Abstract
APO2L/TRAIL (TNF-related apoptosis-inducing ligand) induces death of tumor cells through two agonist receptors, TRAIL-R1 and TRAIL-R2. We demonstrate here that N-linked glycosylation (N-glyc) plays also an important regulatory role for TRAIL-R1-mediated and mouse TRAIL receptor (mTRAIL-R)-mediated apoptosis, but not for TRAIL-R2, which is devoid of N-glycans. Cells expressing N-glyc-defective mutants of TRAIL-R1 and mouse TRAIL-R were less sensitive to TRAIL than their wild-type counterparts. Defective apoptotic signaling by N-glyc-deficient TRAIL receptors was associated with lower TRAIL receptor aggregation and reduced DISC formation, but not with reduced TRAIL-binding affinity. Our results also indicate that TRAIL receptor N-glyc impacts immune evasion strategies. The cytomegalovirus (CMV) UL141 protein, which restricts cell-surface expression of human TRAIL death receptors, binds with significant higher affinity TRAIL-R1 lacking N-glyc, suggesting that this sugar modification may have evolved as a counterstrategy to prevent receptor inhibition by UL141. Altogether our findings demonstrate that N-glyc of TRAIL-R1 promotes TRAIL signaling and restricts virus-mediated inhibition.
Similar content being viewed by others
Main
APO2L/TRAIL (TNF-related apoptosis-inducing ligand) belongs to the TNF subfamily of apoptosis-inducing ligands.1 This promising anti-tumor compound has attracted much interest in oncology due to its ability to trigger selective cell death in a large variety of human tumors.2 Owing to its restricted expression on immune cells, TRAIL plays an important physiological function during both tumor and viral immune surveillance.3 TRAIL triggers apoptosis by binding to one or both of its agonist receptors, namely TRAIL-R1/DR4 and/or TRAIL-R2/DR5.4 TRAIL binding induces the formation of a membrane-bound death-inducing signaling complex (DISC), formed by the recruitment of caspase-8 and caspase-10 to TRAIL-R1 and/or TRAIL-R2 via the adaptor protein FADD.5 Recruitment of initiator caspases to the DISC allows their activation and subsequent release in the cytosol where they cleave and activate effector caspases, including caspase-3, leading to cell dismantling.6 Conversely, binding of TRAIL to its regulatory receptors TRAIL-R3/DcR1 or TRAIL-R4/DcR2 selectively impairs the engagement of the apoptotic machinery.7, 8 Besides regulation provided by differential expression of agonist or antagonist receptors, cell sensitivity to TRAIL can also be controlled by their post-translational modifications. For example, O-glycosylation of the cysteine-rich domains of TRAIL-R2 is required for efficient TRAIL DISC formation and apoptosis.9 However, whether glycosylation-mediated regulation is a conserved feature of the death receptor family remains, so far, unclear.
Cytomegalovirus (CMV/HHV-5, the prototypic β-herpesvirus) establishes lifelong persistence/latency despite a robust host immune response, as do all herpes viruses. CMV infection is essentially benign in healthy, young persons, but causes severe diseases if immunity is compromised or naïve (e.g. transplants, AIDS, congenital infection).10, 11 Both human and mouse CMV dedicate more than half of their ~230 kB DNA genomes to open reading frames that modulate host immune defenses,12, 13 many of which target signaling by TNF family cytokines.14 Our previous work revealed that the HCMV UL141 protein promotes intracellular retention of both TRAIL-R1 and TRAIL-R2,15 a function similarly performed by the MCMV m166 protein to mTRAIL-R,16 contributing to the resistance of CMV-infected cells to killing by TRAIL-expressing NK cells. UL141 is an immunoglobulin (Ig) fold-containing protein that is structurally unrelated to TRAIL. Analysis of the crystal structure of UL141 in complex with TRAIL-R2 revealed that UL141 binds uniquely to TRAIL-R2, only partially overlapping with the TRAIL-binding site.17
In the present study, we show that N-glycosylation (N-glyc) increases TRAIL-induced receptor aggregation and apoptosis by both human TRAIL-R1 and mTRAIL-R. We also provide evidence that CMV UL141 binds with higher affinity to TRAIL-R1 lacking N-glyc, suggesting N-glyc of this death receptor, which is absent in TRAIL-R2, may have evolved in response to virus-imposed pressure.
Results
Cell death induced by human TRAIL-R1 is regulated by N-glycosylation
Despite clear evidence demonstrating that O-glycosylation (O-glyc) is required for efficient TRAIL-R2-induced apoptosis in human tumor cells,9 it remains unclear whether this post-translational modification might similarly regulate apoptosis induced by TRAIL-R1 or by the single rodent agonist receptor ortholog, mTRAIL-R (also coined as mouse killer or MK).18 Specifically, TRAIL-induced cell death was impaired when eight serine and threonine residues of TRAIL-R2 (SSS74,75,77AAA and TTTTT130,131,132,135,143AAAAA) were mutated9 of which S77, T130 and T135 are predicted to be putative O-glyc sites using the bioinformatic tool GlycoEP.19 These O-glyc sites, albeit well conserved in most primate TRAIL-R2 orthologs, are poorly conserved in rodents (Figure 1a and Supplementary Figure S1). Additionally, predicted O-glyc sites in primates are also less conserved in TRAIL-R1 than in TRAIL-R2, with the exception of the threonine stretch at the end of the first cysteine-rich repeat (CRD1) (Figure 1a and Supplementary Figure S1). Interestingly, however, while TRAIL-R2 is devoid of N-glyc sites, one conserved consensus site is present within the first cysteine-rich domains of TRAIL-R1 in six out of seven primate species (Supplementary Figure S1). Conserved putative N-glyc sites are also present in mTRAIL-R (Figure 1a and Supplementary Figure S1), either at the beginning of CRD1 in the '50s loop', which interacts with TRAIL,20 or at the end of CRD1 (Supplementary Figures S1 and S2).
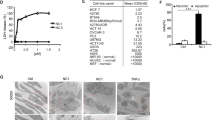
TRAIL-R1 is N-glycosylated in human cancer cell lines. (a) Sequence comparison of the extracellular domain of TRAIL-R1 (TNFRSF10A), TRAIL-R2 (TNFRSF10B) and mouse TRAIL-R. O-glycosylated threonine and serine sites demonstrated to regulate TRAIL-R2-induced apoptosis are shown in orange boxes.9 Serine or threonine sites predicted to be O-glycosylated (GlycoEP server) are shown in red. Predicted N-glycosylation sites (NetNGlyc1.0 and GlycoEP servers) are depicted in green boxes. Conserved cysteines are highlighted in pale gray boxes. (b) Flow cytometric analysis of TRAIL-R1 and TRAIL-R2 expression in two human colorectal cancer cell lines, SW480 and HCT116. Gray-filled histogram, isotype control. White-filled histograms, TRAIL-R1 or TRAIL-R2 staining. (c) Cell viability of SW480 (white circles) and HCT116 (black squares) obtained by methylene blue staining, after stimulation with increasing concentrations of His-hTRAIL, 4C9 (TRAIL-R1 selective ligand) or M1d (TRAIL-R2 selective peptidomimetic) as indicated. Data are the mean±S.D. (n=3). (d) Cell viability of B-cell lymphomas, VAL (white circles), BL2 (white squares), RL (gray squares), SUDHL4 (black squares) and Ramos (black circles) measured by alamar blue staining after exposure of increasing concentrations of His-hTRAIL, 4C9, M1d or Fas ligand. Data are the mean±S.D. (n=3). (e) TRAIL-R1 and TRAIL-R2 immunoblots obtained from SW480 and HCT116 cell lysates after PNGase F treatment. GAPDH was used as a loading control. (f) TRAIL-R1 immunoblot obtained from indicated B-cell lymphoma cell lysates after PNGase F treatment. HSC70, loading control. One representative blot is shown. The black arrow corresponds to WT TRAIL-R1. Gray arrow shows non-glycosylated TRAIL-R1
Since N-glyc plays a regulatory role for other members of the TNF family, including CD95/Fas,21 we investigated the N-glyc status of TRAIL-R1 in a panel of human colorectal cancer and B-cell lymphoma cell lines. The human colon cancer cell lines SW480 and HCT116 express both TRAIL-R1 and TRAIL-R2 (Figure 1b) and are sensitive to TRAIL-induced cell death (Figure 1c). Apoptosis in these cells could be induced either through TRAIL-R1 using the TRAIL mutant ligand 4C922 or through TRAIL-R2 using the TRAIL peptidomimetic M1d23 (Figure 1c). In contrast, several B-lymphomas cells lines (VAL, BL2, RL, SUDHL4, Ramos) engaged apoptosis preferentially through TRAIL-R1 (Figure 1d), irrespective of their relative expression of the TRAIL receptors (Supplementary Figure S3a and ref. 24) or their sensitivity to Fas ligand (Figure 1d). In all of these tumor cell lines TRAIL-R1 was N-glycosylated, as demonstrated by removal of N-linked glycans with peptide N-glycanase F (PNGaseF), which shifted TRAIL-R1 migration to the expected theoretical molecular weight (MW) of 42 kDa (Figures 1e and f). Consistent with the absence of predicted N-glyc sites, TRAIL-R2 expressed in colon cancer cell lines was unaffected by PNGaseF (Figure 1e). In order to determine the impact of N-glycosylation on the apoptotic activity of TRAIL-R1, U2OS cells expressing low levels of TRAIL-R2 and nearly undetectable levels of TRAIL-R1 (Figure 2a) were reconstituted with equivalent levels of wild type (TRAIL-R1WT) or an N-glycosylation-deficient mutant of TRAIL-R1 (TRAIL-R1N156A) (Figure 2a). TRAIL-R1N156A migrated with a lower apparent MW than TRAIL-R1WT in U2OS cells, was insensitive to PNGaseF digestion and migrated identically to TRAIL-R1WT from cell extracts treated with PNGaseF, indicating that TRAIL-R1 contains a single N-linked glycan on N156 (Figure 2b). Contrary to parental U2OS cells, which are poorly sensitive to TRAIL- and Fas ligand-induced cell death (Supplementary Figure S3b), U2OS cells expressing TRAIL-R1WT displayed substantially increased sensitivity to apoptosis or cell death induced by TRAIL, 4C9 or TRAIL-functionalized nanoparticles (NPT)25 (Figure 2c, Supplementary Figures S4a and b). In contrast, cells expressing TRAIL-R1N156A displayed reduced sensitivity to TRAIL agonists (Figure 2c, Supplementary Figures S4a and b). Apoptosis induced by TRAIL-R1 was not affected by benzylGalNac (Supplementary Figure S4c), a general inhibitor of O-glyc, while this treatment does restrict TRAIL-R2 signaling.9 Confirming that benzylGalNac selectively inhibited O-glyc and had no effect on N-glyc, benzylGalNac treatment led to loss of α2,3-linked sialylation on O-glyc proteins, but not on N-glyc proteins, using Maackia amurensis lectin II and Sambucus nigra lectin, respectively (Supplementary Figure S4d).
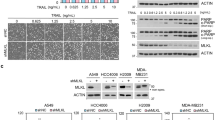
Mutation of the unique TRAIL-R1 N-glycosylation site inhibits TRAIL-induced cell death. (a) Flow cytometric analysis of TRAIL-R1 and TRAIL-R2 in mock-, TRAIL-R1 WT- or TRAIL-R1 N156A-infected U2OS cells. (b) TRAIL-R1 immunoblot obtained from cell extracts treated or not with PNGase F of TRAIL-R1-WT or TRAIL-R1-N156A-infected U2OS cells. HSC70 was used as a loading control. (c) Quantification of apoptosis in U2OS cells expressing or not WT or N156A TRAIL-R1 cells stimulated for 16 h with 1 μg/ml His-hTRAIL or 4C9. Data are the mean±S.D. (n=3), *P <0.05; **P<0.01; NS=not significant. (d) Anti-human IgG Fc capture biosensors were functionalized with TRAIL-R1-Fc WT or TRAIL-R1-Fc N156A recombinant soluble receptors. Binding/dissociation kinetics of indicated Flag-hTRAIL concentrations were monitored by BioLayer Interferometry (BLI) using an Octet Red instrument. Binding constants KD (M), kon (M−1s−1) and koff (s−1) of sensograms are shown below. (e) TRAIL DISC composition in WT or N-glycosylation-deficient TRAIL-R1 (N156A)-expressing U2OS cells was analyzed by immunoblot after His-hTRAIL stimulation (1 μg/ml) and immunoprecipitation using either an anti-histidine or an anti-caspase-8 antibody. The black arrow corresponds to fully glycosylated TRAIL-R1. Gray arrow shows non-glycosylated TRAIL-R1. (f) TRAIL immunofluorescence staining of U2OS cells expressing WT or N156A TRAIL-R1 or not (Mock) stimulated with Flag-hTRAIL crosslinked with M2 for the indicated time at 37 °C, fixed and stained using an anti-M2 antibody. Unstimulated cells were fixed and incubated in the presence of crosslinked TRAIL for 30 min on ice. Cell nuclei were counterstained with DAPI
TRAIL bound to recombinant TRAIL-R1WT-Fc, TRAIL-R1N156A-Fc and TRAIL-R2WT-Fc with similar affinities (8, 8.7 and 5.3 nM, respectively, measured by biolayer interferometry) (Figure 2d and Supplementary Figure S5a). TRAIL-R1-Fc-binding affinities to 293 T cells expressing membrane-bound TRAIL were also similar (Supplementary Figure S5b), indicating that impaired ligand binding is an unlikely cause for the decreased pro-apoptotic activity of TRAIL-R1N156A. Instead, loss of function in U2OS cells expressing TRAIL-R1N156A was associated with reduced caspase-8 and -10 recruitment within the TRAIL DISC (not shown) and restricted receptor clustering, as evidenced by immunofluorescence (Figure 2e).
Contribution of N-glycosylation to TRAIL-R1-induced apoptosis was further investigated in HCT116 and MDA-MB-231 cells double deficient for TRAIL-R1 and TRAIL-R2 (DKO)26 that were reconstituted with WT, N156A or N156Q forms of TRAIL-R1. Surface expression levels of the WT and mutated receptors were comparable for all constructs (Figure 3a) and as expected mutations N156A and N156Q prevented N-glycosylation (Figure 3b). TRAIL-induced cell death was almost fully restored with WT TRAIL-R1 but not with TRAIL-R1 N-glyc mutants (Figure 3c). Consistently, aggregation of TRAIL-R1 and recruitment of caspase-8 and -10 within the TRAIL DISC were decreased in cells expressing N-glyc-deficient TRAIL-R1 (Figures 3d–f). Taken together, these results indicate that N-glyc of TRAIL-R1, although not required for ligand binding, stimulates TRAIL-induced apoptosis through enhanced receptor clustering and DISC formation.
Glycosylation of TRAIL-R1 is required for TRAIL DISC formation. (a) Flow cytometric analysis of TRAIL-R1 and TRAIL-R2 in parental, TRAIL-R1 and TRAIL-R2 double deficient (DKO), DKO reconstituted with TRAIL-R1 WT (DR4recWT), TRAIL-R1 N156A (DR4recN156A) or TRAIL-R1 N156Q (DR4recN156Q), HCT116 or MDA-MB-231 cells. (b) Western blot of TRAIL-R1 and (c) apoptosis analysis in corresponding cells. Data are the mean±S.D. (n=3), *P<0.05; ***P<0.001; NS=not significant. (d) TRAIL DISC composition in WT or N-glycosylation-deficient TRAIL-R1 expressing HCT116 and MDA-MB-231 cells was analyzed by immunoblot after His-hTRAIL stimulation (1 μg/ml) and immunoprecipitation using an anti-histidine antibody. The black arrows correspond to cleaved products of caspase-8, caspase-10 or FLIP. Gray arrows correspond to their proform. Empty arrows show nonspecific (ns) immunoreactive bands. (e) Caspase-8 immunopull-down analysis of TRAIL DISC formation in parental, DR4recWT and DR4recN156A HCT116 cells, after stimulation as above. (f) Immunofluorescence analysis of TRAIL-R1 aggregation after 30 min stimulation with 1 μg/ml His-hTRAIL using an anti-TRAIL-R1 antibody. Cell nuclei were counterstained with DAPI
N-glycosylation of TRAIL-R is evolutionarily conserved
Since mTRAIL-R encodes three potential N-glyc sites in its two CRDs, we determined whether these sites were modified. mTRAIL-R isolated in extracts of L929 fibrosarcoma or B16 melanoma cells migrated at a MW of 55-60 kDa, higher than predicted from its amino acid sequence (42 kDa) (Figure 4a). Treatment of these cells with the N-glycosylation inhibitor tunicamycin or digestion of L929 cell extracts with PNGase F, converted mTRAIL-R to the expected 42 kDa band, demonstrating that these predicted sites are indeed modified by N-glyc (Figures 4a and b).
mTRAIL-R is N-glycosylated and requires enforced receptor aggregation to trigger apoptosis. (a) Mouse tumor cell lines B16 (melanoma) and L929 (fibrosarcoma) were treated with increasing concentrations of tunicamycin (0.25 to 4 μM) for 24 h and protein extracts were analyzed by western blot for mTRAIL-R expression. HSC70 was included as a loading control. (b) Cell lysates from L929 cells were treated with PNGase F and mTRAIL-R migration profile was analyzed by immunoblot as above. (c) Cell viability of L929 cells stimulated for 16 h with increasing concentrations of soluble Flag-mTRAIL crosslinked or not with M2 anti-Flag antibody (200 ng/ml) was measured by methylene blue staining. (d) Cell viability of indicated cell lines in the presence of increasing concentrations of plate-coated anti-mTRAIL-R agonist antibody MD5-1, measured as above. (e) Parental or mTRAIL-R infected B16 cells were stained for mTRAIL-R using the MD5-1 antibody and analyzed by flow cytometry. Gray histogram, isotype control; White, Mock-infected control; Black, cells transduced with a mTRAIL-R retrovirus. Corresponding sensitivity of Mock- (white circles) or mTRAIL-R- transduced B16 cells (black circles) to a fixed concentration of Flag-mTRAIL ligand (100 ng/ml) crosslinked with increasing amounts of anti-Flag M2 antibody. (f) B16 cells infected with WT mTRAIL-R or a chimeric receptor encoding the extracellular domain of human TRAIL-R2 fused to the intracellular domain of mTRAIL-R (h/m-TRAIL-R2) cells were stained for TRAIL-R2 and analyzed by flow cytometry. Gray histograms, isotype control; white, anti-TRAIL-R2 (B-D37) antibody. Right panels: corresponding sensitivity to increasing concentrations of Flag-mTRAIL (white circles or squares) or His-hTRAIL (black circles or squares) was assessed as above. All above data are the mean±S.D. (n=3), **P<0.01; NS=not significant
Mouse tumor cell lines are less sensitive to TRAIL-mediated apoptosis than human ones
Unlike most human tumor cell lines in which cell death can be induced by recombinant His-hTRAIL or by crosslinked Flag-TRAIL, we found that several mouse tumor cell lines were quite resistant to soluble human and mouse TRAIL (Supplementary Figure S6a), despite expressing moderate (B16) to high (L929, EMT6H) levels of mTRAIL-R at both the mRNA and cell-surface protein levels (Supplementary Figures S6b and c). Enforced receptor aggregation induced by crosslinking of a soluble Flag-tagged mTRAIL, however, enhanced apoptosis of L929 cells, but had no effect on B16 cells (Figure 4c and Supplementary Figure S6d). The agonistic anti-mTRAIL-R antibody (clone MD5-1) failed to induce apoptosis when used in solution (Supplementary Figure S6e), but as reported,27 could induce apoptosis of both L929 and EMT6H cells when adsorbed to cell culture dishes (Figure 4d). B16 cells, however, remained resistant even to plate-coated MD5-1 or crosslinked soluble Flag-mTRAIL (Figure 4d and Supplementary Figure S6d).
Although B16 cells expressed detectable mTRAIL-R at the cell surface, its levels were lower than those of L929 and EMT6H cells (Supplementary Figure S6c). Consequently, we tested whether ectopic expression of mTRAIL-R in B16 or L929 cells could enhance their sensitivity to crosslinked Flag-mTRAIL, and found it did for both these cell lines (Figure 4e and Supplementary Figure S6f). In addition, an oligomerized version of mTRAIL (coined Superkiller) also induced more apoptosis of cells engineered to express higher levels of mTRAIL-R, an effect that was further enhanced after treatment with the protein synthesis inhibitor cycloheximide (Supplementary Figure S6g). Nevertheless, increased mTRAIL-R expression was still unable to confer sensitivity of these two tumor cell lines to non-crosslinked TRAIL (Figure 4e and Supplementary Figure S6f). However, expression of a chimeric receptor composed of the extracellular domain of human TRAIL-R2 fused to the intracellular domain of mTRAIL-R was able to induce apoptosis in the absence of TRAIL crosslinking (Figure 4f), suggesting that the inability of soluble TRAIL to induce mTRAIL-R apoptosis in several cell lines upon binding to soluble TRAIL is due to an inability of the receptor ectodomain to mediate higher-order aggregation in the absence of forced crosslinking, and not due to an inherent difference in downstream signaling.
Cell death induced by mouse TRAIL receptor is regulated by N-glycosylation
In order to determine the impact of mTRAIL-R N-glycosylation on TRAIL-induced apoptosis, mTRAIL-R mutants lacking one, two or all three N-glycosylation sites (N99, N122 and/or N150) were generated and expressed in B16 cells. Immunoblot analysis demonstrated that all three asparagines are N-glycosylated in B16 cells (Figure 5a). With the exception of the triple mutant, whose surface expression at the cell surface was reduced by twofold, all single and double mutants were expressed similarly to WT mTRAIL-R in B16 cells (Figure 5b and Supplementary Figure S7a). mTRAIL-R-induced apoptosis (using crosslinked Flag-mTRAIL, 'Superkiller' His-mTRAIL or plate-bound MD5-1) was reduced in mutants N99/122A and N99/122/150A, while other mutants behaved like WT (Figures 5c, d and Supplementary Figure S7b). Mutations N99/122A and N99/122/150A had essentially no impact on ligand binding (Supplementary Figure S7c), but impaired receptor clustering upon ligand binding in both B16 and U2OS cells (Supplementary Figure S7d). In addition, mTRAIL-RN99/122A also showed enhanced internalization compared with WT (Supplementary Figure S8). These results indicate that N-glyc of mTRAIL-R favors TRAIL-induced apoptosis of tumor cells.
N-glycosylation regulates apoptosis induced by mTRAIL-R. (a) Cell lysates of B16 cells expressing mTRAIL-R N-glycosylation mutants were analyzed by immunoblotting for mTRAIL-R and HSC70 expression. (b) Flow cytometric analysis of mTRAIL-R expression levels in B16 cells engineered to express WT or N-glycosylation mutants derivatives using the MD5-1 antibody. Isotype control (thin line), WT mTRAIL-R (pale gray line) and mTRAIL-R glycosylation mutants (black line). (c) Sensitivity of mTRAIL-R N-glycosylation mutants to a fixed Flag-mTRAIL concentration (100 ng/ml) crosslinked with increasing concentrations of M2. (d) Cell viability of indicated cell lines in the presence of increasing concentrations of plate-coated anti-mTRAIL-R agonist antibody MD5-1, measured by methylene blue. Data in panels c and d are the mean±S.D. (n=3), **P<0.01; NS, not significant
The HCMV UL141 glycoprotein alters TRAIL-R1 N-glycosylation
Recently we showed that the HCMV UL141 glycoprotein binds to TRAIL-R1 and TRAIL-R2, restricting their cell-surface expression and desensitizing infected cells to NK cell-mediated killing.15 In agreement with these findings, cell-surface levels of both death receptors was reduced in primary human dermal fibroblasts (neonatal human dermal fibroblasts, NHDF) upon infection with HCMVWT, but not with a mutant virus lacking an intact UL141 open reading frame (HCMVΔUL141) (Figure 6a). As a control, cell-surface expression of MHC-I was shown to be reduced in NHDF infected with both WT and ΔUL141 viruses (Figure 6a). Reduced TRAIL-R1 and TRAIL-R2 surface expression was not due to UL141-mediated inhibition of gene expression or translation, as the total protein levels of both death receptors was in fact markedly increased in cells infected with either HCMVWT or HCMVΔUL141 (Figure 6b and Supplementary Figure S9a). In addition, the size of TRAIL-R1 was slightly reduced upon infection with HCMVWT but not with HCMVΔUL141. A treatment with PNGaseF revealed that this was due to a reduction in the size/composition of N-glyc (Figure 6b and Supplementary Figure S9). Although the N-linked glycan of TRAIL-R1 was sialylated (neuraminidase treatment modestly reduced the MW of TRAIL-R1), desialylation alone did not explain the reduced MW of TRAIL-R1 in HCMVWT-infected cells, as size differences still persisted after neuraminidase treatment (Supplementary Figure S9b). Similar results were obtained upon infection with the mouse virus: an infection with MCMVWT reduced the MW of mTRAIL-R more efficiently than an infection with a virus lacking m166 (MCMVΔm166), the functional ortholog of UL14116 (Supplementary Figures S9c and d). The decreased MW of mTRAIL-R was not solely due to differences in sialylation, because the difference persisted after treatment with neuraminidase and sialidase A (Supplementary Figures S9c and d). Thus, our results show that both human and mouse CMV alter N-glyc of TRAIL-R1 and mTRAIL-R via the UL141 and m166 proteins, respectively. Intesrestingly, N-glyc of TRAIL-R1 appears detrimental for binding to UL141. Indeed, 293 T cells transfected with a GPI-anchored version of UL141, bound TRAIL-R1N156A-Fc significantly better than TRAIL-R1WT-Fc, reaching an affinity comparable to that of TRAIL-R2-Fc, which lacks N-glyc (Figure 6c).
HCMV infection alters TRAIL-R1 electrophoretic mobility and restricts its cell-surface expression. (a) NHDFs were infected with the WT or ΔUL141 HCMV, or mock infected, at a multiplicity of infection (MOI) of ~2. After 72 h cell-surface levels of TRAIL-R1, TRAIL-R2 and MHC1 were analyzed by flow cytometry. Gray histograms show isotype controls. (b) TRAIL-R1 and TRAIL-R2 immunoblots obtained from U2OS Mock, TRAIL-R1 WT (DR4 WT), TRAIL-R1 N156A (DR4 N156A) or cell extracts of NHDF cells infected as above +/− PNGase F treatment. (c) 293 T cells transfected with a UL141-GPI-linked expression plasmid were stained with increasing concentrations of hTRAIL-R1-Fc (WT), hTRAIL-R1-Fc (N156A) or hTRAIL-R2-Fc (WT), as indicated, and binding was monitored by flow cytometry using an anti-hFc. Mean fluorescence intensities (MFI) were plotted with Prism and binding affinities (in nM) to WT TRAIL-R1, WT TRAIL-R2 or N-glycosylation-deficient TRAIL-R1 mutant (N156A) were calculated. WT TRAIL-R1-Fc and TRAIL-R1-Fc (N156A) electrophoretic mobility profile, after Coomassie blue staining, are shown in the inset
Discussion
Until recently, proximal regulation of TRAIL signaling was thought to occur primarily through homotypic protein–protein interactions,7, 28 but recent results have shown that glycosylation of human TRAIL receptors can also plays a role. Indeed, O-glycosylation of TRAIL-R2 is a prerequisite for TRAIL/TRAIL-R2-mediated apoptosis and correlates directly with the cellular expression levels of glucosyltransferases or fucosyltranferases. O-glycosylation is required for receptor clustering, DISC formation, caspase-8 recruitment and downstream signaling.9 However, despite this strong evidence showing that O-glyc can regulate TRAIL-mediated apoptosis through TRAIL-R2, which contains no N-glyc sites, nothing was known with regard to the role that glycosylation might play in regulating TRAIL-R1 signaling. TRAIL-R1 encodes significantly fewer consensus O-glyc sites but harbors an N-glycosylation site on N156 that is located within the first CRD.
Here we provide experimental evidence that the TRAIL apoptotic signaling pathway is regulated by N-glycosylation. We demonstrate that both human TRAIL-R1 and mouse TRAIL receptor are N-glycosylated and that this post-translational modification enhances their ability to trigger apoptosis. Unlike TRAIL-R2,9 however, apoptosis induced by TRAIL-R1 was not affected by the general inhibitor of O-glyc benzylGalNac (Supplementary Figure S4b). As opposed to Fas,21 the N-glyc sites of TRAIL-R1 orthologs are located within the '50's loop', a major TRAIL interaction site. However, our studies show that N-glyc at this location does not affect TRAIL-binding or its steady-state cell-surface expression, excluding these as possibilities for restricted signaling. Instead, our findings support a model where N-glyc alters the distribution of TRAIL-R1 and/or its arrangement at the cell surface before ligand binding, and also regulates receptor clustering/aggregation upon TRAIL stimulation, as this was reduced in cells expressing the N-glyc mutants of both TRAIL-R1 or mTRAIL-R after TRAIL stimulation. These findings suggest that this post-translational modification is an evolutionarily conserved mechanism to modulate TRAIL receptor signaling, perhaps by enhancing homotypic receptor interactions, or potentially regulating associations with yet to be identified cellular partners.
In agreement with a model where N-glyc might regulate heterotypic protein interactions of TRAIL-R1, the HCMV UL141 glycoprotein exhibited ~5 fold higher binding affinity for the non-glycosylated form of TRAIL-R1. This finding is noteworthy, as UL141 potently restricts host innate antiviral defenses by blocking the cell-surface expression of both human TRAIL death receptors.15 UL141 displays lower affinity for N-glyc TRAIL-R1 than O-glyc TRAIL-R2, suggesting that binding of non/under N-glyc TRAIL-R1 by UL141 may be favored in infected cells. This would be consistent with a large proportion of UL141 being localized to the endoplasmic reticulum and endosomal compartment(s) in HCMV-infected cells.15 In this regard, irrespective of whether UL141 has evolved this N-glyc sensitivity in order to bind TRAIL-R1 during its initial synthesis/translation, or to ‘trap’ it at the cell surface or during its recycling, this strategy is likely to provide the virus with an advantage to attenuate innate immune defences involving TRAIL.
Understanding more precisely how O- and N-glycosylation regulate TRAIL receptor trafficking, DISC formation and apoptosis is likely to be important not only to comprehend TRAIL's selective antitumoral properties but also may be key to design novel rational TRAIL-derivative compounds for use in cancer therapy.29 The fact that TRAIL-R1 is N-glycosylated while TRAIL-R2 is O-glycosylated could also provide potential clues to the differential promotion of apoptosis by these two receptors in some tumor cell types.30 However, this preferential engagement is unlikely to be regulated primarily by a loss of TRAIL-R1 N-glyc, as it is fully glycosylated in all the tumor cell lines we have examined, including those inducing cell death primarily through TRAIL-R1 signaling. O- and N-glyc proteins can undergo additional post-translational modifications, including sialylation, fucosylation or terminal glycan branching.31, 32 As recently reported for EGFR33 and Fas,34 these terminal modifications have the potential to affect TRAIL receptor pro-apoptotic signaling. Accordingly, a deficiency in GPD-mannose-4-6-dehydratase, a GDP-mannose converting enzyme essential for de novo fucosylation, was found to inhibit TRAIL- and Fas-ligand-induced cell death.35 Moreover, a galactose-binding lectin, galectin-3, has been reported to both inhibit and enhance TRAIL-mediated apoptosis.36, 37, 38 It will be interesting in the future to test whether this might be related to TRAIL receptor glycosylation status.
Keeping in mind that neoplastic transformation involves drastic changes in glycosylation,39 galectin-3 expression40 and N-terminal sugar modifications,41 all should be considered as potentially important regulators of the TRAIL-mediated tumor killing.
Altogether, our results provide the first evidence that TRAIL-R1 N-glycosylation likely plays an important role in modulating both TRAIL-mediated antiviral responses and tumor immune surveillance.
Material and methods
Cell lines and viruses
The mouse mammary cell line clone EMT6H, which derives from the breast epithelial EMT-6 cells,1 was kindly given by Professor Ali Bettaied (Dijon, France), the human osteosarcoma U2OS cell line was obtained from Professor Serge Lebecque (Lyon, France), the human B Lymphoma cell lines, VAL, RL, SUDHL4 and BL2 were provided by Dr. Thierry Guillaudeux (Rennes, France), the Ramos human B Lymphoma cell line was provided by Dr. Jean-Ehrland Ricci (Nice, France) and the MDA-MB-231 breast carcinoma cell line was obtained from Dr. Patrick Legembre (Rennes, France). HCT116 colon carcinoma cells were from the ATCC (LGC Standards, Molsheim, France). TRAIL-receptor-deficient HCT116 and MDA-MB-231 (DKO) cells were generated using the TALEN approach as described by Dufour et al.26 Primary MEF were generated from WT and mTRAIL-R−/− (a kind gift from Dr Astar Winoto3) d13.5 embryos of C57BL/6 mice. NHDF cells were obtained from Clonetics (San Diego, CA, USA). All other cell lines were purchased from the American type culture collection (ATCC). Mouse B16, L929 cell lines and human B-lymphomas were maintained in RPMI medium (Lonza, Levallois-Perret, France) and primary MEF and NIH-3T3 cells were maintained in DMEM (Gibco, Thermo Fisher Scientific, Courtaboeuf, France). EMT6H cells were cultured in EMEM medium (Lonza). Human colon carcinoma cells HCT116 and SW480, U2OS and NHDF cells were cultured in DMEM medium (Lonza). For NHDF, insulin and bFGF (Sigma-Aldrich, Lyon, France) were added to the media. All cells were cultured with 10% fetal calf serum (Lonza) in 5% CO2. HCMV (Merlin strain) WT and ΔUL141 viruses 4 were generated in NHDF cells and purified by pelleting cell-associated plus cell-free virus through a 20% sorbitol cushion, as described.5 MCMV (K181 BAC-derived strain) WT and Δm166 (containing 2 stop codons inserted after the 44th amino acid by ‘traceless mutagenesis’ of the m166 orf 6) virus stocks were produced in primary MEF as described.6 Cells were infected at an MOI=2 for all experiments.
Real-time PCR assay
RNA was extracted using the RNeasy Mini Kit from Qiagen (Les Ulis, France). M-MLV Reverse Transcriptase (Promega, Charbonnières, France) was used to synthesize cDNAs from total RNA. Real-time PCR was performed in triplicate using the syber green PCR master Mix from Applied Biosystems (Courtaboeuf, France) and analyzed using the 7500 Fast Detection System (Applied Biosystems). Specific forward and reverse primers were: murine TRAIL-R, 5′-CCT CTC GGA AAG GGC ATT C-3′ and 5′-TCC TGCTCGATG ACC AGC T-3′7 and murine cyclophilin A, used as a standardizing control, 5′-GGCCGA TGA CGA GCC C-3′ and 5′-TGT CTT TGG AAC TTT GTC TGC AA-3′.
Ligand production, chemicals and antibodies
Flag-tagged recombinant soluble murine TRAIL, his-tagged human TRAIL and Fas ligand were produced and used as described previously.8 Mouse SuperKiller His-TRAIL (SuperKiller His-mTRAIL) was purchased from Adipogen (Coger, Paris, France). TRAIL-R1 selective TRAIL ligand (4C9) and TRAIL-R2 selective peptidomimetic (M1d) were produced as described earlier.9, 10 Functionalized nanoparticules with TRAIL (NPT) are described elsewere.11 MD5-1 antibody was purchased from eBioscience (Paris, France) and the anti-Flag M2 monoclonal antibody was from Sigma-Aldrich. Crosslinking of TRAIL was achieved by incubating the ligand with anti-Flag M2 antibody for 30 min at 4 °C with mixing prior to cell treatment. For western blot analysis of human proteins, anti-TRAIL-R2 antibody was purchased from Chemicon (Millipore, Molsheim, France), anti-FADD from Transduction Laboratories (BD Biosciences, Le Pont de Claix, France), anti-caspase-8 and anti-caspase-10 were from Medical & Biological Laboratories (Clinisciences, Montrouge, France). Murine DR5 was revealed by western blot using the anti-mouse TRAIL-R2 antibody from Leinco Technologies (St. Louis, MO, USA). Flow cytometry analysis for murine DR5 was performed using the MD5-1 antibody, with a PE-conjugated anti-armenian hamster secondary antibody from BD Biosciences. For human receptors, anti-TRAIL-R1 (wB-S26) and anti-TRAIL-R2 (B-D37) antibodies were provided by Gen-Probe (Diaclone, Besançon, France). The secondary antibody was an Alexa-488-coupled goat anti-mouse from Molecular Probes (Invitrogen, Cergy Pontoise, France). Tunicamycin and swainsonine were from Sigma-Aldrich. PNGaseF was purchased from New England Biolabs (Evry, France). Neuraminidase and sialidase A were from Europa-Bioproducts (Cambridge, England). For apoptosis measurement, Annexin V (556422) and 7AAD (559925) were from BD Biosciences.
Plasmid constructions
The parental murine full-length TRAIL-R and human TRAIL-R1 or TRAIL-R2 retroviral pMSCV vectors were obtained by subcloning using HindIII/XhoI from the pCR3-mTRAIL-R, pCR3-TRAIL-R1 and pCR3-hTRAIL-R2 vectors.12, 13 Chimeric mouse/human TRAIL receptor constructs were generated as follows. First, a modified version of the full-length hTRAIL-R2 harboring two restriction sites, BglII and BamHI, encompassing the transmembrane domain was obtained by synthesis from Genscript (Piscataway, NJ, USA) and subcloned into the pMSCV retroviral vector using HindIII/XhoI to generate pMSCV-hTRAIL-R2-BglII-TM-BamHI. Next, mTRAIL-R extracellular and intracellular domains, ECD and ICD, respectively, were obtained by PCR from pCR3-mTRAIL-R using the following primer sets: mTRAIL-R (ECD) 5′-aaa aga tct AAC CTA GGC CTC TGG ATA GGA G-3′ and 5′-ttt tgg gcc cTC AAA CGC ACT GAG ATC CT-3′; mTRAIL-R (ICD) 5′-aaaaAAGCT TAT GGA GCC TCC AGG ACC CAG-3′ and 5′-ttt tgg atc cAT GCCAAG ATG CCC AAG CCG-3′. PCR products were digested using BglII/ApaI or HindIII/BamHI, respectively, and digestion products were inserted into pMSCV-hTRAIL-R2-BglII-TM-BamHI to generate pMSCV-mTRAIL-R(ECD)-hTRAIL-R2(ICD) and pMSCV-hTRAIL-R2(ECD)-mTRAIL-R(ICD): referred to in the text as h/mTRAIL-R or m/hTRAIL-R2. Human TRAIL-R1 N156A or N156Q and Mouse TRAIL receptor variants N99A, N122A, N150A mutants were created by routine site-directed mutagenesis from the pMSCV-mTRAIL-R vector using the following sets of primers: N156A 5′-ACA CCG CCG CTT CCA ACA ATT TGT TTG CTT GCC-3′ and 5′-TGG AAGCGG CGG TGT AAC CCA CAC CCT CTG T; N156Q, 5′-ACA CCC AAGCTT CCA ACA ATT TGT TTG CTT GCC-3′ and 5′-TGG AAG CTT GGG TGTAAC CCA CAC CCT CTG T; N99A, 5′-CTA CAC CAG CCA TTC CGC CCATTC TCT GGA TTC-3′ and 5′-GAA TCC AGA GAA TGG GCG GAA TGGCTG GTG TAG-3′; N122A, 5′-CGT AGA AAC CCG ATG CGC CAT AAC CAC AAATAC G-3′ and 5′-CGT ATT TGT GGT TAT GGC GCA TCG GGT TTC TAC G-3′; N150A, 5′-CTG CCA GTC ATG CTC TGC ATG CAC TGA CGG GGA AG-3′ and 5′-CTT CCC CGT CAG TGC ATG CAG AGC ATG ACT GGC AG-3′ to generate pMSCV-hTRAIL-R1-N156A, pMSCV-hTRAIL-R1-N156Q, pMSCV-mTRAIL-R-N99A; pMSCV-mTRAIL-R-N122A and pMSCV-mTRAIL-R-N150A. Double and triple mutants were obtained as above through successive site-directed mutagenesis to generate pMSCV-mTRAIL-R-N99/122A; pMSCV-mTRAIL-R-N122/150A, pMSCV-mTRAIL-R-N99/150A and pMSCV-mTRAIL-R-N99/122/150A. The UL141-GFP fusion plasmid was generated as described,5 and the UL141-GPI expression plasmid was generated by PCR amplification of the UL141 ectodomain (FIX strain5) and cloning upstream of the sequence encoding for the addition of the TRAIL-R3 GPI linkage.14 Sequences of all constructs were confirmed by sequencing.
Glycosidase treatments
Cell lysates were prepared using a NP40 lysis buffer containing 1% NP40, 20 mM Tris-HCl pH 7.5, 150 mM NaCl, 10% glycerol and a proteinase inhibitor cocktail (Roche, Meylan, France). Ten micrograms of protein were digested with PNGase F or sialidases (neuraminidase or sialidase A) according to the manufacturer’s recommendations (Europa-Bioproducts, Cambridge, England). Alternatively cells were incubated in the presence of 1 μg/ml tunycamycin before lysis.
Retrovirus production and cell transduction
The generation of viruses has been described previously.15 Viral particles of the various human and mouse TRAIL receptor constructs were produced to transduce mouse B16F10, L929, EMT6H tumor cell lines as well as the human tumor cell line SW480. Transduction was performed for 16 h in six-well plates on 0.1 to 0.3 × 106 cells in the presence of Polybrene (8 μg/ml). Cells were then washed in phosphate-buffered saline, harvested, plated in complete medium containing puromycin (2.5 μg/ml) and incubated for 3 days before amplification and subsequent analysis of the polyclonal populations.
Cell viability assays
TRAIL ligands (Flag-mTRAIL; His-hTRAIL or SuperKiller His-mTRAIL) or MD5-1 were titrated in 96-well plates using 5 × 104 cells per well, incubated at 37 °C for 16 h. In another set of experiments, the crosslinking anti-Flag M2 monoclonal antibody was titrated in the presence of 100 ng/ml Flag-mTRAIL as above. Alternatively, the M2 antibody was used at 1 μg/ml to titrate Flag-mTRAIL. In other experiments, the agonist anti-mTRAIL-R antibody MD5-1 was incubated in protein A-coated plates (Fisher Scientific, Illkirch, France) in PBS overnight at 4 °C with gentle rocking at increasing concentrations. The next day, unbound antibodies were removed and plates were washed three times in sterile PBS before adding 4 × 104 cells per well in complete medium. Plates were then incubated at 37 °C for 16 h with 5% CO2. Cell viability was either measured by methylene blue (adherent cells) or Alamar blue staining (Fischer Scientific).
Apoptosis measurement
Apoptosis was determined by detection of phosphatidylserine externalization after co-labeling with Annexin V-FITC/propidium iodide, according to the manufacturer's instructions. Analyses were performed on a FACScan cytometer (BD Bioscience). Apoptosis is displayed as the percentage of cells presenting a positive staining compared with nontreated cells as control. Each experiment was carried out independently at least three times.
Immunoprecipitations
For TRAIL DISC analysis in human cells expressing the murine TRAIL receptor, 8 × 107 cells in PBS were stimulated in 1 ml of complete medium with 5 μg Flag-TRAIL with or without 10 μg M2 antibody for 60 min at 37 °C. Cells were then washed with PBS before lysis in NP40 lysis buffer containing a protease inhibitor cocktail. Cell lysates were pre-cleared with Sepharose 6B (Sigma-Aldrich) for 1 h, then the DISC was immunoprecipitated overnight with protein G-coated beads (Amersham Biosciences, Les Ullis, France) at 4 °C in the presence of an anti-caspase-8 antibody (C20, SC6136). Beads were washed four times with NP40 lysis buffer, and then immunoprecipitated complexes were eluted in loading buffer (63 mM Tris-HCl pH 6.8, 2% SDS, 0.03% phenol red, 10% glycerol, 100 mM DTT), then boiled for 5 min before analysis by western blot.
Western blotting
Immunoprecipitates or cell lysates were separated by SDS-PAGE and then transferred to PVDF membrane. Nonspecific binding sites were blocked by incubation in PBS containing 0.5% Tween 20 and 5% powdered milk. Immunoblots were incubated with a specific primary antibody and then an HRP-conjugated secondary antibody. Blots were developed using an enhanced chemiluminescence method according to the manufacturer's protocol (Amersham).
Binding studies
Binding data processing was performed with FortéBio Data Analysis Software version 7.1.0.36 with Savitsky-Golay filtering to reduce noise. Association and dissociation data were fit globally (single phase exponential decay function) in Prism version 5.0a software (GraphPad Software, San Diego, CA, USA).
In silico analysis
Sequence alignment across species was performed using CLC Sequence Viewer 6.5.2 software (CLC bio, Aarhus, Denmarkoctet). O- and N-glycosylated sites were predicted using the GlycoEP server (prediction of glycosides in eukaryotic glycoproteins),16 NetNGlyc1.0 and NetOGlyc 3.1 servers available at http://www.imtech.res.in/raghava/glycoep/ and at the CBS (Center for biological sequence analysis (http://www.cbs.dtu.dk/services/NetNGlyc/orNetOGlyc/), respectivley. Representation of TRAIL-R1 and mTRAIL-R 3D structure prediction were inferred from TRAIL-R2 crystallographic structure using PHYRE2 Protein Fold Recognition server,17 at http://www.sbg.bio.ic.ac.uk/phyre2. Evolutionary history of primate and rodent TRAIL agonist receptors was inferred using the Neighbor-Joining method using the software MEGA 6.06 (Molecular Evolutionary Genetics Analysis).
Statistical analysis
Statistical analysis was performed using the Student's t-test. All statistical analyses were performed using Prism version 5.0a software (GraphPad Software, San Diego, CA, USA). *P<0.05 and **P<0.01 were considered significant.
Production of soluble TRAIL receptors and BLI biolayer interferometry analysis
Murine mTRAIL-R variants N99A, N122A, N150A mutants and human TRAIL-R1 variant fused to human Fc IgG1 were created by routine site-directed mutagenesis from pCR3-TRAIL-R1-hFc or pCR3-mTRAIL-R-hFc vectors using the following sets of primers: TRAIL-R1 forward 5′-GGG TGT GGG TTA CAC CGC CGC TTC CAA CAA TTT G-3′, reverse 5′-CAA ATT GTT GGA AGC GGC GGT GTA ACC CAC ACC C-3′ and primer sets for mTRAIL-R described in Plasmid constructions. All constructs were confirmed by sequencing. To produce these soluble recombinants receptors, 6 × 106 293 T cells were seeded in 10 cm tissue culture dish and cultured in DMEM medium (Lonza) with 10% fetal calf serum for 24 h. 293 T cells were then transfected with pCR3-mTRAIL-R-WT-hFc, pCR3-mTRAIL-R-N99/122A-hFc, pCR3-mTRAIL-R-N99/122/150A-Fc, pCR3-TRAIL-R1-WT-hFc, pCR3-TRAIL-R1-N156A-WT-hFc using calcium phosphate transfection method. After 16 h, cells were washed twice with HBSS, then 10 ml of Opti-MEM (Invitrogen) were added in each 10 cm tissue culture dish. Seventy-two hours latter, cell culture supernatant was collected, cleared by centrifugation and filtered. Production of soluble hFc-fused WT or mutant mTRAIL-R or TRAIL-R1 was assessed by western blot using the anti-mouse TRAIL-R2 antibody from Leinco Technologies and the anti-TRAIL-R1antibody (wB-K32) from Gen-Probe (Diaclone, Besançon, France). Purification of hFc fusion proteins was achieved by an overnight pull-down with protein A/G-coated beads (Millipore) at 4 °C with mixing. Beads were washed four times with PBS, and pulled-down proteins was eluted in 100 mM glycine-HCl, pH 2. pH neutralization was achieved by adding 1M Tris, pH 9.0. Quantitation of hFc fusion proteins were determined using an Octet Red System with anti-human IgG quantitation (AHQ) biosensors (FortéBIO). All Octet experiments were designed and analyzed with data acquisition software (7.1) and data analysis software (7.1), respectively. Data were fit with GraphPad version 5.
References
Bodmer JL, Schneider P, Tschopp J . The molecular architecture of the TNF superfamily. Trends Biochem Sci 2002; 27: 19–26.
Gonzalvez F, Ashkenazi A . New insights into apoptosis signaling by Apo2L/TRAIL. Oncogene 2010; 29: 4752–4765.
Falschlehner C, Schaefer U, Walczak H . Following TRAIL's path in the immune system. Immunology 2009; 127: 145–154.
Merino D, Lalaoui N, Morizot A, Solary E, Micheau O . TRAIL in cancer therapy: present and future challenges. Expert Opin Ther Targets 2007; 11: 1299–1314.
Bodmer JL, Holler N, Reynard S, Vinciguerra P, Schneider P, Juo P et al. TRAIL receptor-2 signals apoptosis through FADD and caspase-8. Nat Cell Biol 2000; 2: 241–243.
Ashkenazi A . Directing cancer cells to self-destruct with pro-apoptotic receptor agonists. Nat Rev Drug Discov 2008; 7: 1001–1012.
Merino D, Lalaoui N, Morizot A, Schneider P, Solary E, Micheau O . Differential inhibition of TRAIL-mediated DR5-DISC formation by decoy receptors 1 and 2. Mol Cell Biol 2006; 26: 7046–7055.
Lalaoui N, Morle A, Merino D, Jacquemin G, Iessi E, Morizot A et al. TRAIL-R4 promotes tumor growth and resistance to apoptosis in cervical carcinoma HeLa cells through AKT. PLoS One 2011; 6: e19679.
Wagner KW, Punnoose EA, Januario T, Lawrence DA, Pitti RM, Lancaster K et al. Death-receptor O-glycosylation controls tumor-cell sensitivity to the proapoptotic ligand Apo2L/TRAIL. Nat Med 2007; 13: 1070–1077.
Britt WJ . Vaccines against human cytomegalovirus: time to test. Trends Microbiol 1996; 4: 34–38.
Pereira L, Maidji E, McDonagh S, Tabata T . Insights into viral transmission at the uterine-placental interface. Trends Microbiol 2005; 13: 164–174.
Mohr CA, Cicin-Sain L, Wagner M, Sacher T, Schnee M, Ruzsics Z et al. Engineering of cytomegalovirus genomes for recombinant live herpesvirus vaccines. Int J Med Microbiol 2008; 298: 115–125.
Yu D, Silva MC, Shenk T . Functional map of human cytomegalovirus AD169 defined by global mutational analysis. Proc Natl Acad Sci USA 2003; 100: 12396–12401.
Benedict CA . Viruses and the TNF-related cytokines, an evolving battle. Cytokine Growth Factor Rev 2003; 14: 349–357.
Smith W, Tomasec P, Aicheler R, Loewendorf A, Nemcovicova I, Wang EC et al. Human cytomegalovirus glycoprotein UL141 targets the TRAIL death receptors to thwart host innate antiviral defenses. Cell Host Microbe 2013; 13: 324–335.
Verma S, Loewendorf A, Wang Q, McDonald B, Redwood A, Benedict CA . Inhibition of the TRAIL death receptor by CMV reveals its importance in NK cell-mediated antiviral defense. PLoS Pathogens 2014; 10: e1004268.
Nemcovicova I, Benedict CA, Zajonc DM . Structure of human cytomegalovirus UL141 binding to TRAIL-R2 reveals novel, non-canonical death receptor interactions. PLoS Pathogens 2013; 9: e1003224.
Wu GS, Burns TF, McDonald ER, Meng RD, Kao G, Muschel R et al. Induction of the TRAIL receptor KILLER/DR5 in p53-dependent apoptosis but not growth arrest. Oncogene 1999; 18: 6411–6418.
Chauhan JS, Rao A, Raghava GP . In silico platform for prediction of N-, O- and C-glycosites in eukaryotic protein sequences. PLoS One 2013; 8: e67008.
Hymowitz SG, Christinger HW, Fuh G, Ultsch M, O'Connell M, Kelley RF et al. Triggering cell death: the crystal structure of Apo2L/TRAIL in a complex with death receptor 5. Mol Cell 1999; 4: 563–571.
Shatnyeva OM, Kubarenko AV, Weber CE, Pappa A, Schwartz-Albiez R, Weber AN et al. Modulation of the CD95-induced apoptosis: the role of CD95 N-glycosylation. PLoS One 2011; 6: e19927.
Reis CR, van der Sloot AM, Natoni A, Szegezdi E, Setroikromo R, Meijer M et al. Rapid and efficient cancer cell killing mediated by high-affinity death receptor homotrimerizing TRAIL variants. Cell Death Dis 2010; 1: e83.
Pavet V, Beyrath J, Pardin C, Morizot A, Lechner M-C, Briand J-P et al. Multivalent DR5 peptides activate the TRAIL death pathway and exert tumoricidal activity. Cancer Res 2010; 70: 1101–1110.
Jacquemin G, Granci V, Gallouet AS, Lalaoui N, Morlé A, Iessi E et al. Quercetin-mediated Mcl-1 and survivin downregulation restores TRAIL-induced apoptosis in non-Hodgkin's lymphoma B cells. Haematologica 2012; 97: 38–46.
Zakaria AB, Picaud F, Rattier T, Pudlo M, Dufour F, Saviot L et al. Nanovectorization of TRAIL with single wall carbon nanotubes enhances tumor cell killing. Nano Lett 2015; 15: 891–895.
Dufour F, Rattier T, Constentinescu AA, Zischler L, Morlé A, Ben Mabrouk H et al. TRAIL receptor gene editing unveils TRAIL-R1 as a master player of apoptosis induced by TRAIL and ER stress Oncotarget (in press).
Takeda K, Yamaguchi N, Akiba H, Kojima Y, Hayakawa Y, Tanner JE et al. Induction of tumor-specific T cell immunity by anti-DR5 antibody therapy. J Exp Med 2004; 199: 437–448.
Shirley S, Morizot A, Micheau O . Regulating TRAIL receptor-induced cell death at the membrane: a deadly discussion. Recent Pat Anticancer Drug Discov 2011; 6: 311–323.
Micheau O, Shirley S, Dufour F . Death receptors as targets in cancer. Br J Pharmacol 2013; 169: 1723–1744.
MacFarlane M, Kohlhaas SL, Sutcliffe MJ, Dyer MJ, Cohen GM . TRAIL receptor-selective mutants signal to apoptosis via TRAIL-R1 in primary lymphoid malignancies. Cancer Res 2005; 65: 11265–11270.
Rak JW, Basolo F, Elliott JW, Russo J, Miller FR . Cell surface glycosylation changes accompanying immortalization and transformation of normal human mammary epithelial cells. Cancer Lett 1991; 57: 27–36.
Rillahan CD, Antonopoulos A, Lefort CT, Sonon R, Azadi P, Ley K et al. Global metabolic inhibitors of sialyl- and fucosyltransferases remodel the glycome. Nat Chem Biol 2012; 8: 661–668.
Liu YC, Yen HY, Chen CY, Chen CH, Cheng PF, Juan YH et al. Sialylation and fucosylation of epidermal growth factor receptor suppress its dimerization and activation in lung cancer cells. Proc Natl Acad Sci USA 2011; 108: 11332–11337.
Swindall AF, Bellis SL . Sialylation of the Fas death receptor by ST6Gal-I provides protection against Fas-mediated apoptosis in colon carcinoma cells. J Biol Chem 2011; 286: 22982–22990.
Moriwaki K, Shinzaki S, Miyoshi E . GDP-mannose-4,6-dehydratase (GMDS) deficiency renders colon cancer cells resistant to tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) receptor- and CD95-mediated apoptosis by inhibiting complex II formation. J Biol Chem 2011; 286: 43123–43133.
Mazurek N, Jie Sun Y, Feng Liu K, Gilcrease MZ, Schober W, Nangia-Makker P et al. Phosphorylated galectin-3 mediates tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) signaling by regulating PTEN in human breast carcinoma cells. J Biol Chem 2007; 282: 21337–21348.
Mazurek N, Byrd JC, Sun Y, Hafley M, Ramirez K, Burks J et al. Cell-surface galectin-3 confers resistance to TRAIL by impeding trafficking of death receptors in metastatic colon adenocarcinoma cells. Cell Death Differ 2012; 19: 523–533.
Mazurek N, Byrd JC, Sun Y, Ueno S, Bresalier RS . A galectin-3 sequence polymorphism confers TRAIL sensitivity to human breast cancer cells. Cancer 2011; 117: 4375–4380.
Varki A, Kannagi R, Toole BP . Glycosylation changes in cancer. In: Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, Bertozzi CR et al (eds). Essentials of Glycobiology, 2nd edn. Cold Spring Harbor: New York, 2009.
Eude-Le Parco I, Gendronneau G, Dang T, Delacour D, Thijssen VL, Edelmann W et al. Genetic assessment of the importance of galectin-3 in cancer initiation, progression, and dissemination in mice. Glycobiology 2009; 19: 68–75.
Harduin-Lepers A, Krzewinski-Recchi MA, Colomb F, Foulquier F, Groux-Degroote S, Delannoy P . Sialyltransferases functions in cancers. Front Biosci (Elite Ed) 2012; 4: 499–515.
Acknowledgements
This work is supported by grants from the program 'Investissements d’Avenir' with reference ANR-11-LABX-0021-01-LipSTIC Labex, the Conseil Regional de Bourgogne, the INCa (Institut National du Cancer, POLYNOM-174), the Cancéropôle Grand-Est, la Ligue Nationale Contre le Cancer and the ANR (Agence Nationale de la Recherche, 07-PCV-0031 and SphingoDR). SS, FD, AM and GM were supported by fellowships from the INCa, ANR, the Ministry of Research and Education and the foundation ARC. PS is supported by grants of the Swiss National Science Foundation, DMZ and CAB by the National Institute of Health (AI117530 and AI101423, respectively). CG's group has the label 'Ligue contre le Cancer team'. We are indebted to Pr Ali Bettaieb (EPHE, Dijon, France) for EMT6H cells, Pr Serge Lebecque (INSERM U1052, Lyon, France) for U2OS cells, Dr Thierry Guillaudeux (INSERM U917, Rennes, France) and Dr Jean-Ehrland Ricci (INSERM U1065, Nice, France) for B lymphoma cell lines. We thank the FEDER for their support.
Author contributions
OM and CAB designed research; FD, TR, SS, GP, AAC, AM, ZAB, SC and EH performed experiments; GM, EZ, DMZ, RS, GG, FP, GH, TG, CG, PS, CAB and OM analyzed data; and PS, CAB and OM wrote the paper.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by E Baehrecke
Supplementary Information accompanies this paper on Cell Death and Differentiation website
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
About this article
Cite this article
Dufour, F., Rattier, T., Shirley, S. et al. N-glycosylation of mouse TRAIL-R and human TRAIL-R1 enhances TRAIL-induced death. Cell Death Differ 24, 500–510 (2017). https://doi.org/10.1038/cdd.2016.150
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cdd.2016.150
This article is cited by
-
Therapeutic potential of melatonin in targeting molecular pathways of organ fibrosis
Pharmacological Reports (2024)
-
ER stress-induced cell death proceeds independently of the TRAIL-R2 signaling axis in pancreatic β cells
Cell Death Discovery (2022)
-
Lewis glycosphingolipids as critical determinants of TRAIL sensitivity in cancer cells
Oncogene (2022)
-
TRAIL promotes hepatocellular carcinoma apoptosis and inhibits proliferation and migration via interacting with IER3
Cancer Cell International (2021)
-
Yap-Hippo pathway regulates cerebral hypoxia-reoxygenation injury in neuroblastoma N2a cells via inhibiting ROCK1/F-actin/mitochondrial fission pathways
Acta Neurologica Belgica (2020)