as the major SUMO acceptor site. In agreement with our observations, a proteome-wide analysis recently identified K422 of endogenous FKBP51 as a putative SUMOylated site,40 thus strengthening our findings.
We demonstrate that the inhibitory effect of FKBP51 on GR activity is regulated by SUMO conjugation to FKBP51. We show that impairment of FKBP51 SUMOylation prevents FKBP51-mediated inhibition of GR nuclear translocation, hormone-binding affinity and transcriptional activity (Figure 6). Changes in PIAS4 expression together with environmental stresses such as heat shock modulate FKBP51 SUMOylation and finally impact on GR activity, confirming that SUMO conjugation to FKBP51 determines its effect on GR signaling. Consistent with previous results, expression of PIAS4 has no effect on Dex stimulation of GR reporter genes.41 We show that under this condition PIAS4 potentiates FKBP51’s inhibitory effect on GR transcriptional activity, uncovering a novel regulatory player in GR signaling.
Our findings strongly support that SUMOylation of FKBP51 is necessary to enhance its binding to Hsp90 (Figure 6), as demonstrated by co-immunoprecitation and Hsp90-FKBP51 interaction assays. Our results also provide evidence that this mechanism operates at the endogenous level thus highlighting its physiological relevance. Remarkably, recruitment of FKBP51 as a cochaperone to Hsp90 protein complexes modulates the activity of other nuclear receptors, besides GR.42, 43 It is thus likely that SUMO conjugation to FKBP51 may impact on their activity as well. Further studies will be needed to address the overall significance of this mechanism on other nuclear receptors and in different contexts.
Regarding the FKBP51–Hsp90 interaction, the TPR domain of FKBP51, although necessary, was found to be insufficient for full binding.3 A conserved sequence downstream of the TPR domain together with the last C-terminal amino acids of FKBP51 is critical to this interaction3 and was proposed to adopt alternative conformations that significantly impact on Hsp90 binding. K422 is located in this C-terminal tail region, which has not yet been crystallized.4 It is likely that the modification of K422 by SUMO might modulate the conformation of this region for FKBP51–Hsp90 interaction. Interestingly, in silico analysis reveals that Hsp90 bears SUMO-interacting motifs, which can mediate non-covalent SUMO binding and cooperate in interactions between binding partners and SUMOylated proteins.44, 45 This raises the possibility that SUMO–SIM interactions involving the C-terminal region of FKBP51 are required to allow for efficient binding between FKBP51 and Hsp90. Such complex regulatory mechanisms should be investigated in future studies.
Proteins bearing TPR domains compete with each other for interaction with Hsp90 in order to gain access to the GR heterocomplex and modulate GR activity.5, 32 Therefore, the fact that impairment of FKBP51 SUMOylation prevents both FKBP51 binding to Hsp90 and its inhibitory effect on GR activity while promotes FKBP52 recruitment and GR nuclear translocation provides a mechanistic explanation, suggesting that SUMO conjugation to FKBP51 underlies its preferential recruitment to the GR chaperone complex, which in turn determines the final outcome of GR signaling. Interestingly, we found through in silico analysis that FKBP52 has SUMO conjugation consensus and non-consensus sites. It would be interesting to investigate whether FKBP52 is indeed conjugated to SUMO as well, and whether it affects its function, particularly regarding GR activity.
Considering that the upregulation of FKBP51 activity has been related to abnormal HPA stress responses,2 and given the importance of SUMOylation in regulating the activity of FKBP51 as a GR cochaperone, our findings provide novel insights into the molecular mechanisms implicated in the regulation of the stress response and other processes where the interaction between Hsp90 and FKBP51 is involved.42
Materials and Methods
Cell culture and transfection
HEK293T, Neuro-2a and HT22 cells were grown in DMEM (Gibco/Thermo Fisher Scientific, Waltham, MA, USA) (pH 7.3) supplemented with 10% FBS for HEK293T and Neuro-2a, and 5% FBS for HT22 cells, 2.2 g/l NaHCO3, 10 mM HEPES, 4 mM l-glutamine, 100 U/ml penicillin and 100 mg/ml streptomycin. FKBP51 knock-out (FKBP51−/−) MEFs kindly provided by Dr. MB Cox (University of Texas, El Paso, USA) were grown in DMEM supplemented with 10% FBS and 100 units/ml streptomycin, and have been described before.6
HEK293T cells were transfected with Lipofectamine 2000 (Life Technologies/Thermo Fisher Scientific, Carlsbad, CA, USA) following the manufacturer’s instruction, and were electroporated for in vitro hormone binding, as previously described.46 FKBP51−/− MEFs were electroporated as previously described.46 HT22 and Neuro-2a cells were transfected with Lipofectamine and PLUS reagent (Life Technologies/Thermo Fisher Scientific) following the manufacturer’s instructions. Lipofectamine 2000 was used in siRNA experiments.
Plasmids, siRNAs and reagents
The following plasmids used in this work were previously described, as indicated: FLAG-FKBP51 and FLAG-FKBP52;5 GST-Ubc9;47 HA-GR and the GR reporter plasmid pΔ(TAT)4-Luc (TAT4-luc);18 the NF-κB reporter plasmid (κB-luc);48 and FLAG-Hsp90 and the Gaussia luciferase reporter plasmid.32 The pHC-RED1-C1 plasmid was obtained from Clontech (Mountain View, CA, USA). The following plasmids were kindly provided: HA-SUMO2, V5-Ubc9, and 6 × His-SUMO1, SUMO2 and SUMO3 by Dr. R Hay;49, 50, 51 YFP-GR by Dr. JA Cidlowski;52 FLAG-SENP1 by Dr. E Yeh;53 PIAS4 expression vector by Dr. JA Iñiguez-Lluhí;54 PIAS4 and control shRNA by Dr. MJ Hendzel;55 the GR reporter plasmid MMTV-luc by Dr. R Evans;56 p65 expression vector by Dr. M Karin; the β-galactosidase reporter plasmid by Dr. D Spengler;57 GST-Hsp90 by Dr. K Nagata;58 pGEX-PIAS4 by Dr. SP Goff.59 The FKBP51 point mutants were constructed by site-directed mutagenesis using the following primers to mutate Lys-138 to Arg: 5′-GAGCTCCTTGATTTCAGAGGAGAGGATTTATTTG-3′ and 5′-CAAATAAATCCTCTCCTCTGAAATCAAGGAGCTC-3′; and to mutate Lys-422 to Arg: 5′-AGAAGTTTGCAGAGCAGGATGCCAGAGAAGAGGCC-3′ and 5′-CCATTGCTTTATTGGCCTCTTCTCTGGCATCCTGCTCTG-3′. The 6 × His-FKBP51-FLAG plasmid was constructed by inserting the FKBP51 cDNA sequence into pPROEX HTa vector digested with NcoI and XbaI.
The siRNA sequences targeting mouse PIAS4 were 5′-AAGGCUUUGGUCAAGGAGAAG-3′ (positions 886–906) and 5′-AAGCACGAACTGGTGACCAGGGC-3′ (positions 112–134). A non-silencing RNA duplex (5′-CUUACGCUGAGUACUUCGATT-3′) was used as a control.
Dex, N-ethylmaleimide (NEM), Na2MoO4, PMA, CHX and cAMP were acquired from Sigma-Aldrich (St. Louis, MO, USA).
Western blot assays and antibodies
Cells washed with ice-cold PBS were lysed in 2 × Laemmli sample buffer with 20 mM NEM. Whole-cell lysates were sonicated, heated to 95 °C for 5 min and cooled on ice. Protein samples were resolved by SDS-PAGE and transferred onto nitrocellulose membranes for immunoblotting with the indicated antibodies. Membranes were developed using HRP-conjugated secondary antibodies (Bio-Rad, Hercules, CA, USA) and the enhanced chemiluminescence reagent Super Signal West Dura (Thermo Scientific, Rockford, IL, USA). The primary antibodies used were anti-FKBP51 (BD Biosciences, San Jose, CA, USA), anti-FKBP51 and anti-GAPDH (Abcam, Cambridge, UK), anti-HA (Covance, Madison, WI, USA), anti-FLAG (Sigma-Aldrich), anti-ACTIN, anti-PIAS4, anti-p65 and anti-Hsp90 (Santa Cruz Biotechnology, Dallas, TX, USA).
For Supplementary Figure 2b, HEK293T cells transfected with the indicated plasmids were treated 24 h post transfection with cycloheximide (CHX) (50 μg/ml) for the indicated time periods. Immunoreactive signals were quantified by densitometry using ImageJ software (Open Source Software).
Immunofluorescence analysis
Brains from adult mice (BALB/c) killed by decapitation were sectioned in a cryostat (Microm HM550, Thermo Scientific) at 30 μm thickness. Cryosections were analyzed by immunofluorescence using the following primary antibodies: anti-FKBP51 (BD Biosciences), monoclonal anti-SUMO2/3 antibody (SUMO-2 8A2, DSHB, Iowa, IA, USA), or anti-SUMO2/3 (Abcam) and anti-NeuN; and the corresponding Alexa Fluor secondary antibodies (Invitrogen/Thermo Fisher Scientific, Rockford, IL, USA). Nuclei were stained with DAPI.
HT22 cells were analyzed by immunofluorescence using anti-FKBP51 (Abcam) antibodies and Alexa Fluor 647-conjugated secondary antibodies. Nuclei were stained with DAPI.
All images were captured by an inverted LSM 710 Axio Observer Z1 microscope (Carl Zeiss, Oberkochen, Germany) and analyzed with Fiji (Open Source Software).
Immunoprecipitation assays
HEK293T cells transfected with the indicated plasmids were lysed 48 h post transfection in lysis buffer (50 mM Tris-HCl pH 7.4, 1 mM EDTA, 1% Triton X-100, 150 mM NaCl, 2 mM orthovanadate, 1 mM PMSF, 1 × Protease Inhibitor cocktail) supplemented with 20 mM Na2MoO4 for GR/Hsp90/FKBP51/FKBP52 co-immunoprecipitation assays. For the shRNA experiments (Figure 5f), cells were transfected with the shRNA plasmids and 24 h later with PIAS4 expression vector. Immunoprecipitation was performed using anti-FLAG (Sigma-Aldrich) or anti-HA (Covance) antibodies. For detection of endogenous SUMO-FKBP51 conjugates (Figure 1g), the protocol from Barysch et al.60 was adapted as follows. HEK293T cells were lysed in lysis buffer (50 mM Tris-HCl pH 7.4, 1 mM EDTA, 1% Triton X-100, 150 mM NaCl, 50 mM NEM, 1 mM PMSF, 1 mM DTT, 1X Protease Inhibitor cocktail). Immunoprecipitation was performed using anti-SUMO2/3 antibody (SUMO-2 8A2, DSHB). All samples were analyzed by western blotting.
Luciferase assays
HT22 cells were transfected with TAT4-luc or MMTV-luc together with GR expression vector and the indicated plasmids or siRNA and stimulated 16 h later, or 72 h later when siRNA was co-transfected, as indicated. HEK293T cells were transfected with κB-luc together with the indicated plasmids and stimulated 16 h later as indicated. Luciferase and β-galactosidase activities were determined according to the manufacturer's instructions (Promega, Madison, WI, USA). FKBP51−/− MEF cells were electroporated with TAT4-luc together with GR expression vector and the indicated plasmids and stimulated 24 h later as indicated. Firefly luciferase and Gaussia luciferase activities were determined as described in Schulke et al.32 Luciferase activity was relativized to β-galactosidase or Gaussia luciferase activity.
RT-qPCR analysis
HT22 cells were transfected and stimulated 48 h later as indicated. Total RNA isolation, reverse-transcription and qPCR analysis were performed as previously described,18 using the following primer sequences: for SGK1, 5′-AGCATAACGGGACAACATC-3′ (forward) and 5′-GCATCTCATACAGGACAGC-3′ (reverse); and for GILZ, 5′-GACTTCATGTCTTCAGTGGACA-3′ (forward) and 5′-AATGCGGCCACAGATG-3′ (reverse).
For Figure 4g, HT22 cells were transfected and samples were analyzed 72 h later using the following primers: 5′-CAGAGGTTGAAGACGATTG-3′ (forward) and 5′-CAGGTAGAACACAGCATC-3′ (reverse).
SGK1, GILZ and PIAS4 mRNA levels were relativized to HPRT mRNA levels, using the following primers: 5′-TGGGCTTACCTCACTGCTTTCC-3′ (forward) and 5′-CCTGGTTCATCATCGCTAATCACG-3′ (reverse).
Recombinant proteins
GST, GST-Ubc9, GST-PIAS4 and GST-Hsp90, and 6 × His-FKBP51-FLAG were expressed in Escherichia coli M15 (pREP4) strain by induction with 0.5 mM IPTG and purified with glutathione Sepharose 4B beads (GE Healthcare, Little Chalfont, UK) and Ni-NTA Agarose (QIAGEN, Germantown, MD, USA), respectively. Proteins were analyzed by SDS-PAGE Coomassie R-250 staining for quantity and purity and by western blotting.
In vitro SUMOylation and deSUMOylation assays
In vitro SUMOylation and deSUMOylation assays were performed as previously described.18, 47
GST pull-down assays
GST pull-down assays were performed as previously described.18 Recombinant FKBP51 was previously SUMOylated in vitro, and pull-down assays were performed in the presence of 20 mM NEM and 20 mM Na2MoO4.
Purification of 6 × His-SUMO-conjugated proteins
Purification of 6 × His-SUMO-conjugated proteins was performed as previously described.18 HEK293T and HT22 cells were transfected in 35-mm and 60-mm dishes respectively and harvested 48 h later. For Figure 4d, cells were transfected in 10-cm dishes with the shRNA plasmids and with the indicated expression vectors 24 h later. After 48 h, cells were harvested.
In vitro hormone-binding assays
HEK293T cells were transfected and lysed 48 h later in lysis buffer (5 mM Tris-HCl pH 7.4, 1 mM EDTA, 10% glycerol, 20 mM Na2MoO4, 1 mM PMSF, 1 × Protease Inhibitor cocktail, 2 mM β-mercaptoethanol). Cytosolic fractions were obtained from homogenized lysates after centrifugation at 20 000 × g for 30 min and then at 100 000 × g for 1 h at 4 °C. Protein concentration was quantified by Bradford assay (Bio-Rad, Hercules, CA, USA) and adjusted to 2 μg/μl. An aliquot was taken to analyze by western blotting. Hormone binding assays were performed using increasing amounts of [3H]-Dex (0.1–100 nM) in the absence (total binding) and in the presence of 1000-fold molar excess of unlabeled Dex (non-specific binding). The amount of bound [3H]-Dex was measured in a Beckman Coulter LS6500-S multipurpose scintillation counter (Beckman Coulter, Inc., Brea, CA, USA). Data were fit to one-site binding equation using a non-linear regression method and is expressed as percentage of the Bmax in the absence of transfected FKBP51.
Nuclear translocation assay
Nuclear translocation assays were performed in glass-bottom plates. HT22 cells were transfected and stimulated 48 h later with Dex at time t=0 s in serum-free DMEM without phenol red (Gibco/Thermo Fisher Scientific). Fluorescence imaging was performed with an inverted Axio Observer Z1 LED fluorescence microscope (Carl Zeiss) equipped with a live-imaging system at 37 °C and 5% CO2. Images were collected every 120 s during 3600 s for Figure 5b, and every 9 s during 297 s for Supplementary Figure 5; and captured and analyzed using the image software ZEN Blue pro 2011 (Carl Zeiss, Oberkochen, Germany) and Fiji. Mean fluorescence intensity was quantified from background-subtracted images by drawing a region of interest in the nucleus of each cell (n=4 cells, for each condition for Figure 5b; and n=8 cells, for each condition for Supplementary Figure 5). The mean nuclear fluorescence intensity for each time point is defined as the ratio between mean fluorescence intensity at each time point and mean fluorescence intensity before Dex stimulation.
Protein interaction assays
For Figure 5e (left panel), HEK293T cells were transfected with FLAG-Hsp90 expression vector and harvested in lysis buffer (50 mM Tris-HCl pH 7.4, 1 mM EDTA, 1% Triton X-100, 400 mM NaCl, 2 mM orthovanadate, 1 mM PMSF, 1 × Protease Inhibitor cocktail). FLAG-Hsp90 was immunoprecipitated using anti-FLAG antibody and eluted in lysis buffer containing 100 μg/ml of FLAG peptide (Sigma-Aldrich). FLAG-Hsp90 (500 ng) was incubated for 1 h at RT with in vitro SUMOylated FKBP51 (500 ng) in lysis buffer supplemented with 20 mM Na2MoO4, 1 mM DTT and 20 mM NEM. FKBP51 was immunoprecipitated using anti-FKBP51 antibody. Samples were analyzed by western blotting.
Neurite outgrowth assay
Neurite outgrowth assays were performed on polylysine-coated plates. HT22 and Neuro-2a cells were transfected and stimulated 48 h later with Dex or vehicle as indicated. The medium was removed for differentiation. HT22 cells were differentiated in Neurobasal medium (Gibco/Thermo Fisher Scientific) containing 1 × N2 supplement (Gibco/Thermo Fisher Scientific) for 24 h. Neuro-2a cells were differentiated in medium containing 0.2% FBS and 500 μM cAMP for 24 h. Images were obtained using an inverted Axio Vert A1 microscope (Carl Zeiss) and analyzed using Fiji. Neurite length was quantified with Neurite Simple Tracer plugin in RFP+ cells together with their soma diameter. Cells bearing neurites at least 1.5-fold for HT22 and 2.0-fold for Neuro-2a longer than the soma diameter were considered as differentiated, and the percentage of differentiated RFP+ cells over total RFP+ cells was calculated (n=20 cells, for each condition).
Statistics
Experiments were performed at least in triplicate. The results are presented as mean±S.E.M. Statistics were performed by Student’s t-test (for Figures 4g lower panel and 4i) and analysis of variance (ANOVA) in combination with Tukey's (for Supplementary Figure 4) or Bonferroni’s (for the remaining Figures) tests (GraphPad, GraphPad Software Inc., La Jolla, CA, USA).
Abbreviations
- CHX:
-
cycloheximide
- Dex:
-
dexamethasone
- FKBP51:
-
FK506-binding protein 51
- FKBP52:
-
FK506-binding protein 52
- GILZ:
-
GC-induced leucine zipper
- GR:
-
glucocorticoid receptor
- GCs:
-
glucocorticoids
- Hsp90 :
-
heat-shock protein 90
- HPA:
-
hypothalamus-pituitary-adrenocortical
- MEFs:
-
mouse embryonic fibroblasts
- NEM:
-
N-ethylmaleimide
- NF-κB:
-
nuclear factor κB
- PTM:
-
post-translational modification
- PIAS4:
-
protein inhibitors of activated STATs 4
- SGK1:
-
serum GC kinase 1
- shRNA:
-
short hairpin RNA
- SUMO:
-
small ubiquitin-like modifier
- SIM:
-
SUMO-interacting motif
- SENP:
-
SUMO-specific protease
- TPR:
-
tetratricopeptide repeat
- Ubc9:
-
ubiquitin-conjugating enzyme 9
- wt:
-
wild type
References
de Kloet ER, Joels M, Holsboer F . Stress and the brain: from adaptation to disease. Nat Rev Neurosci 2005; 6: 463–475.
Binder EB . The role of FKBP5, a co-chaperone of the glucocorticoid receptor in the pathogenesis and therapy of affective and anxiety disorders. Psychoneuroendocrinology 2009; 34: S186–S195.
Cheung-Flynn J, Roberts PJ, Riggs DL, Smith DF . C-terminal sequences outside the tetratricopeptide repeat domain of FKBP51 and FKBP52 cause differential binding to Hsp90. J Biol Chem 2003; 278: 17388–17394.
Sinars CR, Cheung-Flynn J, Rimerman RA, Scammell JG, Smith DF, Clardy J . Structure of the large FK506-binding protein FKBP51, an Hsp90-binding protein and a component of steroid receptor complexes. Proc Natl Acad Sci USA 2003; 100: 868–873.
Wochnik GM, Ruegg J, Abel GA, Schmidt U, Holsboer F, Rein T . FK506-binding proteins 51 and 52 differentially regulate dynein interaction and nuclear translocation of the glucocorticoid receptor in mammalian cells. J Biol Chem 2005; 280: 4609–4616.
Touma C, Gassen NC, Herrmann L, Cheung-Flynn J, Bull DR, Ionescu IA et al. FK506 binding protein 5 shapes stress responsiveness: modulation of neuroendocrine reactivity and coping behavior. Biol Psychiatry 2011; 70: 928–936.
Davies TH, Ning YM, Sanchez ER . A new first step in activation of steroid receptors: hormone-induced switching of FKBP51 and FKBP52 immunophilins. J Biol Chem 2002; 277: 4597–4600.
Riggs DL, Roberts PJ, Chirillo SC, Cheung-Flynn J, Prapapanich V, Ratajczak T et al. The Hsp90-binding peptidylprolyl isomerase FKBP52 potentiates glucocorticoid signaling in vivo. EMBO J 2003; 22: 1158–1167.
Storer CL, Dickey CA, Galigniana MD, Rein T, Cox MB . FKBP51 and FKBP52 in signaling and disease. Trends Endocrinol Metab 2011; 22: 481–490.
Fani N, Gutman D, Tone EB, Almli L, Mercer KB, Davis J et al. FKBP5 and attention bias for threat: associations with hippocampal function and shape. JAMA Psychiatry 2013; 70: 392–400.
Hay RT . SUMO: a history of modification. Mol Cell 2005; 18: 1–12.
Geiss-Friedlander R, Melchior F . Concepts in sumoylation: a decade on. Nat Rev Mol Cell Biol 2007; 8: 947–956.
Liberman AC, Druker J, Refojo D, Holsboer F, Arzt E . Glucocorticoids inhibit GATA-3 phosphorylation and activity in T cells. FASEB J 2009; 23: 1558–1571.
Kadmiel M, Cidlowski JA . Glucocorticoid receptor signaling in health and disease. Trends Pharmacol Sci 2013; 34: 518–530.
Antunica-Noguerol M, Aprile-Garcia F, Budziñski ML, Proto-Cassina L, Liberman AC, Arzt E . The interplay between the glucocorticoid receptor activity and post-translational modifications in the immune and neuroendocrine systems. Adv Neuroimmune Biol 2014; 5: 19–32.
Tian S, Poukka H, Palvimo JJ, Janne OA . Small ubiquitin-related modifier-1 (SUMO-1) modification of the glucocorticoid receptor. Biochem J 2002; 367: 907–911.
Holmstrom SR, Chupreta S, So AY, Iniguez-Lluhi JA . SUMO-mediated inhibition of glucocorticoid receptor synergistic activity depends on stable assembly at the promoter but not on DAXX. Mol Endocrinol 2008; 22: 2061–2075.
Druker J, Liberman AC, Antunica-Noguerol M, Gerez J, Paez-Pereda M, Rein T et al. RSUME enhances glucocorticoid receptor SUMOylation and transcriptional activity. Mol Cell Biol 2013; 33: 2116–2127.
Mollapour M, Bourboulia D, Beebe K, Woodford MR, Polier S, Hoang et al. Asymmetric Hsp90 N domain SUMOylation recruits Aha1 and ATP-competitive inhibitors. Mol Cell 2014; 53: 317–329.
Guidotti G, Calabrese F, Anacker C, Racagni G, Pariante CM, Riva MA . Glucocorticoid receptor and FKBP5 expression is altered following exposure to chronic stress: modulation by antidepressant treatment. Neuropsychopharmacology 2013; 38: 616–627.
Bonfiglio JJ, Inda C, Senin S, Maccarrone G, Refojo D, Giacomini D et al. B-Raf and CRHR1 internalization mediate biphasic ERK1/2 activation by CRH in hippocampal HT22 Cells. Mol Endocrinol 2013; 27: 491–510.
Rodriguez MS, Dargemont C, Hay RT . SUMO-1 conjugation in vivo requires both a consensus modification motif and nuclear targeting. J Biol Chem 2001; 276: 12654–12659.
Sampson DA, Wang M, Matunis MJ . The small ubiquitin-like modifier-1 (SUMO-1) consensus sequence mediates Ubc9 binding and is essential for SUMO-1 modification. J Biol Chem 2001; 276: 21664–21669.
Erlejman AG, De Leo SA, Mazaira GI, Molinari AM, Camisay MF, Fontana V et al. NF-kappaB transcriptional activity is modulated by FK506-binding proteins FKBP51 and FKBP52: a role for peptidyl-prolyl isomerase activity. J Biol Chem 2014; 289: 26263–26276.
Saaltink DJ, Vreugdenhil E . Stress, glucocorticoid receptors, and adult neurogenesis: a balance between excitation and inhibition? Cell Mol Life Sci 2014; 71: 2499–2515.
Obradovic D, Gronemeyer H, Lutz B, Rein T . Cross-talk of vitamin D and glucocorticoids in hippocampal cells. J Neurochem 2006; 96: 500–509.
Ma'ayan A, Jenkins SL, Barash A, Iyengar R . Neuro2A differentiation by Galphai/o pathway. Sci Signal 2009; 2: cm1.
Tempe D, Piechaczyk M, Bossis G . SUMO under stress. Biochem Soc Trans 2008; 36: 874–878.
Cai Q, Verma SC, Kumar P, Ma M, Robertson ES . Hypoxia inactivates the VHL tumor suppressor through PIASy-mediated SUMO modification. PLoS One 2010; 5: e9720.
Denny WB, Valentine DL, Reynolds PD, Smith DF, Scammell JG . Squirrel monkey immunophilin FKBP51 is a potent inhibitor of glucocorticoid receptor binding. Endocrinology 2000; 141: 4107–4113.
Tatro ET, Everall IP, Kaul M, Achim CL . Modulation of glucocorticoid receptor nuclear translocation in neurons by immunophilins FKBP51 and FKBP52: implications for major depressive disorder. Brain Res 2009; 1286: 1–12.
Schulke JP, Wochnik GM, Lang-Rollin I, Gassen NC, Knapp RT, Berning B et al. Differential impact of tetratricopeptide repeat proteins on the steroid hormone receptors. PLoS One 2010; 5: e11717.
Anacker C, Cattaneo A, Musaelyan K, Zunszain PA, Horowitz M, Molteni R et al. Role for the kinase SGK1 in stress, depression, and glucocorticoid effects on hippocampal neurogenesis. Proc Natl Acad Sci USA 2013; 110: 8708–8713.
Sarabdjitsingh RA, Isenia S, Polman A, Mijalkovic J, Lachize S, Datson N et al. Disrupted corticosterone pulsatile patterns attenuate responsiveness to glucocorticoid signaling in rat brain. Endocrinology 2010; 151: 1177–1186.
Lang F, Bohmer C, Palmada M, Seebohm G, Strutz-Seebohm N, Vallon V . (Patho)physiological significance of the serum- and glucocorticoid-inducible kinase isoforms. Physiol Rev 2006; 86: 1151–1178.
Yachi K, Inoue K, Tanaka H, Yoshikawa H, Tohyama M . Localization of glucocorticoid-induced leucine zipper (GILZ) expressing neurons in the central nervous system and its relationship to the stress response. Brain Res 2007; 1159: 141–147.
Frodl T, Carballedo A, Hughes MM, Saleh K, Fagan A, Skokauskas N et al. Reduced expression of glucocorticoid-inducible genes GILZ and SGK-1: high IL-6 levels are associated with reduced hippocampal volumes in major depressive disorder. Transl Psychiatry 2012; 2: e88.
Henley JM, Craig TJ, Wilkinson KA . Neuronal SUMOylation: mechanisms, physiology, and roles in neuronal dysfunction. Physiol Rev 2014; 94: 1249–1285.
Datwyler AL, Lattig-Tunnemann G, Yang W, Paschen W, Lee SL, Dirnagl U et al. SUMO2/3 conjugation is an endogenous neuroprotective mechanism. J Cereb Blood Flow Metab 2011; 31: 2152–2159.
Tammsalu T, Matic I, Jaffray EG, Ibrahim AF, Tatham MH, Hay RT . Proteome-wide identification of SUMO2 modification sites. Sci Signal 2014; 7: rs2.
Tirard M, Jasbinsek J, Almeida OF, Michaelidis TM . The manifold actions of the protein inhibitor of activated STAT proteins on the transcriptional activity of mineralocorticoid and glucocorticoid receptors in neural cells. J Mol Endocrinol 2004; 32: 825–841.
Ni L, Yang CS, Gioeli D, Frierson H, Toft DO, Paschal BM . FKBP51 promotes assembly of the Hsp90 chaperone complex and regulates androgen receptor signaling in prostate cancer cells. Mol Cell Biol 2010; 30: 1243–1253.
Hubler TR, Denny WB, Valentine DL, Cheung-Flynn J, Smith DF, Scammell JG . The FK506-binding immunophilin FKBP51 is transcriptionally regulated by progestin and attenuates progestin responsiveness. Endocrinology 2003; 144: 2380–2387.
Lin DY, Huang YS, Jeng JC, Kuo HY, Chang CC, Chao TT et al. Role of SUMO-interacting motif in Daxx SUMO modification, subnuclear localization, and repression of sumoylated transcription factors. Mol Cell 2006; 24: 341–354.
Pfander B, Moldovan GL, Sacher M, Hoege C, Jentsch S . SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature 2005; 436: 428–433.
Gassen NC, Hartmann J, Zschocke J, Stepan J, Hafner K, Zellner A et al. Association of FKBP51 with priming of autophagy pathways and mediation of antidepressant treatment response: evidence in cells, mice, and humans. PLoS Med 2014; 11: e1001755.
Carbia-Nagashima A, Gerez J, Perez-Castro C, Paez-Pereda M, Silberstein S, Stalla GK et al. RSUME, a small RWD-containing protein, enhances SUMO conjugation and stabilizes HIF-1alpha during hypoxia. Cell 2007; 131: 309–323.
Liberman AC, Antunica-Noguerol M, Ferraz-de-Paula V, Palermo-Neto J, Castro CN, Druker J et al. Compound A, a dissociated glucocorticoid receptor modulator, inhibits T-bet (Th1) and induces GATA-3 (Th2) activity in immune cells. PLoS One 2012; 7: e35155.
Desterro JM, Rodriguez MS, Hay RT . SUMO-1 modification of IkappaBalpha inhibits NF-kappaB activation. Mol Cell 1998; 2: 233–239.
Tatham MH, Jaffray E, Vaughan OA, Desterro JM, Botting CH, Naismith JH et al. Polymeric chains of SUMO-2 and SUMO-3 are conjugated to protein substrates by SAE1/SAE2 and Ubc9. J Biol Chem 2001; 276: 35368–35374.
Jaffray EG, Hay RT . Detection of modification by ubiquitin-like proteins. Methods 2006; 38: 35–38.
Schaaf MJ, Cidlowski JA . Molecular determinants of glucocorticoid receptor mobility in living cells: the importance of ligand affinity. Mol Cell Biol 2003; 23: 1922–1934.
Cheng J, Wang D, Wang Z, Yeh ET . SENP1 enhances androgen receptor-dependent transcription through desumoylation of histone deacetylase 1. Mol Cell Biol 2004; 24: 6021–6028.
Chun TH, Itoh H, Subramanian L, Iniguez-Lluhi JA, Nakao K . Modification of GATA-2 transcriptional activity in endothelial cells by the SUMO E3 ligase PIASy. Circ Res 2003; 92: 1201–1208.
Ismail IH, Gagne JP, Caron MC, McDonald D, Xu Z, Masson JY et al. CBX4-mediated SUMO modification regulates BMI1 recruitment at sites of DNA damage. Nucleic Acids Res 2012; 40: 5497–5510.
Hollenberg SM, Evans RM . Multiple and cooperative trans-activation domains of the human glucocorticoid receptor. Cell 1988; 55: 899–906.
Rupprecht R, Arriza JL, Spengler D, Reul JM, Evans RM, Holsboer F et al. Transactivation and synergistic properties of the mineralocorticoid receptor: relationship to the glucocorticoid receptor. Mol Endocrinol 1993; 7: 597–603.
Naito T, Momose F, Kawaguchi A, Nagata K . Involvement of Hsp90 in assembly and nuclear import of influenza virus RNA polymerase subunits. J Virol 2007; 81: 1339–1349.
Yueh A, Leung J, Bhattacharyya S, Perrone LA, de los Santos K, Pu SY et al. Interaction of moloney murine leukemia virus capsid with Ubc9 and PIASy mediates SUMO-1 addition required early in infection. J Virol 2006; 80: 342–352.
Barysch SV, Dittner C, Flotho A, Becker J, Melchior F . Identification and analysis of endogenous SUMO1 and SUMO2/3 targets in mammalian cells and tissues using monoclonal antibodies. Nat Protoc 2014; 9: 896–909.
Acknowledgements
We thank Profs. R Hay and J Iñiguez-Lluhí for fruitful discussions on the results, Dr. AF Schinder for providing the anti-NeuN antibody, Dr. MB Cox for providing FKBP51−/− MEFs and A Attorresi for technical help. This work was supported by grants from the Max Planck Society, Germany; University of Buenos Aires; CONICET; Agencia Nacional de Promoción Científica y Tecnológica, Argentina; and FOCEM-Mercosur (COF 03/11).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by JA Cidlowski
Supplementary Information accompanies this paper on Cell Death and Differentiation website
Rights and permissions
About this article
Cite this article
Antunica-Noguerol, M., Budziñski, M., Druker, J. et al. The activity of the glucocorticoid receptor is regulated by SUMO conjugation to FKBP51. Cell Death Differ 23, 1579–1591 (2016). https://doi.org/10.1038/cdd.2016.44
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cdd.2016.44
This article is cited by
-
Tricyclic antidepressants target FKBP51 SUMOylation to restore glucocorticoid receptor activity
Molecular Psychiatry (2022)
-
Evidence of increased hypoxia signaling in fetal liver from maternal nutrient restriction in mice
Pediatric Research (2020)
-
Relationship Between Homodimeric Glucocorticoid Receptor and Transcriptional Regulation Assessed via an In Vitro Fluorescence Correlation Spectroscopy-Microwell System
Scientific Reports (2018)
-
A comprehensive platform for the analysis of ubiquitin-like protein modifications using in vivo biotinylation
Scientific Reports (2017)