Abstract
Diffuse large B-cell lymphomas (DLBCLs) are a highly heterogeneous group of tumors in which subsets share molecular features revealed by gene expression profiles and metabolic fingerprints. While B-cell receptor (BCR)-dependent DLBCLs are glycolytic, OxPhos-DLBCLs rely on mitochondrial energy transduction and nutrient utilization pathways that provide pro-survival benefits independent of BCR signaling. Integral to these metabolic distinctions is elevated mitochondrial electron transport chain (ETC) activity in OxPhos-DLBCLs compared with BCR-DLBCLs, which is linked to greater protein abundance of ETC components. To gain insights into molecular determinants of the selective increase in ETC activity and dependence on mitochondrial energy metabolism in OxPhos-DLBCLs, we examined the mitochondrial translation pathway in charge of the synthesis of mitochondrial DNA encoded ETC subunits. Quantitative mass spectrometry identified increased expression of mitochondrial translation factors in OxPhos-DLBCL as compared with the BCR subtype. Biochemical and functional assays indicate that the mitochondrial translation pathway is required for increased ETC activity and mitochondrial energy reserves in OxPhos-DLBCL. Importantly, molecular depletion of several mitochondrial translation proteins using RNA interference or pharmacological perturbation of the mitochondrial translation pathway with the FDA-approved inhibitor tigecycline (Tigecyl) is selectively toxic to OxPhos-DLBCL cell lines and primary tumors. These findings provide additional molecular insights into the metabolic characteristics of OxPhos-DLBCLs, and mark the mitochondrial translation pathway as a potential therapeutic target in these tumors.
Similar content being viewed by others
Main
Cells adapt their metabolism to satisfy changing biosynthetic and bioenergetic needs.1, 2 Investigation of metabolic reprogramming in cancer has provided insights into the metabolic control of proliferation and survival.3, 4, 5 Although the initial focus of this field has been aerobic glycolysis (the Warburg effect),6 increasing evidence points to a complex landscape of tumor metabolic circuitries beyond aerobic glycolysis, including varied contribution of mitochondria to tumor metabolism as well as heterogeneity in fuel utilization pathways.7, 8, 9, 10, 11, 12
Diffuse large B-cell lymphomas (DLBCLs) are a genetically heterogeneous group of tumors that can be classified into distinct molecular subtypes based on gene expression profiles. The cell-of-origin (COO) classification defined DLBCL subsets that shared certain components of their RNA profiles with normal germinal center B cells (GCBs) or in vitro-activated B cells (ABCs), and a third undefined subset designated ‘type 3’.13 Using an independent approach, the consensus cluster classification (CCC) framework compared the transcriptional profiles of DLBCL groups with each other without referencing normal B cells.14 CCC identified tumor-intrinsic distinctions in three highly reproducible clusters; the B-cell receptor/proliferation cluster (BCR-DLBCL) showing increased expression of BCR signaling components, the oxidative phosphorylation cluster (OxPhos-DLBCL) marked by increased expression of genes encoding for mitochondrial electron transport chain (ETC) components, and the host response cluster (HR-DLBCL) characterized by a T-cell-rich inflammatory immune cell infiltrate.14, 15 COO and CC classifications capture different aspects of DLBCL biology. For example, CCC-defined BCR-DLBCLs include BCR-dependent tumors of both ABC and GCB COO subtypes,16, 17 whereas CC-classified OxPhos-DLBCLs include BCR-independent tumors.7, 16 Our previous functional analyses of DLBCL subtypes also uncovered quantitative proteome- and metabolome-level signatures associated with differences in nutrient and energy metabolism.7 Specifically, these studies showed that BCR-dependent DLBCLs have greater glycolytic flux typical of the Warburg phenotype. Unlike BCR-DLBCLs, OxPhos-DLBCLs channel the majority of glucose-derived pyruvate into the mitochondria, display elevated ETC activity, ATP production and fatty acid oxidation (FAO). Importantly, these metabolic distinctions are associated with subtype-selective survival mechanisms. Moreover, acute inhibition of BCR signaling in BCR-DLBCLs increased their FAO capacity, thus revealing a reciprocal relationship between BCR signaling and FAO.7 Overall, these findings underscore the utility of capturing and dissecting metabolic distinctions in DLBCL subtypes.
The ETC is comprised of a series of large multi-subunit complexes housed within the mitochondrial inner membrane, which carry out multiple redox reactions that ultimately lead to the reduction of molecular oxygen to water. The initial electron donors for these reactions are supplied by the tricaboxylic acid cycle in the form of NADH and FADH2. Respiratory chain complexes I (NADH dehydrogenase also known as NADH-ubiquinone oxidoreductase), III (ubiquinol-cytochrome c reductase), and IV (cytochrome c oxidase) extrude protons across the inner membrane while transferring electrons. The resulting proton gradient is subsequently coupled with ATP synthesis by the activity of the F0F1 ATP synthase (complex V), completing the process of oxidative phosphorylation (OXPHOS). Except for complex II (succinate dehydrogenase), the protein constituents of the ETC complexes are encoded by two independently transcribed and translated genomes; nuclear and mitochondrial.18, 19 The mitochondrial DNA (mtDNA) encodes 13 subunits contributing to complexes I, III, IV, and V, 22 transfer RNAs, and 2 ribosomal RNAs. The mechanism for decoding the mitochondrial genome requires nuclear-encoded factors, including ribosomes, translation initiation, and elongation factors, and tRNA synthetases that are distinct from the cytoplasmic counterparts dedicated to translation of nuclear transcripts.20 Mutations in mtDNA and mitochondrial translation factors are associated with ETC failure in several human pathologies,20, 21 highlighting the functional relevance of the mitochondrial genome. Functional fidelity of the ETC not only requires the coordinate synthesis of respiratory chain subunits encoded by the nuclear and mitochondrial genomes but also proper assembly and organization of ETC complexes into higher-order supercomplexes in the inner membrane. The ETC structural organization is modulated by dedicated chaperones and assembly factors, mitochondrial membrane morphology, and membrane lipid composition.22, 23 The differences in ETC activity and OXPHOS dependency among DLBCL subtypes has prompted investigation of pathways in charge of synthesis and assembly of respiratory chain complexes and their contribution to metabolic heterogeneity in DLBCL subsets. Here we interrogated the mitochondrial translation machinery and its functional contribution to energy metabolism and survival of OxPhos-DLBCLs versus non-OxPhos/BCR-dependent subtypes.
Results
Increased expression of mitochondrial translation proteins in OxPhos-DLBCLs
Our previous assessment of the mitochondrial proteome in OxPhos- and non-OxPhos/BCR-dependent DLBCLs revealed the enrichment of several ETC subunits and ETC assembly factors in OxPhos-DLBCLs that is consistent with increased ETC activity in this subtype.7 These previous analyses, based on a high-performance, single-dimension liquid chromatography–tandem mass spectrometry platform,24 quantified predominantly nuclear-encoded ETC subunits. Because OxPhos-DLBCLs display increased activity of several ETC complexes that are encoded by both the nuclear and mitochondrial genomes,7 we predicted that the protein-level enrichment of ETC subunits in this subtype would also include mtDNA-encoded subunits. To enable more extensive interrogation of the mitochondrial proteome, we utilized deep efficient peptide sequencing and quantification (DEEP SEQ) mass spectrometry25 in conjunction with isobaric tags for relative and absolute quantification (iTRAQ) labeling. Mitochondria isolated from three independent OxPhos- and three non-OxPhos/BCR-DLBCL cell lines were analyzed using this platform. The DLBCL subtype designation of these cells lines based on the CCC and COO classifications has been previously reported13, 16, 26, 27 (Supplementary Information). The DEEP SEQ platform not only quantified the enrichment of mtDNA-encoded ETC subunits in OxPhos-DLBCLs compared with BCR counterparts but also revealed significantly higher levels of numerous protein components of the mitochondrial translation machinery (Figures 1a–e; Table 1; Supplementary Tables 1 and 2). These analyses indicated higher expression across several classes of mitochondrial translation proteins, including mitochondrial elongation factors (TUFM (Tu translation elongation factor, mitochondrial) and GFM1 (G elongation factor, mitochondrial, 1)), mitochondrial ribosomal proteins (mitochondrial ribosomal protein (MRP) S5, S7, S9, S16, S22, S25, L12, and L46), and proteins in charge of mitochondrial tRNA synthesis and function (DARS2 (aspartyl-tRNA synthetase 2), YARS2 (tyrosyl-tRNA synthetase 2), and PUS1 (pseudouridylate synthase 1); Figures 1a and b; Table 1). The enrichment in mtDNA-encoded ETC subunits and mitochondrial translation proteins in OxPhos-DLBCLs is further consistent with higher expression of proteins involved in mtDNA maintenance (SSBP1 (single-stranded DNA-binding protein 1)) and transcription (TFAM (mitochondrial transcription factor A) and LRPPRC (leucine-rich pentatricopeptide repeat containing protein)) in this DLBCL subtype18, 28 (Figure 1f; Table 1; Supplementary Table 2).
Enrichment of mitochondrial translation components in OxPhos-DLBCL mitochondria. Multiplex quantitative proteomic analysis of isolated mitochondria from three independent OxPhos- (Karpas 422, Pfeiffer, and Toledo) and three non-OxPhos/BCR- (Ly1, DHL4, and DHL6) DLBCL cell lines using DEEP SEQ mass spectrometry. (a) Heat map illustrating increased levels of mitochondrial translation proteins in OxPhos-DLBCL cell lines compared with BCR counterparts. See also Table 1 and Supplementary Table 2 detailing OxPhos/BCR abundance ratios for the indicated proteins and corresponding amino-acid sequences for the identified peptides. (b) Schematic diagram of the mitochondrial translation machinery. Highlighted in green are proteins that were enriched in the OxPhos-DLBCL proteomic analysis shown in a, Table 1, and Supplementary Table 2. (c–f) MS/MS spectra of the indicated unique peptides for GFM1 (c), TUFM (d), MRPS7 (e), and TFAM (f) recorded during DEEP SEQ mass spectrometry analysis of mitochondrial proteins shown as an example of differentially enriched peptides. Ions of b- and y-type are shown in green and yellow, respectively. Relative protein ratios in BCR- and OxPhos-DLBCL cell lines are derived from iTRAQ reporter ion intensities shown in inset mass spectrum. ETC, electron transport chain; mtDNA, mitochondrial DNA; mRNA, messenger RNA; rRNA, ribosomal RNA; tRNA, transfer RNA
Genetic and pharmacologic inhibition of the mitochondrial translation pathway is selectively toxic to OxPhos-DLBCLs
Elevated levels of mitochondrial translation components in OxPhos-DLBCLs warranted examination of their contribution to DLBCL survival. To this end, three distinct components of the mitochondrial translation machinery (GFM1, TUFM, and MRPS7) were depleted using two independent short hairpin RNAs (shRNAs) per gene in four OxPhos- and four non-OxPhos/BCR-DLBCL cell lines, and cell viability was measured. Knockdown of each of these translation components proved selectively toxic to OxPhos-DLBCL cell lines while sparing the non-OxPhos subset (Figures 2a–c; Supplementary Figure 1).
Differential requirement of the mitochondrial translation pathway for the survival of DLBCL subsets. Effect of shRNA-mediated depletion of the mitochondrial translation elongation factors GFM1 (a) and TUFM (b), and mitochondrial ribosomal protein MRPS7 (c) on the viability of the indicated OxPhos- and BCR-DLBCL cell lines. Blue bars show rescue of cell viability upon pretreatment with 0.5 mM N-acetyl cysteine (NAC) before shRNA-mediated knockdown. Cell viability was assessed 24 h after knockdown of the indicated proteins. Error bars±S.E.M., n=3–4 independent experiments per condition. **P<0.01; ***P<0.001, comparing the different shRNAs to control or NAC-treated samples; two-tailed Student’s t-test. See also Supplementary Figure 1
A common outcome of ETC inhibition, for example, following interference with mitochondrial translation, is the generation of superoxide.29, 30 This led us to predict that the toxic effects of the above knockdowns may be, in part, dependent on reactive oxygen species (ROS). Consistent with this idea, pretreatment with the antioxidant N-acetyl cysteine (NAC) before knockdown partially, but significantly, rescued cell death in this setting (Figures 2a–c).
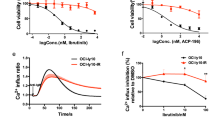
To provide a pharmacologic correlate to these findings, we tested the effect of tigecycline (also known as Tygacil), an FDA-approved glycylcycline antibiotic known to inhibit the prokaryotic ribosome31, 32, 33 and to interfere with mitochondrial translation in eukaryotes without affecting the translation of mitochondrial proteins encoded by nuclear genes (Supplementary Figure 2a).34 The effect of tigecycline on viability of DLBCL subsets was examined at a dose range of 1–5 μM over a 24–72 h time course (Figures 3a–c; Supplementary Figure 2b). OxPhos-DLBCLs were selectively sensitive to tigecycline starting at a dose of 2.5 μM (Figure 3a) with an average viability of 40% and 28%, following 72 h treatment with 2.5 and 5.0 μM tigecycline, respectively (Figure 3c). Of note, this dose range and time course of tigecycline-induced toxicity is in line with pharmacokinetic properties and tolerability of this compound in humans.32 The viability of BCR-DLBCLs was unaffected under these conditions. In comparison, BCR-DLBCLs were selectively sensitive to the glycolytic inhibitor 2-deoxy-d-glucose (Supplementary Figure 3), which is consistent with the glycolytic phenotype of this DLBCL subtype we have previously reported.7 In addition to interfering with OxPhos-DLBCL survival, tigecycline had a select antiproliferative effect in this subtype (Figure 3d). Overall, the above genetic and pharmacologic studies demonstrate that OxPhos-DLBCLs are particularly vulnerable to interference with the mitochondrial translation pathway.
Effect of tigecycline on survival and proliferation of OxPhos-DLBCLs. (a, b) Viability of OxPhos- and BCR-DLBCL cell lines over a 72 h time course, following 2.5 (a) and 5.0 (b) μM tigecycline treatment. (c) Dose–response assessment of viability following 72 h treatment of DLBCL cell lines with tigecycline. (d) Effect of tigecycline on proliferation of OxPhos- and BCR-DLBCL cell lines. Error bars±S.E.M., n=3–7 independent experiments per cell line. ***P<0.001; two-tailed Student’s t-test. See also Supplementary Figures 2 and 3
Bioenergetic consequences of tigecycline treatment in DLBCL subtypes
Inhibition of mitochondrial translation is expected to diminish the activity of respiratory chain complexes that contain mtDNA-encoded subunits. To verify the differential effect of tigecycline on ETC complexes, NADH dehydrogenase (complex I) and succinate dehydrogenase (complex II) enzymatic activities were measured in mitochondria isolated from cells treated with 1 μM tigecycline for 24 h. This dose effectively interferes with translation of mtDNA-encoded complex I subunit NADH dehydrogenase subunit 4 (ND4) but does not have any cytotoxic effects on DLBCL cell lines (Supplementary Figures 2a and b), enabling the examination of tigecycline’s effect on ETC complex activity under conditions where cell survival is not significantly affected. Tigecycline treatment reduced the NADH dehydrogenase enzymatic activity, but did not affect that of succinate dehydrogenase, a respiratory complex that is entirely encoded by the nuclear DNA (Figures 4a and b).
Effect of tigecycline on mitochondrial respiratory complexes. (a, b) Enzymatic activity of NADH dehydrogenase (a) and succinate dehydrogenase (b) in mitochondria isolated from OxPhos- and BCR-DLBCL cell lines following 24 h treatment with 1 μM tigecycline. (c) Changes in mitochondrial respiratory supercomplexes following 24 h treatment with 1 μM tigecycline. Supercomplex (SC) abundance was quantified by densitometry of Blue-Native gels, a representative of which is shown in the left panel. SC abundance was normalized to the abundance of mitochondrial aconitase in each sample (see also Materials and Methods). Error bars±S.E.M., n=5–12 independent experiments. *P<0.05; ***P <0.001; two-tailed Student’s t-test
To further evaluate the biochemical basis of reduced complex I activity in response to tigecycline, we focused on ETC supercomplex assembly. The supramolecular organization of respiratory chain complexes imparts several bioenergetic benefits, including improved functional efficiency.23 In particular, the supercomplex containing complexes I, III, and IV, all of which contain mtDNA-encoded subunits, along with the two electron carriers cytochrome c and ubiquinone preserves the stability of complex I,35, 36, 37 and significantly curtails ROS production from this complex.38 As predicted, tigecycline diminished the abundance of ETC supercomplexes in DLBCL cell lines, and this effect was more prominent in OxPhos- than BCR-DLBCLs (Figure 4c).
To examine the bioenergetic effects of tigecycline in intact cells, we initially focused on mitochondrial spare respiratory capacity (SRC). SRC, the difference between mitochondrial basal and maximal respiration, reflects the mitochondrial reserve capacity to produce energy under cellular stress and increased bioenergetic demand. Alterations in SRC can significantly impact long-term cellular function and survival.39, 40 Compared with BCR-DLBCLs, OxPhos-DLBCLs have higher basal SRC values (Figure 5a), which is consistent with higher ETC activity we have previously reported in this subtype.7 In response to tigecyline, OxPhos-DLBCLs showed significantly larger diminution of SRC (Figure 5b). Consistent with its effect on ETC activity (Figure 4), tigecycline led to increased mitochondrial ROS, which was significantly more prominent in OxPhos-DLBCLs compared with BCR-DLBCLs (Figure 5c). These measurements were carried out following 24 h treatment with 1 μM tigecycline, analogous to the conditions used to measure ETC complex activity and assembly (Figure 4).
Inhibition of the mitochondrial translation pathway reduces spare respiratory capacity and enhances mitochondrial ROS in OxPhos-DLBCLs. (a) Mitochondrial spare respiratory capacity (SRC) in DLBCL subtypes. SRC values shown are the average of all five cell lines per DLBCL subtype indicated on top. (b) Changes in SRC in DLBCL cell lines treated with 1 μM tigecycline for 24 h. (c) Mitochondrial superoxide levels in DLBCL cells that were treated with 1 μM tigecycline for 24 h or exposed to 1 mM NAC before tigecycline treatment. Error bars±S.E.M., n=5–12 independent experiments per condition. ***P<0.001; two-tailed Student’s t-test
Differential toxicity of tigecycline in DLBCL subtypes extends to primary OxPhos- and BCR-DLBCL tumor cells
We next wished to determine whether the differential contribution of the mitochondrial translation pathway to the survival of DLBCL subtypes could be substantiated in primary DLBCLs. For these studies, we used cryopreserved viable tumor cell suspensions from primary DLBCLs that were previously classified as OxPhos- or BCR-DLBCL based on CCC16 (Figure 6a). In concordance with our findings in cell lines, primary OxPhos-DLBCL tumor cells were more sensitive to tigecycline treatment than BCR-DLBCL cells (Figures 6a and b). Overall, these data suggest select dependency of primary OxPhos-DLBCLs on the mitochondrial translation pathway.
Differential sensitivity of primary OxPhos- and BCR-DLBCL tumor cells to tigecycline. DLBCL tumor cells were isolated from two OxPhos- and one non-OxPhos/BCR-DLBCL cryopreserved patient samples and treated with the indicated concentrations of tigecycline. Viability was measured 20 h post treatment. (a) Representative FACS plots of primary tumor cells untreated or treated with 1 μM tigecycline stained with Annexin V and PI. (b) Viability at different doses of tigecycline analyzed as in (a)
Discussion
Our studies provide biochemical and functional evidence for the mitochondrial translation pathway as a survival mechanism that supports a central metabolic feature of OxPhos-DLBCLs; increased mitochondrial energy transduction. Guided by our DEEP SEQ mass spectrometry analysis, we show that genetic or pharmacologic perturbations of the mitochondrial translation apparatus lead to subtype-selective toxicity in OxPhos-DLBCL cell lines and primary OxPhos-DLBCLs. We find that interference with the mitochondrial translation apparatus has a greater bioenergetic impact on OxPhos- than BCR-DLBCLs as evident from a significant reduction in mitochondrial supercomplex abundance, complex I activity, mitochondrial SRC, and elevated mitochondrial ROS.
During normal ETC activity, ~1–2% of the molecular oxygen is converted into superoxide radicals, the precursor for most ROS.41 Mitochondrial ROS can have important signaling functions.42 Defects in the shuttling of electrons through the ETC complexes can lead to increased electron slippage and ROS production depending on the specific site at which electron flow is disrupted.43 This may have deleterious effects depending on the cellular context.43, 44 Increased mitochondrial ROS following tigecycline treatment is likely due to a greater probability of electron slippage when the functional integrity and organization of ETC complexes is diminished, which is consistent with previous studies that examined the consequences of perturbations in mitochondrial translation factors.29, 30 In addition, increased mitochondrial superoxide levels in the face of decreased SRC is in line with other findings that have implicated the mitochondrial SRC as an important determinant of cells capacity to counter oxidative stress.39 In light of these considerations, it is possible that diminished ETC function and SRC are part of a vicious cycle of oxidative stress that contributes to the toxicity associated with inhibition of mitochondrial translation in OxPhos-DLBCLs. This is also consistent with our previous findings that OxPhos-DLBCLs have heightened sensitivity to inhibition of antioxidant pathways.7
The dependency of OxPhos-DLBCLs on the mitochondrial translation pathway co-segregates with increased abundance of mitochondrial translation components, higher ETC activity, and greater reliance on OXPHOS in this subtype compared with BCR/Warburg-type DLBCLs. Within this context, our findings are consistent with an earlier report that identified tigecycline as a compound with activity in acute myeloid leukemia (AML).34 Importantly, increased sensitivity of AML cell lines to tigecycline compared with normal hematopoietic cells was attributed to higher mitochondrial biogenesis and electron transport in AML cells. Tigecycline treatment of AML cells was not associated with an increase in total cellular ROS, although mitochondrial superoxide was not specifically measured in this setting.34 However, our observations are consistent with studies in other cell types that reported increased mitochondrial superoxide content following knockdown of mitochondrial translation factors and examination of MitoSOX Red and DHR123 probe intensities, which specifically measure mitochondrial ROS.29, 45 The basis of the difference between tigecycline’s effect on ROS content in DLBCL and AML cells is unclear. It is also possible that tigecycline may invoke cell context-specific mechanisms that contribute to its toxicity. In the context of OxPhos-DLBCLs, rescue studies using NAC support the idea that oxidative stress is a component of tigecycline toxicity.
Tigecycline selectively inhibits the translation of mtDNA-encoded proteins without affecting global translation34 (Supplementary Figure 2a). Structural studies have identified a tigecyline-binding pocket in the small (30 S) bacterial ribosome subunit and specific interactions with rRNA, which blocks the entry of aminoacyl-tRNA.46 Although the direct target of tigecyline in eukaryotes is not known, initial structural modeling predicts a similar pocket for tigecyline binding may be present in the human small mitoribosome subunit47 (Supplementary Figure 4). However, detailed biochemical and structural studies are required to characterize the target of tigecyline in eukaryotes and to uncover the molecular underpinnings of its specificity towards mitochondrial translation and not the cytosolic translation machinery.
Our results extend the mitochondrial proteomic signature of OxPhos-DLBCLs to numerous components of the mitochondrial translation machinery, consistent with the functional significance of this pathway in OxPhos-DLBCLs. These observations raise the question as to the nature of this programmatic increase in the expression of the mitochondrial translation pathway. It is possible that increased level of mitochondrial translation proteins is transcriptionally regulated. Interestingly, several transcription factors that regulate OXPHOS genes also activate the expression of mitochondrial translation factors (cMYC, estrogen-related receptor (ERR), and ying and yang 1 (YY1)).48 Evidence in human breast cancer biopsies indicates a similar increase in expression of mitochondrial translation factors that appears to be associated with the expression of nuclear respiratory factor 1 (NRF1), and peroxisome proliferator-activated receptor gamma coactivator (PGC) 1-α, which are transcription factors known to stimulate mitochondrial biogenesis.49 In addition, post-transcriptional mechanisms that can account for increased expression of mitochondrial translation proteins cannot be ruled out. The molecular basis for the observed enrichment of mitochondrial translation factors in OxPhos-DLBCLs awaits future studies.
The investigation of metabolic distinctions among DLBCL subtypes has led to the identification of mitochondrial pathways that provide selective pro-survival benefits to OxPhos-DLBCLs. Expanding on our previous studies, which identified mitochondrial FAO as an OxPhos-DLBCL-selective survival pathway,7 we now show selective dependency of this DLBCL subtype on the mitochondrial translation pathway. Identification and targeting of OxPhos-dependent survival mechanisms may have important clinical utility. First, although downstream inhibitors of BCR signaling, including small-molecule inhibitors of spleen tyrosine kinase and Bruton tyrosine kinase kinases, are being evaluated in DLBCL,50, 51 there are currently no clinically approved targeted therapeutic strategies for OxPhos-type DLBCLs. Second, select targeting of OxPhos-type survival mechanisms may have broader implications for resistance mechanisms that enable tumors to escape inhibition of canonical growth factor signaling, including those initiated by BCR, RAS, and BRAF signaling pathways.7, 52, 53 Notably, tigecycline is FDA-approved and is being actively developed for its potential therapeutic benefits in several diseases.54 Our findings warrant investigation of the therapeutic utility of tigecycline and other inhibitors of the mitochondrial translation pathway in DLBCL and other OxPhos-dependent tumors.
Materials and Methods
DLBCL cell lines
DLBCL cell lines used in this study and their consensus cluster assignments have been previously described,27, 55, 56 and further detailed in Supplementary Information.
Mitochondria isolation
Cells were resuspended in mitochondria isolation buffer (MIB; 200 mM mannitol, 70 mM sucrose; 1 mM EGTA; 10 mM HEPES, pH 7.4) containing protease inhibitors, homogenized with 20 strokes of a teflon-glass homogenizer, and resuspended in MIB. The nuclei and cell debris were removed by two consecutive centrifugations at 1000 × g for 5 min and the supernatant containing crude mitochondria was centrifuged twice at 9000 × g for 20 min. The resultant pellet contained mitochondria-enriched heavy membrane (HM) fraction. For enzyme activity assays, the HM fraction was resuspended in MIB.
Sample preparation and iTRAQ labeling
Isolated mitochondria from three independent OxPhos- (Karpas 422, Pfeiffer, and Toledo) and three non-OxPhos/BCR- (Ly1, DHL4, and DHL6) DLBCL cell lines were solubilized in 7.2 M guanidine hydrochloride with 100 mM ammonium bicarbonate and protein concentrations determined by Bradford assay (Bio-Rad, Hercules, CA, USA). Equal amounts of protein were reduced with DTT (10 mM final concentration) for 30 min at 56 ºC, and alkylated with iodoacetamide (22.5 mM) for 30 min at room temperature in the dark. After adding additional DTT (final concentration 20 mM) and diluting guanidine hydrochloride concentration to 1 M with 100 mM ammonium bicarbonate, proteins were digested with trypsin overnight at 37 ºC. Digests were acidified with 10% TFA and desalted by C18. Peptides (50 μg) from the above cell lines were solubilized in 100 μl of 30% 500 mM triethylammonium bicarbonate, pH 8.5/70% ethanol and 1 U of iTRAQ 8-plex reagent was added to each sample (K422-113, Toledo-114, Pfeiffer-115, Ly1-117, DHL4-118, DHL6-119). Reactions were incubated for 1 h at room temperature, combined, dried by vacuum centrifugation, desalted by C18, and dried again.
DEEP SEQ mass spectrometry and data analysis
iTRAQ-labeled peptides (50 μg) were subjected to multidimensional fractionation with a modified NanoAcquity UPLC system (Waters, Milford, MA, USA) consisting of two binary pumps, an autosampler, and an additional 6-port, 2-position valve (Valco, Austin, TX, USA).25 First dimension separations were conducted at high pH (10.0) using a reversed-phase column (200 μm ID fused silica × 20 cm packed with 5 μM XBridge C18). In the second dimension, peptides were fractionated by strong anion-exchange chromatography (200 μm fused silica x 20 cm packed with 5 μM SAX; SEPAX technologies, Neward, DE, USA). Peptides were eluted from first and second dimensions using solutions of acetonitrile and ammonium formate (pH 10), trapped on the final dimension precolumn (200 μm ID fused silica × 4 cm of POROS 10R2) after in-line dilution with 0.1% formic acid, resolved on an analytical column (25 μm ID fused silica packed with 100 cm of 5 μm Monitor C18 (Column Engineering, Ontario, CA, USA), 2–50% B in 580 min, A=0.1% formic acid, B=acetonitrile with 0.1% formic acid) and subjected to MS/MS (5600 Triple TOF mass spectrometer, ABI, Framingham, MA, USA). Replicate analyses were performed each with eight total fractions.
Raw mass spectrometry data files were converted to. mgf using ABSciex MS Data Converter version 1.3 (ABSciex, Framingham, MA, USA) and searched using Mascot version 2.2.1 (Matrix Science, Boston, MA, USA) after recalibration of precursor and product ions using multiplierz scripts.57, 58 Search parameters specified precursor and product ion tolerances of 0.5 Da, trypsin specificity, up to two missed cleavages, fixed carbamidomethylation (C), variable oxidation (M), and fixed iTRAQ modification (N-term, K). Additional multiplierz scripts were used to filter search results to a 1% false discovery rate, remove reverse database hits and identifications with mass deviations >25 p.p.m., and extract iTRAQ reporter ion intensities, which were corrected for isotopic impurities as well as minor variations in source protein concentration. Abundance ratios were derived by summing reporters for all peptides mapping to unique genes across replicates.
RNA interference
Lentiviral vectors containing short hairpins targeting GFM1 (TRCN0000141319 and TRCN0000144593), TUFM (TRCN0000160152 and TRCN0000165471), and MRPS7 (TRCN0000117458 and TRCN0000117459) were purchased from The RNAi Consortium (TRC, The Broad Institute, Cambridge, MA, USA). An empty pLKO.1 vector was used for control. Viral supernatants were used to spinfect 5 × 105 cells for 2 h at 460 × g as previously described.7 The effect of knockdown on cell viability and protein depletion was assessed 24 h after viral infection. Knockdown efficiency was assessed using western blotting.
Tigecycline treatment
Tigecycline (SRP02356t; Sequoia Research Products, Pangbourne, UK) was prepared as a 1 mM stock solution in IMDM (12440-046; Invitrogen, Carlsbad, CA, USA) protected from light as previously described.59 Tigecycline was used at 1–5 μM concentrations for 24–72 h treatment durations as detailed in figure legends.
Viability and proliferation assays
Cell viability was measured using the Annexin V/FITC Apoptosis Detection kit (BD Biosciences, San Jose, CA, USA), followed by flow cytometry. Cell proliferation was measured by EdU uptake. In brief, cells were seeded at 5 × 105 cells per ml in six-well plates and co-treated with 10 μM EdU and 2.5 μM tigecycline for 24 h. A total of 1 × 105 cells were collected and stained using the Click-iT EdU Alexa Fluor 647 Flow Cytometry Assay kit (C10424; Life Technologies, Beverly, MA, USA) following the manufacturer’s instructions. All data points were subtracted from background and normalized to untreated controls.
Biochemical measurement of respiratory chain enzyme activity
NADH dehydrogenase and succinate dehydrogenase activities were measured in 100 and 40 μg, respectively, of isolated mitochondria using immunocapture-based enzyme activity assays (MitoScience, Eugene, OR, USA) as previously described.7
Analysis of mitochondrial respiratory chain supercomplexes
Mitochondrial supercomplexes were examined using Blue-Native electrophoresis as described by Acín-Pérez et al.60 In brief, cells were treated with 1 μM tigecycline for 24 h. Following treatment, 2.5 × 106 cells were collected from each condition, washed twice in PBS, and permeabilized with 0.2% digitonin (D5628-1G; Sigma, St Louis, MO, USA) in PBS for 10 min on ice. Mitochondria were isolated following centrifugation at 10 000 × g for 5 min at 4 ºC, and solubilized in native sample buffer (BN2003; Invitrogen) containing 1% digitonin for 5 min on ice. The lysates were cleared by centrifugation at 18 000 × g for 30 min at 4 ºC. The supernatant was loaded on a 3–12% Invitrogen Blue-Native gel system (BN1001BOX; Invitrogen) and transferred to a PVDF membrane. Supercomplexes were probed using an antibody against a complex III component, ubiquinol-cytochrome c reductase core protein I (UQCRC1; ab110252; Abcam, Cambridge, MA USA). Mitochondrial supercomplexes were quantified by densitometry using ImageJ (National Institutes of Health, Bethesda, MD, USA) and normalized to the mitochondrial aconitase abundance in each sample.
Measurement of mitochondrial SRC
Oxygen consumption rate (OCR) was measured in real time using the XF24e Extracellular Flux Analyzer instrument and the Wave 2.2.0 software (Agilent Technologies, Santa Clara, CA, USA) as previously described.7 Cells were treated with 1 μM tigecycline for 24 h before seeding on XF24e V7 plates coated with 50 μg/ml of poly-l-lysine (P2636; Sigma), and seeded at 3 × 105 cells per well (with the exception of Toledo and Ly1, which were seeded at 4 × 105 and 3.5 × 105 cells per well, respectively) in 600 μl of sodium bicarbonate-free RPMI medium (US Biological, Salem, MA, USA) supplemented with standard concentrations of the amino acids, 10% FBS, 10 mM d-glucose, 5 mM sodium pyruvate, and 5 mM l-glutamine. To adhere cells to poly-l-lysine-coated plates, the plates were centrifuged at 400 r.p.m. and incubated at 37 °C for 10 min. After baseline measurements, the following order of additions were made using the instrument’s individual injection ports: 2.5 μM oligomycin to determine ATP-coupled OCR, 3 μM FCCP to determine maximal OCR, and a combination of 1 μM antimycin A and 2.5 μM rotenone to inhibit mitochondrial respiration. SRC was deduced from the difference between maximal and basal OCR. SRC values for each cell line were normalized to protein content as measured using a BCA assay (23228; Thermo Scientific, Cambridge, MA, USA).
Determination of mitochondrial superoxide content
Mitochondrial superoxide was measured using MitoSOX Red (Molecular Probes, Eugene, OR, USA) as previously described.7 All values were normalized to mitochondrial membrane potential as measured by TMRE (Molecular Probes).
NAC treatment
NAC (Sigma) was used as follows. In knockdown studies, 0.5 mM NAC was added directly to the media following infection of DLBCL cell lines with lentiviral particles bearing shRNA against GFM1, TUFM, and MRPS7. For reversal of tigecycline effect on ROS accumulation (Figure 5c), cells were pretreated with 1 mM NAC before tigecycline treatment.
Analysis of primary DLBCL samples
Cryopreserved viable primary DLBCL samples were obtained according to Institutional Review Board-approved protocols from Brigham and Women’s Hospital and the Dana-Farber Cancer Institute. These anonymous primary tumor specimens were considered discarded tissues that did not require informed consent. Each primary DLBCL sample was previously classified as BCR- or OxPhos-DLBCL using the CCC.16 For assessment of tigecycline sensitivity, cryopreserved viable primary DLBCL samples were purified using a Ficoll gradient as previously described,16 and tumor cells were treated with the indicated concentrations of tigecycline for 20 h before assessment of cell viability.
Statistical analysis
All values are presented as mean±S.E.M. Statistical significance was determined using two-tailed Student’s t-test. Significance indicated by P-values as follows: *P<0.05, **P<0.01, ***P<0.001.
Abbreviations
- DLBCL:
-
diffuse large B-cell lymphoma
- BCR:
-
B-cell receptor
- OxPhos:
-
oxidative phosphorylation
- ETC:
-
electron transport chain
- mtDNA:
-
mitochondrial DNA
- COO:
-
cell-of-origin
- CCC:
-
consensus cluster classification
- FAO:
-
fatty acid oxidation
- iTRAQ:
-
isobaric tags for relative and absolute quantification
- DEEP SEQ:
-
deep efficient peptide sequencing and quantification
- TUFM:
-
Tu translation elongation factor mitochondrial
- GFM1:
-
G elongation factor mitochondria 1
- MRP:
-
mitochondrial ribosomal protein
- shRNA:
-
short hairpin RNA
- ROS:
-
reactive oxygen species
- NAC:
-
N-acetyl cysteine
- SRC:
-
spare respiratory capacity
- AML:
-
acute myeloid leukemia
References
Metallo CM, Vander Heiden MG . Understanding metabolic regulation and its influence on cell physiology. Mol Cell 2013; 49: 388–398.
Stanley IA, Ribeiro SM, Gimenez-Cassina A, Norberg E, Danial NN . Changing appetites: the adaptive advantages of fuel choice. Trends Cell Biol 2014; 24: 118–127.
Boroughs LK, DeBerardinis RJ . Metabolic pathways promoting cancer cell survival and growth. Nat Cell Biol 2015; 17: 351–359.
DeNicola GM, Cantley LC . Cancer's fuel choice: new flavors for a picky eater. Mol Cell 2015; 60: 514–523.
Lunt SY, Vander Heiden MG . Aerobic glycolysis: meeting the metabolic requirements of cell proliferation. Annu Rev Cell Dev Biol 2011; 27: 441–464.
Koppenol WH, Bounds PL, Dang CV . Otto Warburg's contributions to current concepts of cancer metabolism. Nat Rev Cancer 2011; 11: 325–337.
Caro P, Kishan AU, Norberg E, Stanley IA, Chapuy B, Ficarro SB et al. Metabolic signatures uncover distinct targets in molecular subsets of diffuse large B cell lymphoma. Cancer Cell 2012; 22: 547–560.
Mashimo T, Pichumani K, Vemireddy V, Hatanpaa KJ, Singh DK, Sirasanagandla S et al. Acetate is a bioenergetic substrate for human glioblastoma and brain metastases. Cell 2014; 159: 1603–1614.
Obre E, Rossignol R . Emerging concepts in bioenergetics and cancer research: Metabolic flexibility, coupling, symbiosis, switch, oxidative tumors, metabolic remodeling, signaling and bioenergetic therapy. Int J Biochem Cell Biol 2015; 59C: 167–181.
Schug ZT, Peck B, Jones DT, Zhang Q, Grosskurth S, Alam IS et al. Acetyl-CoA Synthetase 2 Promotes Acetate Utilization and Maintains Cancer Cell Growth under Metabolic Stress. Cancer Cell 2015; 27: 57–71.
Weinberg F, Hamanaka R, Wheaton WW, Weinberg S, Joseph J, Lopez M et al. Mitochondrial metabolism and ROS generation are essential for Kras-mediated tumorigenicity. Proc Natl Acad Sci USA 2010; 107: 8788–8793.
Yuneva MO, Fan TW, Allen TD, Higashi RM, Ferraris DV, Tsukamoto T et al. The metabolic profile of tumors depends on both the responsible genetic lesion and tissue type. Cell Metab 2012; 15: 157–170.
Wright G, Tan B, Rosenwald A, Hurt EH, Wiestner A, Staudt LM . A gene expression-based method to diagnose clinically distinct subgroups of diffuse large B cell lymphoma. Proc Natl Acad Sci USA 2003; 100: 9991–9996.
Monti S, Savage KJ, Kutok JL, Feuerhake F, Kurtin P, Mihm M et al. Molecular profiling of diffuse large B-cell lymphoma identifies robust subtypes including one characterized by host inflammatory response. Blood 2005; 105: 1851–1861.
Monti S, Chapuy B, Takeyama K, Rodig SJ, Hao Y, Yeda KT et al. Integrative analysis reveals an outcome-associated and targetable pattern of p53 and cell cycle deregulation in diffuse large B cell lymphoma. Cancer Cell 2012; 22: 359–372.
Chen L, Monti S, Juszczynski P, Ouyang J, Chapuy B, Neuberg D et al. SYK inhibition modulates distinct PI3K/AKT- dependent survival pathways and cholesterol biosynthesis in diffuse large B cell lymphomas. Cancer Cell 2013; 23: 826–838.
Pfeifer M, Grau M, Lenze D, Wenzel SS, Wolf A, Wollert-Wulf B et al. PTEN loss defines a PI3K/AKT pathway-dependent germinal center subtype of diffuse large B-cell lymphoma. Proc Natl Acad Sci USA 2013; 110: 12420–12425.
Gustafsson CM, Falkenberg M, Larsson NG . Maintenance and Expression of Mammalian Mitochondrial DNA. Annu Rev Biochem 2016; 85: 133–160.
Rebelo AP, Dillon LM, Moraes CT, Mitochondrial DNA . transcription regulation and nucleoid organization. J Inherit Metab Dis 2011; 34: 941–951.
Ott M, Amunts A, Brown A . Organization and Regulation of Mitochondrial Protein Synthesis. Annu Rev Biochem 2016; 85: 77–101.
Shahni R, Wedatilake Y, Cleary MA, Lindley KJ, Sibson KR, Rahman S . A distinct mitochondrial myopathy, lactic acidosis and sideroblastic anemia (MLASA) phenotype associates with YARS2 mutations. Am J Med Genet A 2013; 161: 2334–2338.
Baker MJ, Tatsuta T, Langer T . Quality control of mitochondrial proteostasis. Cold Spring Harb Perspect Biol 2011; 3: a00755.
Enriquez JA . Supramolecular organization of respiratory complexes. Annu Rev Physiol 2016; 78: 533–561.
Zhou F, Lu Y, Ficarro SB, Webber JT, Marto JA . Nanoflow low pressure high peak capacity single dimension LC-MS/MS platform for high-throughput, in-depth analysis of mammalian proteomes. Anal Chem 2012; 84: 5133–5139.
Zhou F, Lu Y, Ficarro SB, Adelmant G, Jiang W, Luckey CJ et al. Genome-scale proteome quantification by DEEP SEQ mass spectrometry. Nat Commun 2013; 4: 2171.
Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature 2000; 403: 503–511.
Polo JM, Juszczynski P, Monti S, Cerchietti L, Ye K, Greally JM et al. Transcriptional signature with differential expression of BCL6 target genes accurately identifies BCL6-dependent diffuse large B cell lymphomas. Proc Natl Acad Sci USA 2007; 104: 3207–3212.
Bestwick ML, Shadel GS . Accessorizing the human mitochondrial transcription machinery. Trends Biochem Sci 2013; 38: 283–291.
Echevarria L, Clemente P, Hernandez-Sierra R, Gallardo ME, Fernandez-Moreno MA, Garesse R . Glutamyl-tRNAGln amidotransferase is essential for mammalian mitochondrial translation in vivo. Biochem J 2014; 460: 91–101.
Rorbach J, Richter R, Wessels HJ, Wydro M, Pekalski M, Farhoud M et al. The human mitochondrial ribosome recycling factor is essential for cell viability. Nucleic Acids Res 2008; 36: 5787–5799.
Jenner L, Starosta AL, Terry DS, Mikolajka A, Filonava L, Yusupov M et al. Structural basis for potent inhibitory activity of the antibiotic tigecycline during protein synthesis. Proc Natl Acad Sci USA 2013; 110: 3812–3816.
Muralidharan G, Micalizzi M, Speth J, Raible D, Troy S . Pharmacokinetics of tigecycline after single and multiple doses in healthy subjects. Antimicrob Agents Chemother 2005; 49: 220–229.
Wenzel R, Bate G, Kirkpatrick P . Tigecycline. Nat Rev Drug Discov 2005; 4: 809–810.
Skrtic M, Sriskanthadevan S, Jhas B, Gebbia M, Wang X, Wang Z et al. Inhibition of mitochondrial translation as a therapeutic strategy for human acute myeloid leukemia. Cancer Cell 2011; 20: 674–688.
Acin-Perez R, Bayona-Bafaluy MP, Fernandez-Silva P, Moreno-Loshuertos R, Perez-Martos A, Bruno C et al. Respiratory complex III is required to maintain complex I in mammalian mitochondria. Mol Cell 2004; 13: 805–815.
Diaz F, Fukui H, Garcia S, Moraes CT . Cytochrome c oxidase is required for the assembly/stability of respiratory complex I in mouse fibroblasts. Mol Cell Biol 2006; 26: 4872–4881.
Schagger H, de Coo R, Bauer MF, Hofmann S, Godinot C, Brandt U . Significance of respirasomes for the assembly/stability of human respiratory chain complex I. J Biol Chem 2004; 279: 36349–36353.
Maranzana E, Barbero G, Falasca AI, Lenaz G, Genova ML . Mitochondrial respiratory supercomplex association limits production of reactive oxygen species from complex I. Antioxid Redox Signal 2013; 19: 1469–1480.
Dranka BP, Hill BG, Darley-Usmar VM . Mitochondrial reserve capacity in endothelial cells: The impact of nitric oxide and reactive oxygen species. Free Radic Biol Med 2010; 48: 905–914.
Nicholls DG . Spare respiratory capacity, oxidative stress and excitotoxicity. Biochem Soc Trans 2009; 37: 1385–1388.
Orrenius S, Gogvadze V, Zhivotovsky B . Mitochondrial oxidative stress: implications for cell death. Annu Rev Pharmacol Toxicol 2007; 47: 143–183.
Schieber M, Chandel NS . ROS function in redox signaling and oxidative stress. Curr Biol 2014; 24: R453–R462.
Brand MD . Mitochondrial generation of superoxide and hydrogen peroxide as the source of mitochondrial redox signaling. Free Radic Biol Med 2016 (e-pub ahead of print 13 April 2016; doi:10.1016/j.freeradbiomed.2016.04.001).
Murphy MP . How mitochondria produce reactive oxygen species. Biochem J 2009; 417: 1–13.
Nagar H, Jung SB, Kwon SK, Park JB, Shong M, Song HJ et al. CRIF1 deficiency induces p66shc-mediated oxidative stress and endothelial activation. PLoS One 2014; 9: e98670.
Schedlbauer A, Kaminishi T, Ochoa-Lizarralde B, Dhimole N, Zhou S, Lopez-Alonso JP et al. Structural characterization of an alternative mode of tigecycline binding to the bacterial ribosome. Antimicrob Agents Chemother 2015; 59: 2849–2854.
Amunts A, Brown A, Toots J, Scheres SH, Ramakrishnan V . Ribosome. The structure of the human mitochondrial ribosome. Science 2015; 348: 95–98.
Scarpulla RC, Vega RB, Kelly DP . Transcriptional integration of mitochondrial biogenesis. Trends Endocrinol Metab 2012; 23: 459–466.
Sotgia F, Whitaker-Menezes D, Martinez-Outschoorn UE, Salem AF, Tsirigos A, Lamb R et al. Mitochondria "fuel" breast cancer metabolism: fifteen markers of mitochondrial biogenesis label epithelial cancer cells, but are excluded from adjacent stromal cells. Cell Cycle 2012; 11: 4390–4401.
Friedberg JW, Sharman J, Sweetenham J, Johnston PB, Vose JM, Lacasce A et al. Inhibition of Syk with fostamatinib disodium has significant clinical activity in non-Hodgkin lymphoma and chronic lymphocytic leukemia. Blood 2010; 115: 2578–2585.
Younes A, Thieblemont C, Morschhauser F, Flinn I, Friedberg JW, Amorim S et al. Combination of ibrutinib with rituximab, cyclophosphamide, doxorubicin, vincristine, and prednisone (R-CHOP) for treatment-naive patients with CD20-positive B-cell non-Hodgkin lymphoma: a non-randomised, phase 1b study. Lancet Oncol 2014; 15: 1019–1026.
Corazao-Rozas P, Guerreschi P, Jendoubi M, Andre F, Jonneaux A, Scalbert C et al. Mitochondrial oxidative stress is the Achille's heel of melanoma cells resistant to Braf-mutant inhibitor. Oncotarget 2013; 4: 1986–1998.
Viale A, Pettazzoni P, Lyssiotis CA, Ying H, Sanchez N, Marchesini M et al. Oncogene ablation-resistant pancreatic cancer cells depend on mitochondrial function. Nature 2014; 514: 628–632.
Bucaneve G, Micozzi A, Picardi M, Ballanti S, Cascavilla N, Salutari P et al. Results of a multicenter, controlled, randomized clinical trial evaluating the combination of piperacillin/tazobactam and tigecycline in high-risk hematologic patients with cancer with febrile neutropenia. J Clin Oncol 2014; 32: 1463–1471.
Chen L, Monti S, Juszczynski P, Daley J, Chen W, Witzig TE et al. SYK-dependent tonic B-cell receptor signaling is a rational treatment target in diffuse large B-cell lymphoma. Blood 2008; 111: 2230–2237.
Davis RE, Ngo VN, Lenz G, Tolar P, Young RM, Romesser PB et al. Chronic active B-cell-receptor signalling in diffuse large B-cell lymphoma. Nature 2010; 463: 88–92.
Askenazi M, Parikh JR, Marto JA . mzAPI: a new strategy for efficiently sharing mass spectrometry data. Nat Methods 2009; 6: 240–241.
Parikh JR, Askenazi M, Ficarro SB, Cashorali T, Webber JT, Blank NC et al. multiplierz: an extensible API based desktop environment for proteomics data analysis. BMC Bioinformatics 2009; 10: 364.
Jitkova Y, Gronda M, Hurren R, Wang X, Goard CA, Jhas B et al. A novel formulation of tigecycline has enhanced stability and sustained antibacterial and antileukemic activity. PLoS One 2014; 9: e95281.
Acin-Perez R, Fernandez-Silva P, Peleato ML, Perez-Martos A, Enriquez JA . Respiratory active mitochondrial supercomplexes. Mol Cell 2008; 32: 529–539.
Acknowledgements
We thank Elaura Patton, Meghan Tedoldi, and Heather Sun for technical assistance, Rebecca Acin-Pérez and José Antonio Enriquez for advice on mitochondrial supercomplexes, and Benjamin Szlyk for graphics. This work was supported by the US National Institutes of Health grants R21 CA178860 (NND and JAM), V Foundation for Cancer Research (NND and MAS), F31 CA171400 (IAS), the Swedish Society for Medical Research (SSMF), and The Malin and Lennart Philipson Foundation (EN). We also acknowledge generous support provided through the Dana-Farber Cancer Institute Strategic Research Initiative and CA188881 (JAM).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by N Chandel
Supplementary Information accompanies this paper on Cell Death and Differentiation website
Supplementary information
Rights and permissions
About this article
Cite this article
Norberg, E., Lako, A., Chen, PH. et al. Differential contribution of the mitochondrial translation pathway to the survival of diffuse large B-cell lymphoma subsets. Cell Death Differ 24, 251–262 (2017). https://doi.org/10.1038/cdd.2016.116
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cdd.2016.116
This article is cited by
-
SIRT1 and HSP90α feed-forward circuit safeguards chromosome segregation integrity in diffuse large B cell lymphomas
Cell Death & Disease (2023)
-
A comprehensive analysis of SLC25A1 expression and its oncogenic role in pan-cancer
Discover Oncology (2023)
-
Mitochondrial adaptation in cancer drug resistance: prevalence, mechanisms, and management
Journal of Hematology & Oncology (2022)
-
Fatty acid metabolism in aggressive B-cell lymphoma is inhibited by tetraspanin CD37
Nature Communications (2022)
-
Simultaneous targeting of glycolysis and oxidative phosphorylation as a therapeutic strategy to treat diffuse large B-cell lymphoma
British Journal of Cancer (2022)