Abstract
Amyloid-β (Aβ) produces neurotoxicity in the brain and causes neuronal death, but the endogenous defense mechanism that is activated on Aβ insult is less well known. Here we found that acute Aβ increases the expression of PIAS1 and Mcl-1 via activation of MAPK/ERK, and Aβ induction of PIAS1 enhances HDAC1 SUMOylation in rat hippocampus. Knockdown of PIAS1 decreases endogenous HDAC1 SUMOylation and blocks Aβ induction of Mcl-1. Sumoylated HDAC1 reduces it association with CREB, increases CREB binding to the Mcl-1 promoter and mediates Aβ induction of Mcl-1 expression. Transduction of SUMO-modified lenti-HDAC1 vector to the hippocampus of APP/PS1 mice rescues spatial learning and memory deficit and long-term potentiation impairment in APP/PS1 mice. It also reduces the amount of amyloid plaque and the number of apoptotic cells in CA1 area of APP/PS1 mice. Meanwhile, HDAC1 SUMOylation decreases HDAC1 binding to the neprilysin promoter. These results together reveal an important role of HDAC1 SUMOylation as a naturally occurring defense mechanism protecting against Aβ toxicity and provide an alternative therapeutic strategy against AD.
Similar content being viewed by others
Main
The brain of Alzheimer’s disease (AD) patient is characterized by the accumulation of senile plaques, and amyloid-β peptides (Aβ1-40 and Aβ1-42) are the major components of these senile plaques. Aβ is known to cause lipid peroxidation, free radical production, caspase 3 activation and DNA damage that eventually lead to neuronal death.1, 2, 3 In addition, the Aβ peptide or overexpression of Aβ causes cognitive impairment in animals.4, 5 This cognitive impairment correlates with amyloid plaque formation4, 6 or precedes it.7, 8 Further, naturally secreted Aβ or the Aβ peptide also inhibits long-term potentiation (LTP) in the hippocampus in vivo and disrupts synaptic and network function.9, 10 More recently, we have found that Aβ induces the expression of activated signal transducer and activator of transcription-1 (STAT1) and Aβ induction of STAT1 mediates the memory-impairing effect of Aβ.11 On the other hand, it is conceivable that when Aβ produces its toxicity, neurons would develop defense mechanisms to cope with Aβ toxicity. For example, a non-amyloidogenic neurotrophic peptide sAPPα is shown to activate neuroprotectin D1 and promote cell survival.12 In addition, we have found that Aβ activates the MAPK/ERK-SGK (serum- and glucocorticoid-inducible kinase) signaling pathway for neuroprotection against Aβ insult.13 However, with the role and mechanism of Aβ-induced toxicity been studied extensively, the endogenous protection mechanism induced by Aβ is less well known.
Histone acetylation is one kind of epigenetic regulations that has an important role in a wide range of brain functions and disorders, and histone deacetylases (HDACs) regulate the homeostasis of histone acetylation. The HDAC family contains 18 HDAC proteins that belong to different classifications. Inhibition of class I HDACs (that include HDAC1, HDAC2, HDAC3 and HDAC8) was shown to reverse memory deficits in a mouse model of AD.14 The HDAC inhibitor SAHA and curcumin produce a synergistic effect in protecting against Aβ toxicity.15 These findings support the notion that HDAC inhibitors are neuroprotective and they are potential cognitive enhancers.16, 17, 18 But most of these studies examined the role of one class of HDAC proteins instead of a specific HDAC protein. Besides, they often focus on inhibition of HDAC activity or reduction of HDAC expression, other modifications with HDAC1 is less well studied.
HDAC1 belongs to class I HDAC proteins and it was the first mammalian HDAC protein identified.19 A few reports have shown post-translational modifications of HDAC1 in the cell, such as phosphorylation,20 ubiquitination21 and SUMOylation.22 HDAC1 was found sumoylated at Lys-444 and Lys-476 in the cell, and HDAC1 SUMOylation modulates its biological activity.23 More recently, HDAC1 was found SUMO-modified by both SUMO1 and SUMO2. SUMO1 modification of HDAC1 promotes HDAC1 ubiquitination and degradation, whereas SUMO2 modification of HDAC1 enhances HDAC1 stability in breast cancer cells.24 Despite of the important role of HDAC1 implicated in neurodegenerative diseases and cognitive functions, HDAC1 SUMOylation in the central nervous system has not been studied and its physiological significance is not known either. Protein inhibitor of activated STAT1 (PIAS1) is a SUMO E3 ligase that enhances the SUMOylation of a variety of proteins. In addition, PIAS1 was shown to protect against H2O2-induced cell death,25 indicating that PIAS1 has an anti-apoptotic role. In the present study we aimed to examine the role and mechanism of HDAC1 SUMOylation by PIAS1 in protecting against Aβ toxicity by adopting the APPswe/PS1dE9 (APP/PS1) mice as a mouse model for AD.
Results
Acute Aβ increases the expression of PIAS1 through the mediation of MAPK/ERK and that PIAS1 mediates Aβ induction of Mcl-1 expression
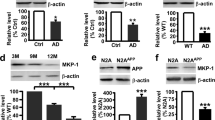
Because PIAS1 was suggested to have a protective role against H2O2-induced cell death25 and the anti-apoptotic gene Mcl-1 was shown to exert a neuroprotective effect in the brain,13, 26 we first examined whether acute Aβ treatment increases the expression of PIAS1 and Mcl-1. Rats were divided to three groups and received 1% NH4OH (control group), Aβ (14 μg) injection and sacrificed 30 min later, and Aβ injection (14 μg) but killed 1 h later. Aβ was directly injected to the CA1 area of rat brain. Their brains were removed and their CA1 tissue was dissected out for western blot analysis. Western blots were carried out using antibodies against PIAS1, Mcl-1, ERK1/2, pERK1/2 and actin. A representative gel pattern is shown in Figure 1a. Results revealed that Aβ injection increased the expression of PIAS1 at both time points examined, but its effect is more significant at 1 h later (Figure 1b). Meanwhile, Aβ increased the phosphorylation level of ERK1 and ERK2, but this effect was significant at 30 min only (Figure 1b). Aβ also increased the expression of Mcl-1, but this effect was apparent at 1 h only (Figure 1b). Because induction of Mcl-1 expression may not take place immediately after PIAS1 expression, in the next experiment, we have examined the effect of Aβ (14 μg) on PIAS1 and Mcl-1 expression at longer time intervals (8, 16 and 48 h), but we also included the 1 h interval as a positive control. Western blots were conducted using anti-PIAS1 and anti-Mcl-1 antibodies. A representative gel pattern is shown in Figure 1c. Results revealed that Aβ increased the expression of PIAS1 at 1, 8 and 16 h later, but the maximum effect occurred at 1 h later. PIAS1 expression was returned to the control level at 48 h later (Figure 1d). But Aβ produced a time-dependent increase in Mcl-1 expression with the maximum effect observed at 48 h later (Figure 1d).
Acute Aβ increases the expression of PIAS1 through the mediation of MAPK/ERK and that PIAS1 mediates Aβ induction of Mcl-1 expression. (a) Western blot was carried out to determine the acute effects of Aβ on expression of PIAS1, pERK1/2, ERK1/2 and Mcl-1 in rat hippocampus 30 min and 1 h later. The four different lanes in each group represent four individual rat brains (CA1 area). (b) Quantified results are shown (n=6 each group; for PIAS1, F2,15=90.57, P<0.001; for pERK1/ERK1, F2,15=166.16, P<0.001; for pERK2/ERK2, F2,15=68.75, P<0.001; for Mcl-1, F2,15=7.26, P<0.01). (c) Time course effect of acute Aβ on PIAS1 and Mcl-1 expression in the hippocampus. The two different lanes in each group represent two individual rat brains (CA1 area). (d) Quantified results are shown (n=5 each group; for PIAS1, F4,20=30.13, P<0.001; for Mcl-1, F4,20=22.27, P<0.001). (e) Effect of ERK inhibition (with sub-threshold concentration of U0126) on Aβ induction of PIAS1 expression in the hippocampus. The two different lanes in each group represent two individual rat brains (CA1 area). (f) Quantified results are shown (n=6 each group; for PIAS1, F3,20=71.39, P<0.001; for pERK1/ERK1, F3,20=260.16, P<0.001; for pERK2/ERK2, F3,20=114.18, P<0.001). (g) Effect of knockdown of PIAS1 (with sub-threshold concentration of PIAS1 siRNA) on Aβ induction of Mcl-1 expression. The three different lanes in each group represent three individual rat brains (CA1 area). (h) Quantified results are shown (n=5 each group; for PIAS1, F3,16=43.14, P<0.001; q=9.19, P<0.001; for Mcl-1, F3,16=86.29, P<0.001; q=13.03, P<0.001). Data are mean±s.e.m. **P<0.01 and #P<0.001
MAPK/ERK-mediated signaling was suggested to mediate neuroprotection against Aβ toxicity,13 and PIAS1 expression is regulated by MAPK/ERK signaling.27 Here we examined whether Aβ induction of PIAS1 expression is mediated through Aβ-induced MAPK/ERK activation. U0126 was used as the inhibitor for MAPK/ERK activation and a sub-threshold concentration of U0126 was used to block Aβ signaling. Rats were divided to four groups and received DMSO (15%)+NH4OH (1%), DMSO+Aβ (14 μg), U0126 (0.7 μg)+NH4OH and U0126 (0.7 μg)+Aβ (14 μg) injections to their CA1 area directly. The time interval between the two injections was 30 min and animals were sacrificed 30 min after the second injection. Western blots were conducted using anti-PIAS1, anti-ERK1/2 and anti-pERK1/2 antibodies. A representative gel pattern is shown in Figure 1e. Results revealed that Aβ consistently increased the expression of PIAS1 and the phosphorylation level of ERK1/2, but these effects were blocked by U0126 pretreatment at a concentration of U0126 (1 μg/μl) that did not produce a significant effect on these measures by itself (Figure 1f). In another experiment, we have found that Aβ induction of PIAS1 expression was also blocked by the PI-3 kinase inhibitor wortmannin (Supplementary Figure S1a). These results suggest that both MAPK/ERK signaling and PI-3 K signaling mediate the effect of Aβ on PIAS1 expression. The above results showed that Aβ increased the expression of PIAS1 and Mcl-1 at several time intervals examined, here we examined whether Aβ induction of Mcl-1 expression is mediated through PIAS1. Rats were divided to four groups and received control siRNA+NH4OH (1%), control siRNA (4 pmol)+Aβ (14 μg), PIAS1 siRNA (4 pmol)+NH4OH and PIAS1 siRNA (4 pmol)+Aβ (14 μg) injections. Similarly, a sub-threshold concentration of PIAS1 siRNA was adopted to block Aβ-induced signaling. The time interval between the two injections was 24 h and animals were killed 24 h after the second injection. Western blots were conducted using anti-PIAS1 and anti-Mcl-1 antibodies. A representative gel pattern is shown in Figure 1g. Results showed that Aβ injection at 24 h later consistently increased the expression of Mcl-1, but this effect was blocked by prior PIAS1 siRNA transfection (Figure 1h). Aβ slightly increased PIAS1 expression, but this effect was prevented by prior PIAS1 siRNA transfection (Figure 1h). The effectiveness of PIAS1 siRNA transfection was confirmed by significant decrease of PIAS1 expression (Figure 1h). The anti-apoptotic effect of Mcl-1 was shown by significant reduction of Aβ-induced caspase 3 activation by a sub-threshold concentration of Flag-Mcl-1 plasmid transfection (0.7 μg/μl) (Supplementary Figure S1b). Because there are other members of the PIAS protein family, including PIAS2, PIAS3 and PIAS4,28 we also examined whether Aβ increased the expression of these PIAS proteins as well. Results revealed that acute injection of Aβ (14 μg) and reverse Aβ (rAβ, rAβ42-1, 14 μg) both did not alter the expression of PIAS2, PIAS3 and PIAS4 (Supplementary Figure S2).
Acute Aβ increases HDAC1 SUMOylation by PIAS1 in the hippocampus
The above results showed that acute Aβ treatment increased PIAS1 expression at 24 h later. Because PIAS1 is a SUMO E3 ligase and HDAC1 is implicated in neurodegenerative diseases, here we examined whether HDAC1 could be sumoylated by PIAS1 in the rat brain and whether HDAC1 SUMOylation is modulated by Aβ treatment. We first carried out the in vitro SUMOylation assay. Recombinant E1, E2, and different His- or GST-tagged proteins were added to the reaction and western blot was carried out. Results revealed that HDAC1 SUMOylation was observed when E1, E2, SUMO1 and HDAC1 proteins were present, but HDAC1 SUMOylation was enhanced when the PIAS1 protein was also added. But HDAC1 SUMOylation was completely blocked by the addition of sentrin-specific protease 1 (SENP1), an enzyme that removes the sumo molecule from sumo-conjugated proteins (Figure 2a). We then carried out the co-immunoprecipitation (co-IP) experiment and the result showed that PIAS1 is associated with HDAC1 endogenously in rat CA1 area (Figure 2b). Next, we examined whether PIAS1 and HDAC1 are present in the same neurons in the hippocampus of the rat brain. Brain sections containing the CA1 region were subjected to immunohistochemistry staining. Results revealed that immunofluorescence for PIAS1 (green), HDAC1 (red) and DAPI (blue) was visualized in neurons in the CA1 area (Figure 2c, upper panel). When these CA1 neurons were visualized at a higher magnification, PIAS1 and HDAC1 were found present and co-localized in the nucleus of the same CA1 neurons (Figure 2c, lower panel). We further examined whether Aβ treatment alters the association between PIAS1 and HDAC1. Results revealed that acute Aβ injection increased the association between PIAS1 and HDAC1 in the CA1 area at 1 h later (Figure 2d). The quantitative result is shown in the right panel. These results suggest that HDAC1 is probably SUMO-modified by PIAS1 in the hippocampus and Aβ may increase the SUMOylation of HDAC1 by PIAS1. To test this hypothesis, we have transfected control siRNA and PIAS1 siRNA (8 pmol) to rat CA1 area and endogenous HDAC1 SUMOylation was examined 48 h later. Results revealed that PIAS1 siRNA transfection significantly decreased the level of endogenous HDAC1 SUMOylation when the cell lysate was immunoprecipitated with anti-HDAC1 antibody and immunoblotted with anti-HDAC1 antibody (Figure 2e, left panel) or anti-SUMO1 antibody (Figure 2e, right panel). Meanwhile, PIAS1 siRNA transfection decreased the level of PIAS1 expression and Mcl-1 expression (Figure 2e, lower-left panel). The quantified results are shown in Figure 2f.
Acute Aβ increases HDAC1 SUMOylation by PIAS1 in the hippocampus. (a) In vitro SUMOylation assay showing HDAC1 SUMOylation by PIAS1. Recombinant E1, E2 proteins (from the SUMO kit) and purified GST-PIAS1, His-SUMO1, His-HDAC1 and GST-SENP proteins were added to the reaction for this assay. (b) Co-IP experiment showing the association between PIAS1 and HDAC1 in rat hippocampus. (c) Immunohistochemistry showing PIAS1 and HDAC1 are both present in the nucleus of the same neurons in CA1 area of rat brain. N=3. Scale bar, 20 μm (upper panel); 10 μm (lower panel). (d) Co-IP experiment showing the association between PIAS1 and HDAC1 with and without acute Aβ treatment with quantified results (n=3 each group, t1,4=6.5, P<0.01). (e) Effects of knockdown of PIAS1 on endogenous HDAC1 SUMOylation, Mcl-1 and PIAS1 expression in rat hippocampus. (f) Quantified results are shown (n=5 each group; for HDAC1 SUMOylation, t1,8=4.02, P<0.01; for Mcl-1, t1,8=11.69, P<0.001; for PIAS1, t1,8=9.6, P<0.001). (g) Effects of Aβ and reverse Aβ on HDAC1 SUMOylation in rat hippocampus (n=5 each group; F2,12=62.07, P<0.001). (h) Effect of knockdown of PIAS1 (with sub-threshold concentration of PIAS1 siRNA) on Aβ induction of HDAC1 SUMOylation in rat hippocampus. (i) Quantified results are shown (n=5 each group; for HDAC1 SUMOylation, F3,16=44.88, P<0.001; for PIAS1, F3,16=49.61, P<0.001). rAβ: reverse Aβ. Data are mean±s.e.m. *P<0.05, **P<0.01 and #P<0.001
We next examined whether acute Aβ enhances HDAC1 SUMOylation by PIAS1. Rats were divided to three groups and received NH4OH (1%), reverse Aβ (14 μg) and Aβ (14 μg) injection to their CA1 area directly. Animals were killed 1 h later and their CA1 tissue was subjected to HDAC1 SUMOylation determination. Results revealed that Aβ, but not rAβ, markedly increased the level of HDAC1 SUMOylation in the hippocampus (Figure 2g). The quantified result is shown in the right panel. We next examined whether Aβ induction of HDAC1 SUMOylation is mediated by PIAS1. A sub-threshold concentration of PIAS1 siRNA was adopted to block Aβ signaling. Rats were divided to four groups and received the following treatments: control siRNA (4 pmol)+NH4OH (1%), control siRNA+Aβ (14 μg), PIAS1 siRNA (4 pmol)+NH4OH and PIAS1 siRNA (4 pmol)+Aβ (14 μg). The time interval between the two injections was 47 h, and animals were killed 1 h after the second injection. Their brains were removed and their CA1 tissue was subjected to HDAC1 SUMOylation determination. The cell lysate was subjected to western blot analysis using anti-PIAS1 antibody. Results revealed that Aβ injection consistently increased the level of HDAC1 SUMOylation in the hippocampus at 1 h later, but this effect was blocked by prior PIAS1 siRNA transfection at a concentration of PIAS1 siRNA that did not apparently affect HDAC1 SUMOylation alone (4 pmol) (Figure 2h). In addition, acute Aβ consistently increased the level of PIAS1 expression, but this effect was also significantly diminished by prior PIAS1 siRNA transfection. A representative gel pattern is shown in Figure 2h. The quantified results are shown in Figure 2i. PIAS1 siRNA at 4 pmol also reduced the level of PIAS1 expression, but the difference of PIAS1 expression between control siRNA+Aβ group and PIAS1 siRNA+Aβ group was much higher than that between control siRNA group and PIAS1 siRNA group (Figure 2i).
We next examined whether increased HDAC1 SUMOylation was induced only within a short time after Aβ treatment to serve as an endogenous defense mechanism. A separate group of animals was similarly injected with NH4OH (1%) or Aβ (14 μg) to their CA1 area, but they were sacrificed 14 days later. This time interval allows the toxicity of Aβ been accumulated in the cell and it may be too late to induce endogenous neuroprotection. Results revealed that Aβ significantly decreased the level of HDAC1 SUMOylation at 14 days later (Supplementary Figure S3).
Identification of candidate SUMO sites on HDAC1 in the cell and in the hippocampus
The above results indicated that HDAC1 could be SUMO-modified by PIAS1. Here we examined the candidate SUMO sites on HDAC1. We first adopted the bioinformatics method and used the SUMO 2.0 Software for analysis (The CUCKOO Workgroup, Hefei, China). According to the prediction from this software, there are four lysine residues that show high score and two of them (Lys-444 and Lys-476) fit to the sequence of consensus SUMO-substrate motif. Three lysine residues show medium score and two lysine residues show low score (Table 1). Result from another study also reveals that Lys-444 and Lys-476 are the candidate sumo sites on HDAC1 in the cell;23 therefore we have generated individual HDAC1 sumo-mutants against each of these nine residues and the double sumo-mutant (K444RK476R) for further studies. The Flag-tagged HDAC1WT plasmid and each individual HDAC1 mutant plasmid was co-transfected with EGFP-PIAS1WT plasmid and Myc-SUMO1WT plasmid to HEK293T cells and the cell lysates were immunoblotted with anti-Flag antibody. Western blots using antibodies against EGFP and Myc were carried out as loading controls. Results revealed that two adjacent SUMO-HDAC1 bands were observed when the Flag-HDAC1WT plasmid was transfected. But the lower SUMO-HDAC1 band was not present when Flag-HDAC1K444R was transfected compared with Flag-HDAC1WT transfection (Figure 3a). Although the upper SUMO-HDAC1 band was missing when Flag-HDAC1K476R was transfected compared with Flag-HDAC1WT transfection (Figure 3b). To further examine whether Lys-444 and Lys-476 are the two major candidate SUMO sites on HDAC1, we have transfected Flag-tagged HDAC1WT plasmid, each single HDAC1 mutant plasmid and HDAC1 double mutant plasmid together with EGFP-PIAS1 plasmid and Myc-SUMO1 plasmid to HEK293T cells. The cell lysates were immunoprecipitated and immunoblotted with anti-Flag antibody. Results similarly revealed that when HDAC1K444R was transfected, the lower SUMO-HDAC1 band was not observed, as indicated by the lower arrowhead (Figure 3c, left panel). When Flag-HDAC1K476R was transfected, the upper SUMO-HDAC1 band was not present, as indicated by the upper arrowhead (Figure 3c, left panel). But when Flag-HDAC1K444RK476R was transfected, the intensity of the SUMO-HDAC1 band was further reduced compared with that of each single mutant transfection, yet the HDAC1 SUMOylation signal did not completely disappear (Figure 3c, left panel). The same results were obtained when cell lysates were immunoblotted with anti-SUMO1 antibody (Figure 3c, right panel). Western blots with antibodies against EGFP and Myc were carried out as loading controls (Figure 3c, lower-left panel). Because ubiquitination also takes place at the lysine residues and HDAC1 ubiquitination has been reported,21 next we examined whether HDAC1 ubiquitination may also occur at Lys-444 and Lys-476. The Flag-tagged HDAC1WT plasmid or HDAC1K444RK476R plasmid was co-transfected with the His-ubiquitin plasmid to HEK293T cells and cell lysates were immunoprecipitated with anti-Flag antibody and immunoblotted with anti-Flag antibody or anti-His antibody. Results revealed that similar bands and intensities of ubiquitinated HDAC1 were observed between the Flag-HDAC1WT group and Flag-HDAC1K444RK476R group when immunoblotted with either antibody (Figure 3d).
Identification of candidate SUMO sites on HDAC1 in the cell and in the hippocampus. (a and b) EGFP-PIAS1WT and Myc-SUMO1WT were co-transfected with Flag-HDAC1WT or different Flag-HDAC1 lysine mutant plasmids to HEK293T cells. HDAC1 SUMOylation was examined by immunoblotting (IB) with anti-Flag antibody. Arrow indicates Lys-444 band in a and Lys-476 band in b. (c) EGFP-PIAS1WT and Myc-SUMO1WT were co-transfected with Flag-HDAC1WT, Flag-HDAC1K444R, Flag-HDAC1K476R or Flag-HDAC1 double sumo-mutant plasmid to HEK293T cells and HDAC1 SUMOylation was examined by immunoprecipitation (IP) with anti-Flag antibody and IB with anti-Flag antibody (left) or anti-SUMO1 antibody (right). The upper arrow indicates the Lys-476 band and the lower arrow indicates the Lys-444 band. (d) Flag-HDAC1WT or Flag-HDAC1K444RK476R plasmid was co-transfected with His-Ubiquitin to HEK293T cells and cell lysates were immunoprecipitated with anti-Flag antibody and immunoblotted with anti-Flag or anti-His antibody. Experiments in a–d are in two repeats. (e) Flag-vector, Flag-HDAC1WT (with or without the addition of sumo1 mutant protein to the SUMOylation reaction), Flag-HDAC1K444R, Flag-HDAC1K476R or Flag-HDAC1K444RK476R plasmid was transfected to rat CA1 area and SUMOylation assay was carried out 48 h later to determine HDAC1 SUMOylation at Lys-444 and Lys-476 in the hippocampus. (left) Immunoblotted with anti-HDAC1 antibody. (right) Immunoblotted with anti-SUMO1 antibody. (f) Quantified results are shown (n=4 each group; F5,18=119.04, P<0.001). (g) PBS, BDNF, IGF-1 or CRF was injected to rat CA1 area and HDAC1 SUMOylation was determined 45 min (for IGF-1) or 1 h (for BDNF and CRF) later. (h) Quantified results are shown (n=4 each group; F3,12=1262.4, P<0.001). Data are mean±s.e.m. #P<0.001
The above results indicated that Lys-444 and Lys-476 are the two major candidate SUMO sites on HDAC1 in HEK293T cells. Next, we examined whether HDAC1 SUMOylation also occurs at these two residues in the brain. Rats were divided to six groups and received the following plasmid transfection to their CA1 area: Flag-vector, Flag-HDAC1WT, Flag-HDAC1K444R, Flag-HDAC1K476R, Flag-HDAC1K444RK476R and Flag-HDAC1WT with purified SUMO1 mutant protein added to the reaction. Animals were killed 48 h after plasmid transfection. Their brains were removed and their CA1 tissue was dissected out and subjected to HDAC1 SUMOylation assay. Cell lysates were immunoprecipitated with anti-HDAC1 antibody and immunoblotted with anti-HDAC1 antibody. Results revealed that enhanced HDAC1 SUMOylation was observed when the Flag-HDAC1WT plasmid was transfected compared with the control group. The intensity of the SUMO-HDAC1 band was reduced when either mutant plasmid was transfected, but HDAC1 SUMOylation was completely blocked when the double sumo-mutant plasmid was transfected. The SUMOylation signal was also absent when the Flag-HDAC1WT plasmid was transfected and the SUMO1 mutant protein added to the reaction (Figure 3e, left panel). Similar results were obtained when cell lysates were immunoblotted with anti-SUMO1 antibody (Figure 3e, right panel). The quantified result is shown in Figure 3f.
BDNF, IGF-1 and CRF increase HDAC1 SUMOylation in the hippocampus
Lastly, we examined whether HDAC1 SUMOylation could be regulated by endogenous molecules that are known to have neurotrophic or neuroprotective effect. Brain-derived neurotrophic factor (BDNF) was shown to have neuroprotective and therapeutic effects in animal models of AD and in other neurological disorders.29 Insulin-like growth factor-1 (IGF-1) was shown to protect hippocampal neurons from Aβ toxicity and it also rescues hippocampal neurons that are pre-exposed to amyloidogenic peptides.30 The neuropeptide corticotropin-releasing factor (CRF) was known to enhance cognitive function and Bdnf gene expression in animals;31 therefore we examined the effects of these three molecules on HDAC1 SUMOylation. Rats were divided to five groups and received the following injections to their CA1 area directly: PBS, BDNF (0.7 μg), IGF-1 (70 pg) and CRF (70 ng). Animals were killed 45 min after IGF-1 injection and 1 h after BDNF and CRF injection and their CA1 tissue was subjected to HDAC1 SUMOylation assay. Result revealed that BDNF, IGF-1 and CRF treatments all markedly increased the level of HDAC1 SUMOylation in rat hippocampus (Figure 3g). The quantified result is shown in Figure 3h. We also examined the effect of dexamethasone, a synthetic glucocorticoid, on HDAC1 SUMOylation. Results revealed that dexamethasone injection to rat hippocampus (21 μg) did not affect HDAC1 SUMOylation (Supplementary Figure S4).
HDAC1 SUMOylation decreases its association with CREB, increases CREB DNA binding to the Mcl-1 promoter and mediates Aβ induction of Mcl-1 expression
The above results demonstrated HDAC1 SUMOylation both in the cell and in the rat brain, and that HDAC1 SUMOylation is regulated by a few endogenous stimuli. In this series of experiments, we examined the molecular mechanism of HDAC1 SUMOylation. HDAC1 is known to form multiprotein complexes containing HDAC1-HDAC1 homodimers or HDAC1-HDAC2 heterodimers. The HDAC1-HDAC1 homodimers, together with the Sin3 core complex and other proteins, form one type of co-repressor complex.32 In addition, HDAC1 was found associated with CREB and HDAC1 disrupts CREB phosphorylation through interaction with protein phosphotase 1. Further, HDAC inhibitors potentiate CREB activity by prolonging CREB phosphorylation.33 These results together suggest that HDAC1 suppresses CREB activity and CREB-mediated gene transcription. On the basis of these observations, we hypothesized that HDAC1 SUMOylation dissociates HDAC1 with CREB and releases CREB from the co-repressor complex that allows CREB to bind to the DNA promoter for transcriptional regulation of gene expression. This hypothesis was examined here. Rats were divided to four groups and received the following plasmid transfections to the CA1 area: Flag-vector, Flag-HDAC1WT, Flag-HDAC1K444RK476R and Flag-HDAC1WT-SUMO1 fusion plasmid. Their CA1 tissue was dissected out and the cell lysates were immunoprecipitated with anti-Flag antibody and immunoblotted with anti-CREB antibody. Results revealed that HDAC1 is associated with CREB in the hippocampus. This association was increased by HDAC1 sumo-mutant transfection, but was decreased by HDAC1-SUMO1 fusion plasmid transfection (Figure 4a, left panel). The quantified result is shown in Figure 4b. Cell lysates were also immunoprecipitated and immunoblotted with anti-Flag antibody to verify plasmid transfection and expression in the CA1 area. To confirm that the shifted band in the Flag-HDAC1-SUMO1 transfection group is indeed the sumo-HDAC1 band, the cell lysate was also immunoprecipitated with anti-Flag antibody and immunoblotted with anti-SUMO1 antibody. A single band at the same molecular weight of sumo-HDAC1 band was observed in the Flag-HDAC1-SUMO1 transfection group only (Figure 4a, right panel).
HDAC1 SUMOylation reduces its association with CREB, increases CREB DNA binding to Mcl-1 promoter and mediates Aβ induction of Mcl-1 expression. (a) Flag-vector, Flag-HDAC1WT, Flag-HDAC1K444RK476R or Flag-HDAC1WT-SUMO1 fusion plasmid was transfected to rat CA1 area and cell lysates were subjected to IP with anti-Flag antibody and IB with anti-Flag and anti-CREB antibodies (left). The same cell lysates were also subjected to IP with anti-HDAC1 antibody and IB with anti-SUMO1 antibody (right). (b) Quantified results are shown (n=3 each group; F3,8=374.94, P<0.001). (c) The same plasmids described in a were transfected to rat CA1 area and the CA1 tissue was subjected to CREB DNA binding assay, and IP and IB with anti-Flag antibody. (d) Quantified results are shown (n=5 each group; F3,16=84.76, P<0.001). (e) The same plasmids described above were transfected to rat CA1 area and the CA1 tissue was subjected to ChIP PCR assay for determination of CREB binding to Mcl-1 promoter. Cell lysates were also IP and IB with anti-Flag antibody. Experiments are in two repeats. (f) Control siRNA or CREB siRNA was transfected to rat CA1 area and the expression of Mcl-1 and CREB was determined by western blot. (g) Quantified results are shown (n=6 each group; t1,10=11.52, P<0.001 for Mcl-1 and t1,10=13, P<0.001 for CREB). (h) The plasmids described above were transfected to rat CA1 area. Aβ was injected to CA1 area 24 h after plasmid transfection and the CA1 tissue was dissected out 24 h after Aβ injection (Flag-vector+NH4OH served as the control group). Mcl-1 expression was determined by western blot. Cell lysates were also subjected to IP and IB with anti-Flag antibody. (i) Quantified results are shown (n=5 each group; F4,20=83.54, P<0.001). Data are mean±s.e.m. *P<0.05 and #P<0.001
If CREB is released from the co-repressor complex upon HDAC1 SUMOylation as we hypothesized, it is conceivable that more 'free CREB' becomes available for DNA binding. This issue was examined here. Rats were divided to four groups and received the same plasmid transfections as described above. Their CA1 tissue was dissected out and the cell lysates were subjected to CREB DNA-binding assay. Results revealed that transfection of Flag-HDAC1WT plasmid decreased CREB DNA-binding compared with the control group. CREB DNA-binding was further reduced by Flag-HDAC1K444RK476R transfection compared with the Flag-HDAC1WT group, but it was enhanced by Flag-HDAC1-SUMO1 fusion plasmid transfection compared with the Flag-HDAC1WT group (Figure 4c). The quantified result is shown in Figure 4d. Cell lysates were immunoprecipitated and immunoblotted with anti-Flag antibody for verification of plasmid transfection and expression in the CA1 area. Western blot against CREB showed equal amount of CREB expression in the lysate of each sample (Figure 4c). These results revealed that HDAC1 SUMOylation increased CREB DNA-binding in the hippocampus, but it is not known whether HDAC1 SUMOylation enhances CREB binding to the DNA promoter directly. To examine this issue, we have conducted ChIP PCR to determine CREB binding to the Mcl-1 promoter in the hippocampus. Rats were divided to four groups and received the same plasmid transfections as described above. The cell lysates of their CA1 tissue were subjected to ChIP PCR assay. Results revealed endogenous CREB binding to the Mcl-1 promoter in the Flag-vector transfection group. Transfection of Flag-HDAC1WT decreased this binding and transfection of Flag-HDAC1K444RK476R further decreased it. But transfection of Flag-HDAC1-SUMO1 fusion plasmid increased CREB binding to the Mcl-1 promoter compared with that in the control group and Flag-HDAC1WT group (Figure 4e). Immunoprecipitation and immunoblotting with anti-Flag antibody revealed similar amount of plasmid transfection and expression in the CA1 area (Figure 4e, lower panel). These results together suggest that HDAC1 SUMOylation increases CREB binding to the Mcl-1 promoter in the CA1 area. Next we examined whether CREB regulates Mcl-1 expression in the hippocampus. Rats were divided to two groups and received control siRNA or CREB siRNA (8 pmol) transfections to their CA1 area. Animals were killed 48 later and their CA1 tissue was dissected out and subjected to western blot analysis for Mcl-1 and CREB expression. A representative gel pattern is shown in Figure 4f. Quantitative analyses of the results revealed that CREB siRNA transfection markedly decreased the expression of both Mcl-1 and CREB (Figure 4g).
The above results together indicated that CREB regulates Mcl-1 expression and HDAC1 SUMOylation enhances CREB binding to the Mcl-1 promoter in the hippocampus, but it is not known whether HDAC1 SUMOylation mediates Aβ induction of Mcl-1 expression. This issue was examined here. Rats were divided to five groups and received the following transfections and injections: Flag-vector+NH4OH (1%), Flag-vector+Aβ (14μg), Flag-HDAC1WT+Aβ (14 μg), Flag-HDAC1K444RK476R+Aβ (14μg) and Flag-HDAC1WT-SUMO1+Aβ (14 μg). The time interval between the two injections was 24 h and animals were killed 24 h after the second injection. Their brains were removed and their CA1 tissue was dissected out and subjected to western blot analysis for Mcl-1 expression. A representative gel pattern is shown in Figure 4h. Quantitative analyses of the results revealed that Aβ consistently increased Mcl-1 expression compared with the control group. Transfection of Flag-HDAC1WT plasmid-blocked Aβ induction of Mcl-1. Transfection of Flag-HDAC1 sumo-mutant plasmid further decreased Aβ induction of Mcl-1 compared with the Flag-HDAC1WT+Aβ group, but transfection of Flag-HDAC1WT-SUMO1 fusion plasmid enhanced Aβ induction of Mcl-1 expression compared with the Flag-HDAC1WT+Aβ group (Figure 4i). The cell lysates were immunoprecipitated and immunoblotted with anti-Flag antibody to confirm plasmid transfection and expression in the CA1 area (Figure 4h).
HDAC1 SUMOylation rescues synaptic and memory deficits in APP/PS1 mice
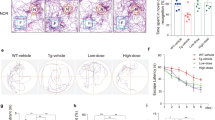
The above results demonstrated that HDAC1 SUMOylation allows more CREB to bind to the Mcl-1 promoter and mediates Aβ induction of Mcl-1 expression. In this series of experiments, we carried out functional assays to examine whether HDAC1 SUMOylation rescues synaptic deficit and memory impairment in APP/PS1 mice (9 months old). We first conducted the Morris water maze experiment. The APP/PS1 mice were divided to three groups and received lenti-vector transduction, lenti-Flag-HDAC1WT vector transduction and lenti-Flag-HDAC1WT-SUMO1 fusion vector transduction to their CA1 area. Another wild-type group of mice received lenti-vector transduction and served as the control. Water maze training started 10 days after lenti-vector transduction. Results revealed that APP/PS1 mice showed significant impairment in spatial acquisition compared with wild-type animals. APP/PS1 mice receiving lenti-Flag-HDAC1WT vector transduction showed similar spatial learning deficit as that of APP/PS1 mice, but transduction of lenti-Flag-HDAC1WT-SUMO1 fusion vector successfully rescued spatial learning deficit in APP/PS1 mice (Figure 5a). Similar results were obtained for the retention test (Figure 5b). We wished to also examine whether HDAC1 SUMOylation rescues memory deficit in a different learning task. Results revealed that the APP/PS1 mice receiving lenti-vector and lenti-Flag-HDAC1WT vector transduction both showed less freezing responses compared with wild-type animals on the retention measure of contextual fear conditioning learning task. This retention deficit in APP/PS1 mice was completely rescued by lenti-Flag-HDAC1WT-SUMO1 vector transduction (Supplementary Figure S5). We further examined HDAC1 SUMOylation level in these animals. Results revealed that HDAC1 SUMOylation was significantly decreased in APP/PS1 mice compared with wild-type mice, but lenti-Flag-HDAC1WT-SUMO1 transduction to APP/PS1 mice reversed this phenomenon compared with APP/PS1 mice. Overexpression of HDAC1WT has a similar effect as that of HDAC1WT-SUMO1 overexpression (Figure 5c). The quantified result is shown in Figure 5d. To verify that the above lenti-Flag-vectors were indeed transducted and expressed in the CA1 area, animals were killed and their CA1 tissue was subjected to immunoprecipitation with anti-Flag antibody and immunoblotting with anti-Flag antibody. Results revealed that the transducted HDAC1 vector is expressed in both lenti-Flag-HDAC1 overexpression groups as indicated by the apparent Flag-tagged bands, but for the lenti-Flag-HDAC1WT-SUMO1 overexpression group, the Flag-tagged band is shifted up, indicating the expression of sumoylated HDAC1 (Figure 5e). The above results showed that SUMOylation of HDAC1 rescued learning and memory deficits in APP/PS1 mice. Next we examined the effect of HDAC1 SUMOylation on synaptic plasticity in APP/PS1 mice by adopting the LTP paradigm. The hippocampal tissue slice of animals receiving the same lenti-vector transductions as described in Figure 5a were subjected to extracellular recording of field excitatory post-synaptic potential (fEPSP). Results revealed that the induction and expression of LTP was significantly impaired in the CA1 area of APP/PS1 mice compared with wild-type mice. Transduction of lenti-Flag-HDAC1WT vector in APP/PS1 mice more apparently impaired late phase LTP induction and LTP expression compared with wild-type mice. But transduction of lenti-Flag HDAC1WT-SUMO1 vector in APP/PS1 mice completely rescued the impairment in both LTP induction and expression in APP/PS1 mice (Figure 5f).
HDAC1 SUMOylation rescues synaptic and memory deficits in APP/PS1 mice. APP/PS1 mice (9 months old) receiving lenti-vector, lenti-Flag-HDAC1WT vector or lenti-Flag-HDAC1WT-SUMO1 vector, and WT mice receiving lenti-vector tranductions were subjected to (a) water maze learning (n=9–10 each group; F3,34=36, P<0.001) and (b) probe trial test (F3,34=12.36, P<0.01). (c) Hippocampal HDAC1 SUMOylation level was determined in these animals and (d) the results are quantified (n=3 each group; F3,8=14.15, P<0.01). (e) The CA1 tissue of animal from a and b was dissected out and the cell lysate was subjected to IP and IB with anti-Flag antibody for verification of lenti-Flag-HDAC1 transductions (n=2 each group). (f) Hippocampal tissue slice of animals receiving the same lenti-vector transductions as described in a were subjected to LTP recording under HFS paradigm (n=5 each group; F3,16=5.71, P<0.01). HFS, high-frequency stimulation. Lenti-F-HDAC1WT: Lenti-Flag-HDAC1WT. Data are mean±s.e.m. *P<0.05, **P<0.01 and #P<0.001
HDAC1 SUMOylation reduces amyloid plaque and apoptotic cells in APP/PS1 mice
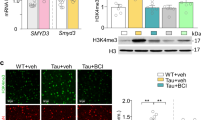
The brains of AD patients and in mouse model of AD are characterized by amyloid plaque accumulation followed by neuronal death. Next, we examined whether HDAC1 SUMOylation has a rescuing effect against these pathologies in APP/PS1 mice. Different APP/PS1 mice were used for this purpose. Antibody against Aβ was used to detect the presence of Aβ and thioflavin S staining was used to detect Aβ aggregation and amyloid plaque formation. Results from immunohistochemistry indicated the presence of Aβ (Figure 6a, left panel) and amyloid plaque (Figure 6a, middle panel) in the CA1 area of APP/PS1 mice, and these two images overlapped (Figure 6a, right panel). ProteoStat dye staining was also used to confirm the presence of amyloid plaque and its co-localization with Aβ in the same area (Figure 6b). Further immunohistochemical results of thioflavin-S staining revealed that transduction of lenti-Flag-HDAC1WT vector to APP/PS1 mice showed slightly higher amount of amyloid plaque, as indicated by the arrowheads, as that of APP/PS1 mice receiving lenti-vector transduction (Figure 6c, lower-left panel versus upper-right panel), but amyloid plaque was significantly reduced in APP/PS1 mice receiving lenti-Flag-HDAC1WT-SUMO1 vector transduction compared with APP/PS1 mice receiving lenti-vector transduction (Figure 6c, lower-right panel versus upper-right panel). The quantified result is shown in the right panel. Similar results were obtained when ProteoStat dye staining was used as a measure of amyloid plaque (Figure 6d). Moreover, lenti-Flag-HDAC1WT-SUMO1 vector transduction also showed a rescuing effect in reducing amyloid plaque in older APP/PS1 mice (12 months old), but this effect was less significant due to more amyloid plaque formation in animals at this age (Supplementary Figure S6).
HDAC1 SUMOylation reduces amyloid plaque and the number of apoptotic cells in APP/PS1 mice. APP/PS1 mice (9 months old) receiving lenti-vector, lenti-Flag-HDAC1WT vector or lenti-Flag-HDAC1WT-SUMO1 vector, and WT mice receiving lenti-vector tranductions were subjected to (a) immunohistochemistry with antibody against Aβ and thioflavin S staining were carried out in CA1 tissue of APP/PS1 mice (9 months old) (n=3). Scale bar, 100 μm. (b) Immunohistochemistry with antibody against Aβ and ProteoStat dye staining were carried out in the same mice (n=3). Scale bar, 100 μm. Animals receiving the same lenti-vector transductions described in Figure 5a were also subjected to (c) thioflavin S staining and quantified (n=4 each group; F3,12=80.81, P<0.001). Arrowheads indicate cells showing thioflavin S staining. Scale bar, 200 μm and (d) ProteoStat dye staining and quantified (n=4 each group; F3,12=201.15, P<0.001). Arrowheads indicate cells showing ProteoStat dye staining. Scale bar, 200 μm and (e) TUNEL staining and quantified (n=4 each group; F3,12=60.4, P<0.001). Cells in brown color indicated by arrowheads are the apoptotic cells. Scale bar, 50 μm. (f) Flag-vector, Flag-HDAC1WT plasmid, Flag-HDAC1 sumo-mutant plasmid or Flag-HDAC1-SUMO1 fusion plasmid was transfected to rat CA1 area and ChIP PCR for HDAC1 binding to the neprilysin promoter was determined. Plasmid transfection and expression was confirmed by immunoprecipitation and immunoblotting using anti-Flag antibody (lower panel). Experiments are in duplicates. Lenti-F-HDAC1WT: Lenti-Flag-HDAC1WT. Data are mean±s.e.m. #P<0.001
We next examined whether HDAC1 SUMOylation also reduces neuronal cell death in APP/PS1 mice. Separate animals were divided to the same groups as described above and TUNEL staining was used to measure cell apoptosis. Results revealed that the number of apoptotic cells was significantly higher in APP/PS1 mice receiving lenti-vector transduction compared with wild-type animals also receiving lenti-vector transduction. This measure was further slightly increased in APP/PS1 mice receiving lenti-Flag-HDAC1WT transduction compared with that of APP/PS1 mice receiving lenti-vector transduction. But the number of apoptotic cells was dramatically decreased in APP/PS1 mice receiving lenti-Flag-HDAC1WT-SUMO1 vector transduction compared with that of APP/PS1 mice receiving lenti-vector transduction (Figure 6e). In studying the mechanism of the rescuing effect of HDAC1 SUMOylation on neuronal death, we have examined the effect of HDAC1 SUMOylation on HDAC1 binding to the promoter of neprilysin, an Aβ-degrading enzyme, in the CA1 area using ChIP PCR assay. Results revealed that transfection of Flag-HDAC1WT plasmid to rat CA1 area increased HDAC1 binding to the neprilysin promoter compared with the Flag-vector transfected group. This effect was further enhanced upon Flag-HDAC1 sumo-mutant transfection, but it was apparently diminished upon Flag-HDAC1WT-SUMO1 fusion plasmid transfection (Figure 6f). Plasmid transfection and expression was confirmed by immunoprecipitation and immunoblotting using anti-Flag antibody (Figure 6f, lower panel).
Discussion
The present study examined the physiological role of HDAC1 SUMOylation in the hippocampus and the results revealed that HDAC1 SUMOylation functions as an endogenous defense mechanism protecting against Aβ-toxicity by releasing CREB from the HDAC1 repressor complex and by increasing CREB binding to the Mcl-1 promoter to enhance Mcl-1 expression. These results are consistent with the report that PIAS1 has an anti-apoptotic role.25 But they are incongruent with the studies showing that PIAS1 is pro-apoptotic.34, 35 One possible explanation for this discrepancy is that for the study showing an anti-apoptotic role of PIAS1, low concentration of PIAS1 plasmid DNA was transfected to HEK293T cells; however, for the studies showing a pro-apoptotic role of PIAS1, inducible PIAS1 expression was adopted. But other mechanisms may also account for this discrepancy. The present results are also consistent with the finding that Aβ activation of MAPK/ERK has a neuroprotective role13 and the notion that ERK1/2 activation acts as a defense mechanism against neuronal damage.36 This result is also congruent with the finding that PIAS1 expression is regulated by MAPK/ERK-mediated signaling.27 In addition, we have found that Aβ also activates Akt, and inhibition of PI-3 K signaling blocked Aβ induction of PIAS1 expression. These results suggest that Aβ activation of PI-3 K signaling also mediates neuroprotection against Aβ toxicity through enhanced PIAS1 expression. These results are consistent with the report that MAPK/ERK and PI3-K activation both mediate the neuroprotective effect of BDNF against glutamate neurotoxicity.37 Moreover, we have found that Aβ does not increase the expression of PIAS2, PIAS3 and PIAS4. This result indicated that Aβ specifically induces PIAS1 expression. But this result does not exclude the possibility that HDAC1 could also be SUMO-modified by PIAS2, PIAS3 or PIAS4. Whether PIAS2, PIAS3 and PIAS4 are induced by other stimuli that also enhance the SUMOylation of HDAC1 remain to be investigated.
The present result is consistent with the previous finding that HDAC1 is SUMO-modified at Lys-444 and Lys-476 in the cell.23 It is also consistent with the result from SUMO Software prediction because Lys-444 and Lys-476 both fit to the sequence of consensus SUMO substrate motif. But in our study, transfection of HDAC1K444RK476R did not completely block HDAC1 SUMOylation in HEK293T cells (Figure 3c). It is possible that there are more than two SUMO residues on HDAC1 in HEK293T cells. On the other hand, our result is incongruent with the finding that SUMO1 modification of HDAC1 promotes HDAC1 ubiquitination24 because blockade of HDAC1 SUMOylation did not affect HDAC1 ubiquitination in the brain. This discrepancy may be partially explained by different cells adopted in these studies and HDAC1 SUMOylation may produce more complicated effects in the brain than in the cell line. In addition, HDAC1 was found phosphorylated by protein kinase A and casein kinase II in vitro38 and HDAC1 phosphorylation at Ser-421 and Ser-423 promotes HDAC1 enzyme activity and complex formation.20 In examination of the relationship between HDAC1 SUMOylation and HDAC1 phosphorylation in the brain, our result showed that HDAC1 SUMOylation is not dependent on HDAC1 phosphorylation (Supplementary Figure S7). This result is consistent with an earlier report showing that overexpression of HDAC1 phosphorylation mutant does not affect HDAC1 SUMOylation in NIH 3T3 cells.22 This result suggests that HDAC1 phosphorylation and HDAC1 SUMOylation may be induced by different stimuli. In studying the molecular mechanism underlying the neuroprotective effect of HDAC1 SUMOylation, we have found that HDAC1 SUMOylation releases CREB from the repressor complex that allows more 'free CREB' to bind to the Mcl-1 promoter and to enhance Mcl-1 expression. These events result in decreased caspase 3 activity in Aβ-treated animals and decreased neuronal death in APP/PS1 mice. HDAC1 is known to interact with other transcription factors as well in addition to CREB, such as NF-κB39 and Sp1,40 and HDAC1 interaction with these transcription factors negatively regulates gene expression. SUMOylation of HDAC1 may also change its association with these transcription factors that leads to enhanced expression of certain anti-apoptotic genes and/or decreased expression of certain pro-apoptotic genes. HDAC1 SUMOylation also decreased the amount of amyloid plaque in APP/PS1 mice. HDAC1 is known to compete with amyloid precursor protein intracellular domain (AICD) for binding to the promoter of an Aβ degrading enzyme neprilysin41 and an Aβ clearance protein transthyretin.42 The transthyretin mRNA level is higher in APP695 cell line compared with that of control cell line due to higher expression level of AICD. But transthyretin mRNA expression is further increased in APP695 cells treated with the HDAC inhibitor valproic acid,42 suggesting that HDAC binding to the transthyretin promoter suppresses transthyretin expression. Our results revealed that HDAC1 sumo-mutant transfection increased, whereas HDAC1-SUMO1 fusion plasmid transfection decreased HDAC1 binding to the neprilysin promoter. These results suggest that sumoylated HDAC1 diminishes HDAC1 suppression of neprilysin expression. Upregulation of neprilysin consequently reduces Aβ accumulation and amyloid plaque formation. But other possibilities may also exist to account for the effect of HDAC1 SUMOylation in reducing amyloid plaque. In addition to HDAC1, other HDAC family proteins were found to be SUMO-modified as well. For example, the SUMO E3 ligase RanBP2 was found to promote the SUMOylation of HDAC4 and blockade of HDAC4 SUMOylation slightly impairs the transcriptional repression and enzyme activity of HDAC4.43 Our preliminary result showed that HDAC2 could also be SUMO-modified by PIAS1 in rat hippocampus and HDAC2 SUMOylation is increased after water maze training (Supplementary Figure S8). Because HDACs, particularly HDAC2, are involved in cognitive impairments under Aβ insult, in AD patients and in animal models of AD,18, 44, 45 it is worth examining whether SUMOylation of HDAC proteins other than HDAC1 is also involved in neuroprotection against Aβ toxicity. Moreover, because HDAC proteins are implicated in other brain disorders as well,46 it is worth to also examine the role of SUMOylation of other HDAC proteins possibly involved in these disorders.
In addition to HDAC1, PIAS1 is known to SUMO-modify many other proteins. We also examined whether the Aβ-dependent induction of PIAS1 affects the SUMOylation of other target proteins. We have chosen STAT1, p65 subunit of NF-κB, p53 and c-Myc as the alternative targets of PIAS1. The results revealed that Aβ induction of PIAS1 also increased the SUMOylation of STAT1 and p65 (Supplementary Figure S9a and S9b), but it did not affect the SUMOylation of p53 (Supplementary Figure S9c). c-Myc and c-Myc SUMOylation were not detected in the brain (Supplementary Figure S9d). STAT1 SUMOylation and NF-κB translocation to the nucleus were shown to facilitate spatial learning.47, 48 Whether SUMOylation of STAT1 and NF-κB protects against Aβ-toxicity requires further investigation.
Accumulative evidence has indicated that dysregulation of several sumoylated proteins is involved in the pathogenesis of AD.49 On the other hand, overexpression of SUMO3 in HEK293T cells was shown to reduce Aβ production whereas SUMO3 mutant transfection increases Aβ production.50 SUMOylation of APP was found to negatively regulate the level of Aβ aggregation and overexpression of Ubc9 and SUMO1 decreases Aβ aggregation in cells transfected with mutant APP.51 These results together suggest that up-regulated SUMOylation may be neuroprotective against AD. This speculation is supported by our finding that HDAC1 SUMOylation rescued the behavioral and synaptic deficits in APP/PS1 mice. It also reduced amyloid plaque and the number of apoptotic cells in APP/PS1 mice. But in another study, the global SUMO1 or SUMO2/3 conjugation levels were found unaltered in Tg2576 mice compared with wild-type mice.52 This latter result may not be inconsistent with the above results because SUMOylation of specific target proteins may be confounded by global SUMOylation. It is worth examining whether HDAC1 SUMOylation also protects against Aβ toxicity in different animal models of AD in future studies.
In examination of a few endogenous molecules, we have found that BDNF, IGF-1 and CRF all enhanced the SUMOylation of HDAC1. Previous report revealed that the neuroprotective effect of BDNF in animal models of AD is mediated through amyloid-independent mechanisms.29 The present result partially supports this notion by showing a novel protective mechanism of BDNF acting on enhanced SUMOylation of HDAC1. IGF-1 belongs to another family of growth factors that influence neuronal development and with its receptors enriched in the hippocampus.53 A previous study has shown that IGF-1 could protect and rescue hippocampal neurons against Aβ toxicity and amyloid-related neurotoxicity.30 In a more recent study, a human cortex-derived neural stem cell line modified to express IGF-1 was shown to exhibit neuroprotective capacity in vitro and persists in targeted brain regions in an animal model of AD.54 In studying the molecular mechanism of IGF-1, we have found that IGF-1 increased the SUMOylation of HDAC1 in rat hippocampus. This result suggests that HDAC1 SUMOylation is a novel mechanism underlying the neuroprotective action of IGF-1. Because we have demonstrated that Aβ induction of PIAS1 expression is also regulated through PI3-K signaling, this result is consistent with the notion that the neurotrophic effect of IGF-1 is mediated through PI3-K/Akt pathway upon activation of the IGF-1 receptors.55 Furthermore, the neuropeptide CRF was shown to induce synaptic plasticity and enhance cognitive function in animals and the memory-enhancing effect of CRF is mediated, at least in part, through increased Bdnf gene expression.31, 56 Here we have demonstrated that CRF-enhanced HDAC1 SUMOylation and this may provide a novel molecular mechanism of CRF. In a recent study, we have found that IGF-1 and CRF both enhance the SUMOylation of MeCP2 that results in alleviation of Rett syndrome in Mecp2 conditional knockout mice.57 The present result implicates that IGF-1 and CRF may also alleviate the pathology of AD through enhanced SUMOylation of HDAC1. But dexamethasone, a synthetic glucocorticoid that binds to the glucocorticoid receptor, did not affect HDAC1 SUMOylation. This result suggests that different steroid hormones may have different mechanisms in regulation of protein SUMOylation.
In summary, we have presently found that HDAC1 could be SUMO-modified by PIAS1 at Lys-444 and Lys-476. Acute Aβ-treatment significantly increases the level of HDAC1 SUMOylation in the hippocampus through MAPK/ERK-mediated signaling at 1 h, but not 14 days later. Enhanced HDAC1 SUMOylation decreases its association with CREB, increases CREB binding to the Mcl-1 promoter and increases Mcl-1 expression. HDAC1 SUMOylation also decreases HDAC1 binding to the neprilysin promoter (Figure 7). These mechanisms together reduce the number of apoptotic cells and the amount of amyloid plaque in the CA1 area of APP/PS1 mice. These mechanisms also rescue the cognitive impairment and synaptic deficit in APP/PS1 mice. Thus, HDAC1 SUMOylation functions as an endogenous defense mechanism protecting against Aβ-toxicity. Stimuli such as BDNF, IGF-1 and CRF that increase the level of HDAC1 SUMOylation without altering the HDAC1 expression level may serve as an alternative therapeutic strategy against AD.
An illustration shows the relationship among Aβ, HDAC1 SUMOylation, HDAC1 association with CREB and other proteins in the repressor complex, CREB binding to the Mcl-1 promoter, Mcl-1 gene expression and HDAC1 binding to the neprilysin promoter in the hippocampus. This illustration also indicates that a few endogenous stimuli could activate HDAC1 SUMOylation in the brain
Materials and methods
Animals
Adult male Sprague-Dawley rats (250–300 g) and adult male APP/PS1 transgenic mice were used in this study. The APP/PS1 mice were purchased from Jackson Laboratory (Bar Harbor, ME, USA) (strain name: B6.Cg-Tg (APPswe,PSEN1dE9)85Dbo/Mmjax, stock number: 005864). They were all bred at the Animal Facility of the Institute of Biomedical Sciences (IBMS), Academia Sinica in Taiwan. All the animals were housed and maintained on a 12/12 h light/dark cycle (light on at 0630 hours) with food and water continuously available. Experimental procedures followed the Guidelines of Animal Use and Care of the National Institute of Health and were approved by the Animal Committee of IBMS, Academia Sinica.
Hippocampal lysate and cell lysate preparation
Animals were killed by decapitation, and their hippocampal tissue was dissected out. Rat hippocampal tissue and HEK293T cells were lysed by brief sonication in lysis buffer containing 50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 2 mM EDTA and 1% IGEPAL CA-630. One tablet of protease inhibitor cocktail (Catalog No. 05892791001, cOmplete ULTRA Tablets, Mini, EDTA-free, EASYpack, Roche, Mannheim, Germany) and one tablet of phosphatase inhibitor (Catalog No. 04906837001, PhosSTOP, Roche) were added to each 10 ml of the lysis buffer.
Immunoprecipitation (IP) and western blot
For IP PIAS1, HDAC1, HDAC2 and Flag, the clarified lysate (0.5 mg) was immunoprecipitated with 3 μl of rabbit anti-PIAS1 antibody (Catalog No. 2474-1, Epitomics, Burlingame, CA, USA), 3 μl of mouse anti-HDAC1 antibody (Catalog No. 5356, Cell Signaling, Danvers, MA, USA) or 3 μl of rabbit anti-HDAC1 N-terminal antibody (Catalog No. A0238, ABclonal, College Park, MD, USA), 3 μl of mouse anti-HDAC2 antibody (Catalog No. 5113, Cell Signaling) and 3 μl of mouse anti-Flag M2 antibody (Catalog No. F1804, Sigma-Aldrich, St. Louis, MO, USA) at 4 °C for overnight. Three microliters of rabbit or mouse IgG was used in the control group. The protein A or G magnetic beads (30 μl, 50% slurry, GE Healthcare, Barrington, IL, USA) were added to the IP reaction product to catch the immune complex at 4 °C for 3 h. The immune complex on beads were washed three times with washing buffer containing 20 mM HEPES (pH 7.4), 150 mM NaCl, 1 mM EDTA, 1% IGEPAL CA-630, 1 mM DTT, 50 mM β-glycerophosphate, 50 mM NaF, 10 mg/ml PMSF, 4 μg/ml aprotinin, 4 μg/ml leupeptin and 4 μg/ml pepstatin before boiling at 95 °C for 10 min and subjected to 8%, 10% or 15% SDS-PAGE followed by transferring onto the PVDF membrane (Millipore, Bedford, MA, USA). Western blot was conducted using the following antibodies: rabbit anti-PIAS1 (1:10 000, Catalog No. 2474-1, Epitomics), rabbit anti-PIAS2 (1:1000, Catalog No. ab58404, Abcam, Cambridge, UK), rabbit anti-PIAS3 (1:1000, Catalog No. 4164, Cell Signaling), rabbit anti-PIAS4 (1:1000, Catalog No. 4392, Cell Signaling), rabbit anti-MAPK (ERK1/2) (1:5000, Catalog No. 4695, Cell Signaling), rabbit anti-phospho-MAPK (pERK1/2) (1:5000, Catalog No. 4376, Cell Signaling), rabbit anti-Akt (1:2000, Catalog NO. 9272, Cell Signaling), rabbit anti-phospho308-Akt (1:2000, Catalog No. 4056, Cell Signaling), rabbit anti-Mcl-1 (1:500, Catalog No. sc-819, Santa Cruz Biotechnology, Dallas, TX, USA), mouse anti-HDAC1 (1:1000, Catalog No. 5356S, Cell Signaling), rabbit anti-HDAC1 N-terminal (1:1000, Catalog No. A0238, ABclonal), rabbit anti-CREB (1:1000, Catalog No. 9197, Cell Signaling), rabbit anti-SUMO1 (1:4000, Catalog No. 40120 SUMOlink kit, Active Motif, Carlsbad, CA, USA), mouse anti-Myc (1:5000, Catalog No. 05-419, Millipore), mouse anti-GFP (1:5000, Catalog No. 11814460001, Roche), mouse anti-Flag M2 (1:5000, Catalog No. F1804, Sigma-Aldrich), mouse anti-His (1:5000, Catalog No. OB05, Millipore) and mouse anti-actin (1:200000, Catalog No. MAB1501, Millipore) antibodies. The secondary antibody used was HRP-conjugated goat-anti-rabbit IgG antibody or goat-anti-mouse IgG antibody (1:6500, Catalog No. 111-035-003 or 115-035-003, Jackson ImmunoResearch). Membrane was developed by reacting with chemiluminescence HRP substrate (Millipore) and exposed to the LAS-3000 image system (Fujifilm, Tokyo, Japan) for visualization of protein bands. The protein bands were quantified using the NIH Image J Software (National Institute of Health, MD, USA).
Plasmid construction, cell culture and DNA transfection
For construction of the Flag-tagged Hdac1 plasmid, full-length Hdac1 was cloned by amplifying the rat hippocampal Hdac1 cDNA (accession # NM_001025409) with primers 5′-ATCGGGATCCATGGCGCAGACTCAGGGC-3′ (forward) and 5′-ATCGCTCGAGTCAGGCCATCTTGACCTCTTC-3′ (reverse). The PCR product was sub-cloned between the BamHI and XhoI sites of the mammalian expression vector pCMVTag2B. Flag-tagged Hdac1 mutant plasmids were generated using the QuickChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA, USA). For construction of the EGFP-tagged pias1 and His-tagged ubiquitin plasmids, the procedure used was the same as that described previously.25 For construction of the Myc-tagged sumo1 plasmid, the procedure used was the same as that described elsewhere.57 For construction of the Flag-Mcl-1 plasmid, the procedure used was the same as that described previously.13 For construction of the Flag-tagged Hdac1-sumo1 fusion plasmid, the previously cloned Flag-tagged Hdac1 plasmid and Myc-tagged sumo1 plasmid were used as templates and the Hdac1 sequence was amplified with primers 5′-ATCGGGATCCATGGCGCAGACTCAGGGCA-3′ (forward) and 5′-ATCGGATATCGGCCATCTTGACCTCTTCT-3′ (reverse). The sumo1 sequence was amplified with primers 5′-ATCGGTCGACATGTCTGACCAGGAGGCAA-3′ (forward) and 5′-ATCGGGGCCCCTAAACCGTCGAGTGACCC-3′ (reverse). The Hdac1 PCR product was sub-cloned between the BamHI and EcoRV sites of the mammalian expression vector pCMVTag2B. The sumo1 PCR product was sub-cloned between the SalI and ApaI sites downstream of the Hdac1 sequence from the previously cloned Flag-tagged Hdac1 plasmid. HEK293T cells were maintained in Dulbecco’s modified Eagle’s medium containing 10% fetal bovine serum and incubated at 37 °C in a humidified atmosphere with 5% CO2. Plasmid transfection was made by using the Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA, USA) in 12-well culture plates according to the manufacturer’s instructions. IP and western blot were conducted 48 h after plasmid transfection.
Lentiviral vector construction and preparation
For construction of the pLenti-Tri-cistronic-GFP lentiviral vector, the GFP construct was cloned by amplifying the GFP gene from pLenti-CMV-GFP-2A-Puro-Blank (ABM, Richmond, BC, Canada) and sub-cloned into the pLenti-Tri-cistronic lentiviral vector (ABM) between the ScaI and KpnI sites downstream of the PGK promoter. The primers used for GFP vector were 5′-ATCGAGTACTGCCACCATGGAGATCGAGTGCCGCATC-3′ (forward) and 5′-ATCGGGTACCGGCGAAGGCGATGGGGGTC-3′ (reverse). For construction of the pLenti-Flag-tagged Hdac1WT GFP and pLenti-Flag-tagged Hdac1WT-sumo1 GFP lentivitral vectors, full-length Hdac1 and Hdac1-sumo1 fusion sequences were sub-cloned into the pLenti-Tri-cistronic-GFP lentiviral vector by amplifying the rat Flag-Hdac1 plasmid and rat Flag-Hdac1-sumo1 fusion plasmid, respectively, with different primers. The forward primer used for obtaining the rat Flag-tagged Hdac1 lentivitral vector was: 5′-TCGCCCGGGGCCACCATGGATTACAAGGATGACGACGATAAGATGGCGCAGACTCAGGGC-3′ and the reverse primer used was: 5′-ATCGCCTAGGGGCCATCTTGACCTCTTCT-3′. The PCR product was sub-cloned between the XmaI and AvrII sites of the pLenti-Tri-cistronic-GFP lentiviral vector downstream of the mini CMV promoter. The forward primer used for obtaining the rat Flag-tagged Hdac1-sumo1 lentivitral vector was: 5′-TCGCCCGGGGCCACCATGGATTACAAGGATGACGACGATAAGATGGCGCAGACTCAGGGC-3′ and the reverse primer used was: 5′-ATCGCCTAGGCTAAACCGTCGAGTGACCC-3′. The PCR product was sub-cloned between the XmaI and AvrII sites of the pLenti-Tri-cistronic-GFP lentiviral vector downstream of the mini CMV promoter. For lentivirus packaging, HEK293LTV cells (Cell Biolabs, San Diego, CA, USA) were transfected with 1.5 μg of psPAX2 (Addgene plasmid #12260), 0.5 μg of pMD2.G (Addgene plasmid #12259), 2 μg of pLenti-Flag-tagged Hdac1WT GFP or 2 μg of pLenti-Flag-tagged Hdac1WT-sumo1 GFP or 2 μg of pLenti-Tri-cistronic-GFP lentiviral vector coding for GFP as control using 10 μl of Lipofectamine 2000 (Invitrogen) in six-well cell culture dish. Lentiviral particles were collected using the speedy lentivirus purification solution (ABM) according to the manufacturer's protocols. Cell culture medium containing lentiviral particles can be harvested for two to three times at 12 h interval until 36 h after transfection, and it was kept at 4 °C for the collecting period. The collected culture medium was further clarified by centrifugation at 2500 × g for 10 min and filtrated through a 0.45 μm syringe filter. The speedy lentivirus purification solution (ABM) was added into filtrated supernatant (1:9, v/v) containing lentiviral particles and mixed thoroughly by inversion. The lentiviral supernatant was centrifuged at 5000 × g at 4 °C for 10 min. Supernatant was then discarded and the viral pellet was re-suspended in ice-cold PBS. After titration, the viral stock was stored at −80 °C in aliquots. The lentivirus titer was determined by lentivirus qPCR Titer Kit (ABM) according to the manufacturer's protocols. The titer of the pLenti-Flag-tagged Hdac1WT GFP lenti-vector and pLenti-Flag-tagged Hdac1WT-sumo1 GFP lenti-vector transducted to mice was 1 × 108 IU/ml.
In vitro SUMOylation assay for recombinant HDAC1 protein
The in vitro SUMOylation assay was performed in a total volume of 20 μl containing 3 μg of His-tagged recombinant HDAC1 protein (Catalog No. BML-SE456-0050, Enzo Life Sciences, Farmingdale, NY, USA), 3 μl of GST-tagged recombinant PIAS1 protein (Catalog No. BML-UW9960, Enzo Life Sciences), 1 μg of GST-tagged recombinant SENP1 enzyme (Catalog No. BML-UW9760-0100, Enzo Life Sciences), 1 μl of E1 activating enzyme, 1 μl of E2 conjugating enzyme and 0.5 μl of SUMO1 protein in SUMOylation buffer provided in the SUMO link kit. In vitro SUMOylation assay was performed by using the SUMO link kit according to the manufacturer’s instructions (Active Motif). Reactions were incubated at 30 °C for 3 h, stopped by adding sample buffer, and products were examined by SDS-PAGE separation.
HDAC1 SUMOylation in the brain
Hippocampal CA1 tissue lysate was prepared in the same way as that prepared for western blot analysis. For IP HDAC1 and HDAC2, the clarified lysate (0.5 mg) was immunoprecipitated with 3 μl of mouse anti-HDAC1 antibody (Catalog No. 5356, Cell Signaling) or 3 μl of rabbit anti-HDAC1 N-terminal antibody (Catalog No. A0238, ABclonal), or 3 μl of mouse anti-HDAC2 antibody (Catalog No. 5113, Cell Signaling) at 4 °C overnight. The protein A or G magnetic beads (30 μl, 50% slurry, GE Healthcare) were added to the IP reaction product to catch the immune complex at 4° C for 3 h. The immune complex on beads were washed three times with washing buffer containing 20 mM HEPES (pH 7.4), 150 mM NaCl, 1 mM EDTA, 1% IGEPAL CA-630, 1 mM DTT, 50 mM β-glycerophosphate, 50 mM NaF, 10 mg/ml PMSF, 4 mg/ml aprotinin, 4 mg/ml leupeptin and 4 mg/ml pepstatin and subjected to SUMOylation reaction with the addition of recombinant PIAS1 protein (3 μl, Catalog No. BML-UW9960, Enzo Life Sciences), E1 (1 μl), E2 (1 μl) and the SUMO1 (0.5 μl) proteins provided in the kit. SUMOylation assay was performed using the SUMO link kit according to the manufacturer’s instructions (Active Motif, Carlsbad, CA, USA) and boiled in Laemmli sample buffer at 95 °C for 10 min. The SUMOylation reaction product was subjected to 8% SDS-PAGE followed by transferring onto the PVDF membrane (Millipore). The membrane was immunoblotted with anti-HDAC1 antibody (1:1000, Catalog No. 5356, Cell Signaling), anti-HDAC1 N-terminal antibody (1:1000, Catalog No. A0238, ABclonal), anti-HDAC2 antibody (1:1000, Catalog No. 5113, Cell Signaling) or anti-SUMO1 antibody (1:4000, Catalog No. 40120, Active Motif). For determination of endogenous HDAC1 SUMOylation after PIAS1 siRNA transfection, no E1, E2, SUMO1 and PIAS1 proteins were added to the IP reaction product. The remaining procedures were the same as that described above.
HDAC1 SUMOylation in HEK293T cells
For HDAC1 SUMOylation determination in HEK293T cells, different Flag-tagged HDAC1 plasmids were co-transfected with EGFP-PIAS1 and Myc-SUMO1 plasmids to HEK293T cells. Forty-eight hours later, the cell lysate was subjected to western blot analyses by using anti-Flag (1:5000, Catalog No. F1804, Sigma-Aldrich) and anti-SUMO1 (1:4000, Catalog No. 40120 SUMOlink kit, Active Motif) antibodies.
Biotinylated oligonucleotides pull-down assay for CREB DNA-binding activity
DNA oligonucleotides containing two CRE elements (underlined) (5′-AGAGATTGCCTGACGTCAGAGAGCTAGGATTGCCTGACGTCAGAGAGCTAG-3′ for the sense strand and 5′-CTAGCTCTCTGACGTCAGGCAATCCTAGCTCTCTGACGTCAGGCAATCTCT-3′ for the antisense strand) were conjugated with a 5′-biotin on the sense strand according to the method described elsewhere.58 Both complementary oligonucleotides were re-suspended in the annealing buffer (10 mM Tris (pH 8.0), 50 mM NaCl, 1 mM EDTA). For annealing the sense and antisense oligonucleotides, 10 μl each of the complementary oligonucleotides together with 80 μl of the annealing buffer were mixed in a 0.5 ml microtube and the tube was placed in a heating block at 90 °C. The heating block was allowed to gradually cool down to room temperature and stored on ice or at −20 °C until use. For the CREB pull-down assay, the clarified hippocampal CA1 tissue lysate (0.4 mg) was added with 6 μl duplex oligonucleotides (100 μM) and poly dI-dC (1 μg/ml, GE Healthcare) at 4 °C for overnight. The streptavidin agarose beads (10 μl, Sigma-Aldrich) were added to the pull-down reaction product to catch the CREB-DNA oligonucleotide complex at 4 °C for 3 h. The pull-down reaction complex on beads was then washed three times with PBS and boiled in Laemmli sample buffer at 95 °C for 10 min. For analysis of CREB DNA-binding activity, the pull-down assay product was subjected to 8% SDS-PAGE followed by transferring onto the PVDF membrane and immunoblotted with anti-CREB antibody (1:2000, Catalog No. 9197, Cell Signaling).
Chromatin immunoprecipitation (ChIP) assay
ChIP assay was performed according to the protocol of Millipore ChIP assay kit (Catalog No. 17-10085). For plasmid DNA transfection, 0.7 μl plasmid DNA complex (1.5 μg/μl) was injected to the rat CA1 area bilaterally 48 h before killing. CA1 tissues were washed using × 1 ice-cold PBS and fixed with 1% formaldehyde by adding formaldehyde to the × 1 ice-cold PBS for 10 min. After adding glycine to quench the un-reacted formaldehyde, tissue were homogenized and re-suspended in cell lysis buffer plus protease inhibitor cocktail II, then changed to nuclear lysis buffer plus protease inhibitor cocktail II for sonication. The chromatin was immunoprecipitated using rabbit anti-CREB antibody (Cell Signaling) and rabbit anti-HDAC1 N-terminal antibody (ABclonal). DNA purified from the immunoprecipitated samples was subjected to PCR reaction. The forward primer for the Mcl-1 promoter is: 5′-CGGAAGAGCCACGGAGTGG-3′ (nucleotide −200 to −182) and the reverse primer for the Mcl-1 promoter is: 5′-CCAGACTCGAGGCAGGCG-3′ (nucleotide −68 to −51). The forward primer for the neprilysin promoter is: 5′-GACACGGTTTTCATTGTCCA-3′ (nucleotide −1264 to −1245) and the reverse primer for the neprilysin promoter is: 5′-TGAGACTCAGCAGGCAGGTA-3′ (nucleotide −1107 to −1088). The PCR product for the Mcl-1 promoter is 150 bps and that for the neprilysin promoter is 177 bps in length. The PCR products were separated by agarose gel electrophoresis.
Extracellular field potentiation recording
APP/PS1 mice transducted with lenti-vector, lenti-Flag-HDAC1WT vector or lenti-Flag-HDAC1WT-SUMO1 vector in their CA1 area were subjected to electrophysiological recording. Wild-type mice transducted with lenti-vector served as the control. Animals were sacrificed and their brain slices were transferred to an immersion-type recording chamber, perfused with ACSF containing 100 μM picrotoxin at a rate of 2 ml/min at room temperature. An incision was made between the CA1 and CA3 areas to remove afferent input from CA3. For extracellular field potential recording, a glass pipette filled with 3 M NaCl was positioned in the CA1 stratum radiatum area to record fEPSP. Bipolar stainless steel stimulating electrodes (Frederick Haer Company, Bowdoin, ME, USA) were placed in the striatum radiatum to stimulate the Schaffer collateral pathway. Stable baseline fEPSP activity was recorded by applying a short-duration current stimulation pulse (~40 μs) at a predetermined intensity every 15 s for 20 min. LTP was induced using the high-frequency stimulation paradigm by delivering three 100 Hz tetani (1 s) with an inter-tetanus interval of 60 s.
Drugs
U0126 was purchased from Millipore (Catalog No. 662005). Aβ (1-42) and reverse Aβ (42-1) were purchased from Anaspec (Catalog No. AS-20276 and AS-27275, Fremont, CA, USA). BDNF was purchased from PeproTech (Catalog No. 450-02, Rocky Hill, NJ, USA). Wortmannin, IGF-1, CRF and dexamethasone were purchased from Sigma-Aldrich. IGF-1 and CRF were dissolved in PBS immediately before use. U0126, wortmannin and dexamethasone was dissolved in 15% DMSO and diluted with PBS before injection. BDNF was dissolved in PBS. Aβ and reverse Aβ were dissolved in 1% NH4OH before injection.
Intra-hippocampal drug infusion, plasmid DNA transfection and siRNA injection
Rats were anesthetized with pentobarbital (40 mg/kg) and subjected to stereotaxic surgery. Two 23-gauge, stainless steel, thin-wall cannulae were implanted bilaterally to the CA1 area of rat brain at the following coordinates: 3.5 mm posterior to the bregma, ±2.5 mm lateral to the midline and 3.4 mm ventral to the skull surface. After recovery from the surgery, Aβ (20 μg/μl), U0126 (1 μg/μl), wortmannin (1 μg/μl), BDNF (1 μg/μl), IGF-1 (100 ng/ml), CRF (100 ng/μl) and dexamethasone (30 ng/μl) were directly injected to the CA1 area at a rate of 0.1 μl/min. A total of 0.7 μl was injected to each side. For transient Hdac plasmid DNA transfection, 0.7 μl plasmid DNA complex (1.5 μg/μl) was injected directly to CA1 area bilaterally in the rat brain using the non-viral transfection agent polyethyleneimine (PEI) which we have previously demonstrated that it does not produce toxicity to hippocampal neurons.59 Before injection, plasmid DNA was diluted in 5% glucose to a stock concentration of 2.77 μg/μl. Branched PEI of 25 kDa (Sigma) was diluted to 0.1 M concentration in 5% glucose and added to the DNA solution. Immediately before injection, 0.1 M PEI was added to reach a ratio of PEI nitrogen per DNA phosphate equals to 10. The mixture was subjected to vortex for 30 s and allowed to equilibrate for 15 min. For siRNA injection, 0.7 μl of PIAS1 siRNA (8 pmol), CREB siRNA (8 pmol) or control siRNA was transfected to CA1 area bilaterally in the rat brain also using the transfection agent PEI. The sense and antisense sequences used for PIAS1 siRNA were adopted from that of a previous study.47 The sequence for PIAS1 siRNA sense strand is: 5′-UCCGGAUCAUUCUAGAGCUtt-3′ and that for PIAS1 siRNA antisense strand is: 5′-AGCUCUAGAAUGAUCCGGAtt-3′. The sequence for CREB siRNA sense strand is: 5′-GCACUUAAGGACCUUUACUtt-3′ and that for CREB siRNA antisense strand is: 5′- AGUAAAGGUCCUUAAGUGCtt-3′.The Silencer Negative Control number 1 siRNA was used as a control. They were all synthesized from Ambion, Thermo Fisher Scientific (Waltham, MA, USA). The inner diameter of the injection needle is 0.31 mm and the wall thickness of the injection needle is 0.12 mm. The injection needle was left in place for 5 min to limit the diffusion of injected agent. Animals were sacrificed 45 min after IGF-1 injection and 1 h after CRF, dexamethasone and BDNF injection. Animals were killed 48 h after plasmid and siRNA transfection and they were killed at different time points after Aβ injection. Their brains were removed and cut by a brain slicer. Their CA1 tissue was further punched out by using a stainless punch with 2 mm inner diameter. Tissues were frozen at −80 °C until biochemical experimentation.
Water maze learning
The water maze used was a plastic, circular pool, 1.2 m in diameter and 25 cm in height that was filled with water (25±2 °C) to a depth of 16 cm. A circular platform of 8 cm in diameter was placed at a specific location away from the edge of the pool. The top of the platform was submerged 0.6 cm below the water surface. Water was made cloudy by adding milk powder. Distinctive, visual cues were set on the wall.
For spatial learning, animals were subjected to three trials a day with one given early in the morning, one given in the early afternoon and another one given in the late afternoon. The training procedure lasted for 5 days and a total of 15 trials were given. For these trials, animals were placed at different starting positions spaced equally around the perimeter of the pool in a random order. Animals were given 60 s to find the platform. If an animal could not find the platform, it was guided to the platform and was allowed to stay on the platform for 20 s. The time that each animal took to reach the platform was recorded as the escape latency. The total distance that animals traveled in the target region as well as their swim speed was also recorded. A probe trial of 60 s was given on day 6 to test their memory retention. Animals were placed in the pool with the platform been removed and the time they spent in each quadrant (target quadrant, left quadrant, opposite quadrant and right quadrant) was recorded.
Contextual fear conditioning learning
Fear conditioning learning was preformed 7–10 days after the probe trial test. One day before conditioning, mice were placed in the conditioning chamber (46 × 30 × 46 cm, L × W × H) for 5 min for adaptation. Twenty-four hours later, these animals were placed into the same chamber for fear conditioning training. After 3 min of free exploration, they were trained with 5 foot shocks (0.5 mA, 1 s) randomly during the following 90 s. At the end of the footshock, immediate freezing response was measured and calculated as the percentage of time that animals spent freezing during the 30 s period. Twenty-four hours later, these animals were placed into the same chamber (but no footshock was delivered) for the retention test. Freezing response was measured and calculated as the percentage of time that animals spent freezing during the 5-min recording period.
Immunohistochemistry
For immunohistochemical staining of PIAS1 and HDAC1 in CA1 area of the rat brain, rats were anesthetized with pentobarbital (100 mg/kg, i.p.) and perfused with ice-cold phosphate-buffered saline followed by 4% paraformaldehyde. Brains were removed and post fixed in 30% sucrose/4% paraformaldehyde solution for 20–48 h. Brains were then frozen, cut into 30-μm sections on a cryostat and mounted on gelatin-coated slides. Brain sections were rinsed with 1 × PBS for 10 min and antigen was retrieved with 0.1 M citric acid/0.1 M sodium citrate buffer at 95 °C for 45 min followed by 1 × PBS for 10 min for three times. The sections were pre-incubated in a blocking solution containing 3% normal goat serum, 3% BSA and 0.5% Triton X-100 in 1 × PBS for 1 h. For visualization of endogenous PIAS1 and HDAC1 in hippocampal CA1 neurons, brain sections were incubated with rabbit anti-PIAS1 antibody (1:200, Catalog No. 2474-1, Epitomics) and mouse anti-HDAC1 antibody (1:200, Catalog No. 5356, Cell Signaling) at 4 °C overnight. Brain sections were then washed with 1 × PBS for 10 min for three times and then incubated with goat anti-rabbit secondary antibody conjugated with Alexa Fluor 488 (1:500, Catalog No. 111-545-003, Jackson ImmunoResearch, West Grove, PA, USA) and Dylight 549-conjugated goat anti-mouse antibody (1:500, Catalog No. 115-505-003, Jackson ImmunoResearch) for 1 h and then washed with 1 × PBS for 10 min for three times. For immunofluorescence detection of the nucleus, tissue sections were added with 20 μl of the DAPI Fluoromount-G mounting medium (SouthernBiotech, Birmingham, AL, USA). Photomicrographs were taken using a Zeiss LSM700 Stage confocal microscope (Carl Zeiss, Oberkochen, Germany).
For immunofluorescence detection of amyloid plaque in the CA1 area, mice were anesthetized with pentobarbital (100 mg/kg, i.p.) followed by the same procedures described above. For visualization of endogenously expressed amyloid plaque in APP/PS1 mice, brain sections containing the CA1 region were incubated with mouse anti-human Aβ antibody (1:250, Catalog No. sc-58508, Santa Cruz Biotechnology) at 4 °C overnight. Brain sections were then washed with 1 × PBS for 10 min for three times and incubated with Dylight 549-conjugated goat anti-mouse antibody (1:500, Catalog No. 115-505-003, Jackson ImmunoResearch) or FITC-conjugated goat anti-mouse antibody (1:500, Catalog No. 115-095-062, Jackson ImmunoResearch) for 1 h and further washed with 1 × PBS for 10 min for three times. The ProteoStat Amyloid Plaque Detection Kit (Enzo Life Sciences) was then used to detect amyloid plaque formation. To verify that the endogenously expressed Aβ forms aggregates, brain sections containing the CA1 region were subjected to 0.01% thioflavin S staining (Sigma-Aldrich) for 15 min and washed with 1 × PBS for 10 min for three times. These sections were then mounted with 20 ml of the DAPI Fluoromount-G mounting medium (SouthernBiotech). Photomicrographs were taken using a Zeiss LSM700 Stage confocal microscope. The number of plaques showing thioflavin-S staining and ProteoStat dye were counted separately in an area of 1.28 mm2.
Caspase 3 activity assay
Caspase 3 activity was performed according to the protocol of Caspase 3 assay kit from Abcam (Catalog No. ab39383). The hippocampal tissue was homogenized in cell lysis buffer to lyse for 10 min. Tissue lysate was mixed with reaction buffer containing DTT and the DEVD-AFC substrate at 37 °C for 2.5 h. The samples were read in a Gemini EM fluorescence microplate reader system (Device, Sunnyvale, CA, USA) using a filter with the excitation wavelength set at 400 nm and emission wavelength set at 505 nm.
TUNEL assay
TUNEL assay was performed according to the protocol of ApopTag Plus Peroxidase In Situ Apoptosis Kit to detect the apoptotic cell (Catalog No. S7101, Millipore). As mentioned previously, brain sections were fixed in 4% paraformaldehyde and retrieved with 0.1 M citric acid/0.1 M sodium citrate buffer at 95 °C for 15 min followed by proteinase K (20 μg/ml) for 10 min at room temperature. Brain sections were permeabilized with pre-cooled ethanol/acetic acid (2:1) for 5 min at −20 °C, followed by reacting with 3% H2O2 for 5 min at room temperature to quench the endogenous peroxidase. Brain sections were then incubated with the TdT enzyme for 1 h at 37 °C followed by incubation with anti-digoxigenin peroxidase conjugate for 30 min at room temperature. Apoptotic nuclei developed brown color with DAB peroxidase substrate. The slides were then counterstained with 0.5% (w:v) methyl green for visualization of both non-apoptotic and apoptotic cells. The number of apoptotic cells was counted separately by using a Leica DM IL LED light microscope in an area of 653 × 489 μm2 from a given hippocampal tissue section.
Statistical analysis
Behavioral data were analyzed with one-way or two-way analysis of variance (ANOVA) with repeated measure followed by post-hoc Newman–Keuls multiple comparisons (represented by q-value). Biochemical data were analyzed with the Student’s t-test or one-way ANOVA followed by Newman–Keuls comparisons. Electrophysiological data were analyzed with two-way ANOVA with repeated measure followed by Newman–Keuls comparisons. Values of P<0.05 were considered statistically significant (*P<0.05, **P<0.01, #P<0.001).
References
Butterfield DA, Drake J, Pocernich C, Castegna A . Evidence of oxidative damage in Alzheimer's disease brain: central role for amyloid beta-peptide. Trends Mol Med 2001; 7: 548–554.
Dickson DW . Apoptotic mechanisms in Alzheimer neurofibrillary degeneration: cause or effect? J Clin Invest 2004; 114: 23–27.
Hardy J, Selkoe DJ . The amyloid hypothesis of Alzheimer's disease: progress and problems on the road to therapeutics. Science 2002; 297: 353–356.
Chen G, Chen KS, Knox J, Inglis J, Bernard A, Martin SJ et al. A learning deficit related to age and beta-amyloid plaques in a mouse model of Alzheimer's disease. Nature 2000; 408: 975–979.
Cleary JP, Walsh DM, Hofmeister JJ, Shankar GM, Kuskowski MA, Selkoe DJ et al. Natural oligomers of the amyloid-beta protein specifically disrupt cognitive function. Nat Neurosci 2005; 8: 79–84.
Hsiao K, Chapman P, Nilsen S, Eckman C, Harigaya Y, Younkin S et al. Correlative memory deficits, Abeta elevation, and amyloid plaques in transgenic mice. Science 1996; 274: 99–102.
Hsia AY, Masliah E, McConlogue L, Yu GQ, Tatsuno G, Hu K et al. Plaque-independent disruption of neural circuits in Alzheimer's disease mouse models. Proc Natl Acad Sci USA 1999; 96: 3228–3233.
Kamenetz F, Tomita T, Hsieh H, Seabrook G, Borchelt D, Iwatsubo T et al. APP processing and synaptic function. Neuron 2003; 37: 925–937.
Freir DB, Holscher C, Herron CE . Blockade of long-term potentiation by beta-amyloid peptides in the CA1 region of the rat hippocampus in vivo. J Neurophysiol 2001; 85: 708–713.
Walsh DM, Klyubin I, Fadeeva JV, Cullen WK, Anwyl R, Wolfe MS et al. Naturally secreted oligomers of amyloid beta protein potently inhibit hippocampal long-term potentiation in vivo. Nature 2002; 416: 535–539.
Hsu WL, Ma YL, Hsieh DY, Liu YC, Lee EHY . STAT1 negatively regulates spatial memory formation and mediates the memory-impairing effect of amyloid-beta. Neuropsychopharmacology 2014; 39: 746–758.
Lukiw WJ, Bazan NG . Survival signalling in Alzheimer's disease. Biochem Soc Trans 2006; 34: 1277–1282.
Hsu WL, Chiu TH, Tai DJ, Ma YL, Lee EHY . A novel defense mechanism that is activated on amyloid-beta insult to mediate cell survival: role of SGK1-STAT1/STAT2 signaling. Cell Death Differ 2009; 16: 1515–1529.
Kilgore M, Miller CA, Fass DM, Hennig KM, Haggarty SJ, Sweatt JD et al. Inhibitors of class 1 histone deacetylases reverse contextual memory deficits in a mouse model of Alzheimer's disease. Neuropsychopharmacology 2010; 35: 870–880.
Meng J, Li Y, Camarillo C, Yao Y, Zhang Y, Xu C et al. The anti-tumor histone deacetylase inhibitor SAHA and the natural flavonoid curcumin exhibit synergistic neuroprotection against amyloid-beta toxicity. PLoS One 2014; 9: e85570.
Abel T, Zukin RS . Epigenetic targets of HDAC inhibition in neurodegenerative and psychiatric disorders. Curr Opin Pharmacol 2008; 8: 57–64.
Chuang DM, Leng Y, Marinova Z, Kim HJ, Chiu CT . Multiple roles of HDAC inhibition in neurodegenerative conditions. Trends Neurosci 2009; 32: 591–601.
Graff J, Tsai LH . The potential of HDAC inhibitors as cognitive enhancers. Annu Rev Pharmacol Toxicol 2013; 53: 311–330.
Taunton J, Hassig CA, Schreiber SL . A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science 1996; 272: 408–411.
Pflum MK, Tong JK, Lane WS, Schreiber SL . Histone deacetylase 1 phosphorylation promotes enzymatic activity and complex formation. J Biol Chem 2001; 276: 47733–47741.
Li Y, Li X, Guo B . Chemopreventive agent 3,3'-diindolylmethane selectively induces proteasomal degradation of class I histone deacetylases. Cancer Res 2010; 70: 646–654.
Colombo R, Boggio R, Seiser C, Draetta GF, Chiocca S . The adenovirus protein Gam1 interferes with sumoylation of histone deacetylase 1. EMBO Rep 2002; 3: 1062–1068.
David G, Neptune MA, DePinho RA . SUMO-1 modification of histone deacetylase 1 (HDAC1) modulates its biological activities. J Biol Chem 2002; 277: 23658–23663.
Citro S, Jaffray E, Hay RT, Seiser C, Chiocca S . A role for paralog-specific sumoylation in histone deacetylase 1 stability. J Mol Cell Biol 2013; 5: 416–427.
Chiou HY, Liu SY, Lin CH, Lee EHY . Hes-1 SUMOylation by protein inhibitor of activated STAT1 enhances the suppressing effect of Hes-1 on GADD45alpha expression to increase cell survival. J Biomed Sci 2014; 21: 53.
Mori M, Burgess DL, Gefrides LA, Foreman PJ, Opferman JT, Korsmeyer SJ et al. Expression of apoptosis inhibitor protein Mcl1 linked to neuroprotection in CNS neurons. Cell Death Differ 2004; 11: 1223–1233.
Liu SY, Ma YL, Lee EHY . NMDA receptor signaling mediates the expression of protein inhibitor of activated STAT1 (PIAS1) in rat hippocampus. Neuropharmacology 2013; 65: 101–113.
Rytinki MM, Kaikkonen S, Pehkonen P, Jaaskelainen T, Palvimo JJ . PIAS proteins: pleiotropic interactors associated with SUMO. Cell Mol Life Sci 2009; 66: 3029–3041.
Nagahara AH, Merrill DA, Coppola G, Tsukada S, Schroeder BE, Shaked GM et al. Neuroprotective effects of brain-derived neurotrophic factor in rodent and primate models of Alzheimer's disease. Nat Med 2009; 15: 331–337.
Dore S, Kar S, Quirion R . Insulin-like growth factor I protects and rescues hippocampal neurons against beta-amyloid- and human amylin-induced toxicity. Proc Natl Acad Sci USA 1997; 94: 4772–4777.
Ma YL, Chen KY, Wei CL, Lee EHY . Corticotropin-releasing factor enhances brain-derived neurotrophic factor gene expression to facilitate memory retention in rats. Chin J Physiol 1999; 42: 73–81.
Delcuve GP, Khan DH, Davie JR . Roles of histone deacetylases in epigenetic regulation: emerging paradigms from studies with inhibitors. Clin Epigenet 2012; 4: 5.
Canettieri G, Morantte I, Guzman E, Asahara H, Herzig S, Anderson SD et al. Attenuation of a phosphorylation-dependent activator by an HDAC-PP1 complex. Nat Struct Biol 2003; 10: 175–181.
Leitao BB, Jones MC, Brosens JJ . The SUMO E3-ligase PIAS1 couples reactive oxygen species-dependent JNK activation to oxidative cell death. FASEB J 2011; 25: 3416–3425.
Liu B, Shuai K . Induction of apoptosis by protein inhibitor of activated Stat1 through c-Jun NH2-terminal kinase activation. J Biol Chem 2001; 276: 36624–36631.
Hetman M, Gozdz A . Role of extracellular signal regulated kinases 1 and 2 in neuronal survival. Eur J Biochem 2004; 271: 2050–2055.
Almeida RD, Manadas BJ, Melo CV, Gomes JR, Mendes CS, Graos MM et al. Neuroprotection by BDNF against glutamate-induced apoptotic cell death is mediated by ERK and PI3-kinase pathways. Cell Death Differ 2005; 12: 1329–1343.
Cai R, Kwon P, Yan-Neale Y, Sambuccetti L, Fischer D, Cohen D . Mammalian histone deacetylase 1 protein is posttranslationally modified by phosphorylation. Biochem Biophys Res Commun 2001; 283: 445–453.
Ashburner BP, Westerheide SD, Baldwin AS Jr. . The p65 (RelA) subunit of NF-kappaB interacts with the histone deacetylase (HDAC) corepressors HDAC1 and HDAC2 to negatively regulate gene expression. Mol Cell Biol 2001; 21: 7065–7077.
Doetzlhofer A, Rotheneder H, Lagger G, Koranda M, Kurtev V, Brosch G et al. Histone deacetylase 1 can repress transcription by binding to Sp1. Mol Cell Biol 1999; 19: 5504–5511.
Belyaev ND, Kellett KAB, Beckett C, Makova NZ, Revett TJ, Nalivaeva NN et al. The transcriptionally active amyloid precursor protein (APP) intracellular domain is preferentially produced from the 695 isoform of APP in a β-secretase-dependent pathway. J Biol Chem 2010; 285: 41443–41454.
Kerridge C, Belyaev ND, Nalivaeva NN, Turner AJ . The Aβ-clearance protein transthyretin, like neprilysin, is epigenetically regulated by the amyloid precursor protein intracellular domain. J Neurochem 2014; 130: 419–431.
Kirsh O, Seeler JS, Pichler A, Gast A, Muller S, Miska E et al. The SUMO E3 ligase RanBP2 promotes modification of the HDAC4 deacetylase. EMBO J 2002; 21: 2682–2691.
Graff J, Rei D, Guan JS, Wang WY, Seo J, Hennig KM et al. An epigenetic blockade of cognitive functions in the neurodegenerating brain. Nature 2012; 483: 222–226.
Selenica ML, Benner L, Housley SB, Manchec B, Lee DC, Nash KR et al. Histone deacetylase 6 inhibition improves memory and reduces total tau levels in a mouse model of tau deposition. Alzheimers Res Ther 2014; 6: 12.
Kazantsev AG, Thompson LM . Therapeutic application of histone deacetylase inhibitors for central nervous system disorders. Nat Rev Drug Discov 2008; 7: 854–868.
Tai DJ, Hsu WL, Liu YC, Ma YL, Lee EHY . Novel role and mechanism of protein inhibitor of activated STAT1 in spatial learning. EMBO J 2011; 30: 205–220.
Meffert MK, Chang JM, Wiltgen BJ, Fanselow MS, Baltimore D . NF-κB functions in synaptic signaling and behavior. Nat Neurosci 2003; 6: 1072–1078.
Lee L, Sakurai M, Matsuzaki S, Arancio O, Fraser P . SUMO and Alzheimer's disease. Neuromol Med 2013; 15: 720–736.
Li Y, Wang H, Wang S, Quon D, Liu YW, Cordell B . Positive and negative regulation of APP amyloidogenesis by sumoylation. Proc Natl Acad Sci USA 2003; 100: 259–264.
Zhang YQ, Sarge KD . Sumoylation of amyloid precursor protein negatively regulates Abeta aggregate levels. Biochem Biophys Res Commun 2008; 374: 673–678.
McMillan LE, Brown JT, Henley JM, Cimarosti H . Profiles of SUMO and ubiquitin conjugation in an Alzheimer's disease model. Neurosci Lett 2011; 502: 201–208.
Kar S, Baccichet A, Quirion R, Poirier J . Entorhinal cortex lesion induces differential responses in [125I]insulin-like growth factor I, [125I]insulin-like growth factor II and [125I]insulin receptor binding sites in the rat hippocampal formation. Neuroscience 1993; 55: 69–80.
McGinley LM, Sims E, Lunn JS, Kashlan ON, Chen KS, Bruno ES et al. Human cortical neural stem cells expressing insulin-like growth factor-1: a novel cellular therapy for Alzheimer's disease. Stem Cells Transl Med 2016; 5: 379–391.
Dupraz S, Grassi D, Karnas D, Nieto Guil AF, Hicks D, Quiroga S . The insulin-like growth factor 1 receptor is essential for axonal regeneration in adult central nervous system neurons. PLoS One 2013; 8: e54462.
Wang HL, Tsai LY, Lee EHY . Corticotropin-releasing factor produces a protein synthesis—dependent long-lasting potentiation in dentate gyrus neurons. J Neurophysiol 2000; 83: 343–349.
Tai DJ, Liu YC, Hsu WL, Ma YL, Cheng SJ, Liu SY et al. MeCP2 SUMOylation rescues Mecp2-mutant-induced behavioural deficits in a mouse model of Rett syndrome. Nat Commun 2016; 7: 10552.
Chen YC, Hsu WL, Ma YL, Tai DJ, Lee EHY . CREB SUMOylation by the E3 ligase PIAS1 enhances spatial memory. J Neurosci 2014; 34: 9574–9589.
Chao CC, Ma YL, Lee EHY . Brain-derived neurotrophic factor enhances Bcl-xL expression through protein kinase casein kinase 2-activated and nuclear factor kappa B-mediated pathway in rat hippocampus. Brain Pathol 2011; 21: 150–162.
Acknowledgements
This work was supported by a Grant from the Ministry of Science and Technology (MOST 104-2321-B-001-061) and research fund from IBMS, Academia Sinica, Taiwan. We thank the Animal Core and Confocal Core Facility of IBMS.
Author contributions
CCT and WLH have helped design and perform most of the experiments, made the constructs and lentivectors, analyzed the data. YLM has helped with the animal surgery and behavioral experiments as well as data analysis. CSJ has helped with the electrophysiological recording experiments.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by N Bazan
Supplementary Information accompanies this paper on Cell Death and Differentiation website
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
About this article
Cite this article
Tao, C., Hsu, W., Ma, Y. et al. Epigenetic regulation of HDAC1 SUMOylation as an endogenous neuroprotection against Aβ toxicity in a mouse model of Alzheimer’s disease. Cell Death Differ 24, 597–614 (2017). https://doi.org/10.1038/cdd.2016.161
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cdd.2016.161
This article is cited by
-
Abnormal protein post-translational modifications induces aggregation and abnormal deposition of protein, mediating neurodegenerative diseases
Cell & Bioscience (2024)
-
Rhes, a striatal enriched protein, regulates post-translational small-ubiquitin-like-modifier (SUMO) modification of nuclear proteins and alters gene expression
Cellular and Molecular Life Sciences (2024)
-
ZNF787 and HDAC1 Mediate Blood–Brain Barrier Permeability in an In Vitro Model of Alzheimer’s Disease Microenvironment
Neurotoxicity Research (2024)
-
PIAS1 alleviates diabetic peripheral neuropathy through SUMOlation of PPAR-γ and miR-124-induced downregulation of EZH2/STAT3
Cell Death Discovery (2021)
-
Galectin-3 promotes Aβ oligomerization and Aβ toxicity in a mouse model of Alzheimer’s disease
Cell Death & Differentiation (2020)