Abstract
Coxsackievirus infection induces an abnormal accumulation of ubiquitin aggregates that are generally believed to be noxious to the cells and have a key role in viral pathogenesis. Selective autophagy mediated by autophagy adaptor proteins, including sequestosome 1 (SQSTM1/p62) and neighbor of BRCA1 gene 1 protein (NBR1), are an important pathway for disposing of misfolded/ubiquitin conjugates. We have recently demonstrated that SQSTM1 is cleaved after coxsackievirus infection, resulting in the disruption of SQSTM1 function in selective autophagy. NBR1 is a functional homolog of SQSTM1. In this study, we propose to test whether NBR1 can compensate for the compromise of SQSTM1 after viral infection. Of interest, we found that NBR1 was also cleaved after coxsackievirus infection. This cleavage took place at two sites mediated by virus-encoded protease 2Apro and 3Cpro, respectively. In addition to the loss-of-function, we further investigated whether cleavage of SQSTM1/NBR1 leads to the generation of toxic gain-of-function mutants. We showed that the C-terminal fragments of SQSTM1 and NBR1 exhibited a dominant-negative effect against native SQSTM1/NBR1, probably by competing for LC3 and ubiquitin chain binding. Finally, we demonstrated a positive, mutual regulatory relationship between SQSTM1 and NBR1 during viral infection. We showed that knockdown of SQSTM1 resulted in reduced expression of NBR1, whereas overexpression of SQSTM1 led to increased level of NBR1, and vice versa, further excluding the possible compensation of NBR1 for the loss of SQSTM1. Taken together, the findings in this study suggest a novel mechanism through which coxsackievirus infection induces increased accumulation of ubiquitin conjugates and subsequent viral damage.
Similar content being viewed by others
Main
Autophagy is a conserved biological process and has long been regarded as a non-selective, bulk degradative pathway to maintain the homeostasis of cellular environment under stress and starvation.1 However, accumulating evidence indicates that autophagy-mediated degradation is more selective than originally thought. Several autophagy adapter proteins, including sequestosome 1 (SQSTM1/p62) and neighbor of BRCA1 gene 1 protein (NBR1), have been identified and found to be essential in mediating selective autophagy.2, 3 They share a similar domain architecture containing an N-terminal Phox/Bem1p (PB1) domain, a microtubule-associated protein light chain (LC3)-interacting region (LIR), and a C-terminal ubiquitin association (UBA) domain, which allow them to recognize and specifically target substrates to autophagosome for degradation. Although the mechanisms underlying specific substrate selection remain not completely understood, increasing evidence supports that ubiquitination serves as a recognition signal for autophagic degradation of misfolded proteins and damaged organelles.4
The accumulation of damaged organelles and protein aggregates are believed to be harmful to the cells and cannot be efficiently removed by the proteasomes.4, 5, 6 Our previous studies have demonstrated an aberrant accumulation of misfolded/ubiquitin protein aggregates in coxsackievirus-infected cells and mouse hearts, suggesting that defective protein degradation may have a role in viral pathogenesis.7, 8, 9 Coxsackievirus type B3 (CVB3), an enterovirus belonging to the family Picornaviridae, is a common cause of viral myocarditis and several other diseases in human.10 We have recently demonstrated that autophagy adapter protein SQSTM1 is cleaved through the proteolytic activity of viral protease 2Apro.11 The cleavage of SQSTM1 results in the dissociation of the N-terminal PB1 domain from the C-terminal UBA and LIR domains, and subsequently the loss-of-function of full-length SQSTM1 in mediating selective autophagy.11
NBR1 is a functional homolog of SQSTM1. Despite the difference of its primary sequence and size from those of SQSTM1, NBR1 shares a similar domain structure with SQSTM1, both containing PB1, ZZ (zinc-finger domain), LIR and UBA domains.12 Depending on the characteristics of the substrates, NBR1 and SQSTM1 work either independently or cooperatively with each other. It was reported that degradation of bacteria, such as Salmonella and Listeria, requires SQSTM1, but not NBR1.13 In contrast, NBR1 alone is sufficient to target peroxisomes to lysosomes for degradation in the absence of SQSTM1.14 NBR1 can also work in concert with SQSTM1 to remove ubiquitinated cargoes.12 The observation that deletion of SQSTM1 fails to induce the accumulation of ubiquitinated proteins implies a possible compensatory role for NBR1 in the compromise of SQSTM1 function.3, 15
The purpose of this study was to determine whether NBR1 could compensate for disrupted SQSTM1 function following coxsackievirus infection. Here we showed that NBR1 was also cleaved during CVB3 infection by virus-encoded protease 2Apro and 3Cpro. We further provided evidence that cleavage of both SQSTM1 and NBR1 not only led to the loss of their native function, but also resulted in the generation of cleavage mutants that display dominant-negative activities. Finally, we demonstrated a mutual regulatory relationship between SQSTM1 and NBR1 during viral infection. Our data suggest a new mechanism through which coxsackievirus infection induces abnormal accumulation of ubiquitin conjugates.
Results
Cleavage of NBR1 following CVB3 infection
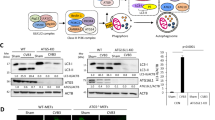
We have previously demonstrated that SQSTM1 is cleaved during coxsackievirus infection, resulting in the disruption of the function of SQSTM1.11 As NBR1 may have a compensatory role for the loss of SQSTM1, we examined NBR1 expression after CVB3 infection. We found that NBR1 was also potentially cleaved after CVB3 infection, generating at least two cleavage fragments (∼100 and ∼50 kDa, respectively) using an anti-N-terminal NBR1 antibody (Figure 1a). Gene expression of NBR1 appeared unaltered after CVB3 infection (Figure 1b).
Cleavage of NBR1 following CVB3 infection. (a) Protein level of NBR1 after CVB3 infection. HeLa cells were sham-infected with PBS or infected with CVB3 for indicated times at an multiplicity of infection (MOI) of 10. Western blot analysis was performed to examine the expression of NBR1 using anti-N-terminal NBR1 antibody, which recognizes amino acids 1∼150 (Santa Cruz Biotechnology), viral capsid protein VP1 and β-actin (loading controls). (b) Gene expression of NBR1 after CVB3 infection. HeLa cells were either sham-infected or infected with CVB3 for 7 h. Gene expression of NBR1 was examined by real-time quantitative RT-PCR and normalized to GAPDH mRNA (mean±S.D., n=3). (c) Cleavage of NBR1 after CVB3 infection. HeLa cells were transiently transfected with a plasmid-expressing HA-tagged NBR1 for 24 h, followed by CVB3 infection for 7 h. Western blotting was carried out to examine protein expression of exogenous NBR1 using anti-HA antibody, which recognizes the N-terminus of NBR1. For size comparison, western blotting of endogenous NBR1 using anti-N-terminal NBR1 antibody was presented on the left lane. Protein expression of β-actin was measured as the loading control. pi, post-infection
To verify the cleavage of NBR1, cells were transiently transfected with an HA-tagged NBR1 construct, followed by CVB3 infection. The cleavage products of exogenous NBR1 detected by an anti-HA antibody (Figure 1c) corresponded well with those of the endogenous NBR1 (Figure 1a), confirming that NBR1 is cleaved after CVB3 infection.
Cleavage of NBR1 is mediated by coxsackieviral proteases 2Apro and 3Cpro
To explore whether coxsackievirus proteases are responsible for NBR1 cleavage, we performed in vitro cleavage assays using purified coxsackieviral 2Apro and 3Cpro. We showed that incubation of cell lysates with purified 2Apro and 3Cpro resulted in the time-dependent generation of NBR1 cleavage products of ∼50 and ∼100 kDa, respectively, similar to those observed in CVB3-infected cells (Figures 2a and b). 2Apro mutant failed to induce the cleavage of NBR1 (Figure 2a). These results suggest that the cleavage of NBR1 is mediated by both coxsackieviral 2Apro and 3Cpro.
Cleavage of NBR1 by coxsackieviral 2Apro and 3Cpro. (a and b) Effect of viral protease 2Apro and 3Cpro on NBR1 cleavage by in vitro cleavage assay. Fifty microgram of protein extracts from HeLa cells was incubated with 0.1 μg of purified coxsackieviral 2Apro (a), 0.1 μg of catalytic 2Apro mutant (a) or 0.1 μg of 3Cpro (b) for increasing periods of time as indicated. (c) Effect of general caspase inhibition on CVB3-induced NBR1 cleavage. HeLa cells were infected with CVB3 for 7 h with or without pan caspase inhibitor Z-VAD-FMK (50 μM). Western blotting was performed to examine protein expression of NBR1 using anti-N-terminal NBR1 antibody, VP1, cleaved caspase-3 and β-actin (loading control). ‘–’, vehicle control
As CVB3 infection results in caspase activation,16 we further examined whether caspase activation has a role in the cleavage of NBR1. Figure 2c showed that treatment with a pan caspase inhibitor Z-VAD-FMK inhibited CVB3-induced caspase-3 cleavage, but did not block the production of NBR1 cleavage fragments, indicating that cleavage of NBR1 is independent of caspase activity.
Identification of the cleavage sites on NBR1
Using a computer server for prediction of cleavage sites by enteroviral proteases (NetPicoRNA V1.0 algorithm), we found two potential cleavage sites by viral protease 3Cpro around the C-terminus of NBR1: residue 682 (FALPE/GPLG) and residue 612 (EEENE/GAGF), which result in the production of a protein of ∼100 kDa, consistent with the high molecular weight fragment detected in CVB3-infected cells. To determine the precise cleavage site, we constructed two NBR1 point mutants (NBR1-E682K and NBR1-E612K, in which glutamic acid (E) residue was replaced with lysine (K) residue). Figure 3a showed that the ∼100 kDa cleavage fragment disappeared, while the ∼50 kDa fragment remained in cells expressing NBR1-E682K, indicating that E682 is a cleavage site on NBR1 after CVB3 infection (Figure 3a).
Identification of the cleavage sites on NBR1. (a and b) Identification of NBR1 cleavage sites. HeLa cells were transiently transfected with HA-NBR1-WT, HA-NBR1-G402E (glycine (G) at amino-acid 402 was replaced by glutamic acid (E)) (a), or HA-NBR1-E682K (glutamic acid (E) at amino-acid 682 was replaced by lysine (K)) (b) for 24 h, followed by sham or CVB3 infection for 7 h. Western blotting was performed to detect various forms of NBR1 using anti-HA antibody. Protein level of β-actin was examined as a loading control. (c) Schematic diagram of the structural domains and the identified cleavage sites on NBR1. PB1, Phox/Bem1p domain; ZZ, zinc-finger domain; CC, coiled-coil domain; LIR, LC3-interacting region; UBA, ubiquitin association domain
Amino-acid sequence alignment of NBR1 with other known viral protease 2Apro substrates, including SQSTM1, reveals a potential cleavage motif (MKNTG at residues 398–402) on NBR1. To determine whether NBR1 is cleaved at T/G, we constructed a point mutation plasmid NBR1-G402E, in which glycine (G) residue at position 402 was replaced with glutamic acid (E) residue. As shown in Figure 3b, the cleavage fragment (∼50 kDa) disappeared and the level of the ∼100 kDa fragment was reduced in cells expressing mutant NBR1 compared with cells expressing wild-type NBR1. This result suggests that NBR1 is primarily cleaved between T401 and G402, likely via the action of viral protease 2Apro. Schematic diagram presented in Figure 3c shows NBR1 structural domains, the identified cleavage sites on NBR1 and the resulting cleavage fragments.
Dominant-negative effects of the C-terminal cleavage fragments of SQSTM1 and NBR1
We have previously shown that cleavage of SQSTM1 as a consequence of CVB3 infection leads to the loss-of-function of full-length SQSTM1.11 Here we further investigated whether the cleavage products have toxic gain-of-function properties. In this study, we focused on the C-terminal fragment of SQSTM1 (SQSTM1-C) as it retains intact LIR and UBA domains, but lacks the critical structures at the N-terminus, which are essential for its function in selective autophagy (Figure 4a). Our hypothesis was that the C-terminal cleavage product of SQSTM1 acts as a dominant-negative mutant by competing with the native SQSTM1 for LC3 and ubiquitin chain binding.
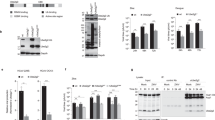
Dominant-negative effect of SQSTM1-C on native SQSTM1-mediated selective autophagy. (a) Schematic diagram of the structural domains and the identified cleavage site on SQSTM1. PB1, Phox/Bem1p domain; ZZ, zinc-finger domain; TB, tumor necrosis-associated factor 6 binding domain; CC, coiled-coil domain; LIR, LC3-interacting region; KIR, keap1-interacting region; UBA, ubiquitin association domain. (b and c) SQSTM1-C inhibits native SQSTM1-mediated formation of insoluble ubiquitin conjugates. HeLa cells were transiently transfected with an empty vector, SQSTM1-WT, SQSTM1-C alone or SQSTM1-WT together with SQSTM1-C for 48 h as indicated, followed by isolation of Triton X-100-soluble and -insoluble fractions. The levels of ubiquitin conjugates in Triton X-100-soluble (b) and -insoluble fractions (c) were analyzed by western blotting using anti-ubiquitin antibody. Protein expression levels of β-actin (b) and α-actinin (c) were examined as loading controls for Triton X-100-soluble and -insoluble fractions, respectively. (d) SQSTM1-C blocks the function of native SQSTM1 in promoting protein aggregate formation. HeLa cells were co-transfected with GFP-SQSTM1-WT, together with pcDNA empty vector, Flag-SQSTM1-WT, Flag-SQSTM1-N or Flag-SQSTM1-C for 48 h. Immunocytochemical staining was performed using anti-Flag antibody. The nucleus was counterstained with DAPI. Cell images were captured by confocal microscopy. The percentage represents the ratio of cells with GFP-SQSTM1 puncta relative to Flag-SQSTM1-WT, -N or -C co-expressing cells (mean±S.D., n=3–5 images, with 35 to 90 co-expressing cells in total in each image). *P=0.002 compared with SQSTM-WT control, by Student’s t-test. (e) Expression of SQSTM1-C results in an increased accumulation of endogenous full-length SQSTM1 and NBR1. HeLa cells were transiently transfected with empty vector or SQSTM1-C for 48 h. Western blot analysis was performed to examine the expression of endogenous SQSTM1 and NBR1, as well as SQSTM1-C as indicated. Expression of β-actin was measured as the loading control
Misfolded proteins and damaged organelles in the cells usually undergo ubiquitination and are present in detergent-insoluble fractions of the cell extracts. Similar to our previous observation,11 Triton-insoluble ubiquitin conjugates were accumulated in cells overexpressing SQSTM1-WT (Figures 4b and c). Interestingly, we found that SQSTM1-WT-mediated insoluble aggregate formation was blocked in the presence of SQSTM1-C (Figures 4b and c). Consistent with this observation, we showed that SQSTM1-WT was translocated from the Triton-insoluble fractions to Triton-soluble parts in cells overexpressing SQSTM1-C (Figures 4b and c).
We further explored the impacts of SQSTM1-C on the ability of native SQSTM1 to form aggregates. HeLa cells were co-transfected with GFP-SQSTM1-WT together with Flag-SQSTM1-WT, Flag-SQSTM1-N, or Flag-SQSTM1-C. Figure 4d showed that the number of cells with GFP puncta was greatly reduced in cells co-expressing Flag-SQSTM1-C compared with cells co-expressing Flag-SQSTM1-WT, that is, 29.93% versus 79.19% (P<0.05). SQSTM1-N appeared to have no effects on WT-SQSTM1-induced protein aggregate formation, that is, 74.82% versus 79.19% (P>0.05; Figure 4d). Together, our results suggest a dominant-negative effect of SQSTM1-C on the function of native SQSTM1 in ubiquitin aggregate formation.
Finally, we examined the effects of the SQSTM1-C on selective autophagy. As both SQSTM1 and NBR1 are targets of selective autophagy themselves, we then assessed protein levels of endogenous SQSTM1 and NBR1 in cells expressing SQSTM1-C. The results shown in Figure 4e demonstrated that expression of SQSTM1-C led to an accumulation of full-length SQSTM1 and NBR1, indicating an inhibitory effect of the SQSTM1-C on selective autophagy.
Similarly, we found that overexpression of 3C-NBR1-C (3Cpro-induced C-terminal fragment of NBR1) resulted in an increased accumulation of endogenous SQSTM1 (Figure 5). However, this effect appeared to be specific for 3C-NBR1-C as 2A-NBR1-C (2Apro-induced C-terminal fragment of NBR1) failed to induce the accumulation of SQSTM1 (Figure 5).
Overexpression of the C-terminal 3Cpro-mediated cleavage product of NBR1 causes an accumulation of native SQSTM1. HeLa cells were transiently transfected with an empty vector, Flag-2A-NBR1-C (C-terminal cleavage fragment of NBR1 induced by 2Apro), Flag-3C-NBR1-C (C-terminal cleavage fragment of NBR1 induced by 3Cpro), alone or in combination for 48 h. Western blot analysis was performed to examine the expression of endogenous SQSTM1, NBR1-C and β-actin (loading control)
Mutual regulation of SQSTM1 and NBR1 expression
The similarities in the structure and function between SQSTM1 and NBR1 prompted us to question whether SQSTM1 and NBR1 are reciprocally regulated during CVB3 infection and if depletion of SQSTM1 results in a compensatory upregulation of NBR1 protein. The data presented in Figure 6 demonstrated that overexpression of SQSTM1 (or NBR1) resulted in increased protein level of NBR1 (or SQSTM1) (both non-cleaved and cleaved forms; Figures 6a and c), whereas knockdown of SQSTM1 (or NBR1) led to reduced protein expression of NBR1 (or SQSTM1) (both non-cleaved and cleaved forms; Figures 6b and d). To further determine whether the mutual regulation of SQSTM1 and NBR1 occurs at the transcriptional level, we examined gene expression of SQSTM1 and NBR1. We showed that the mRNA level of SQSTM1 (or NBR1) was not significantly altered when NBR1 (or SQSTM1) was overexpressed or knocked down (Figures 6e–h), suggesting that SQSTM1 and NBR1 are reciprocally regulated at the post-transcriptional level. Together, these results suggest that the loss-of-function of one autophagic adapter protein during CVB3 infection is unlikely to be compensated by the other.
Reciprocal regulation of SQSTM1 and NBR1. (a and b) Effects of overexpression or knockdown of SQSTM1 on NBR1 protein expression. HeLa cells were transiently transfected with Flag-SQSTM1 construct (a) or SQSTM1 siRNA (si-SQSTM1, Dharmacon, 100 nM) (b) for 24 h, followed by CVB3 infection for 7 h. Western blotting was performed to examine the expression of SQSTM1 using an anti-Flag antibody, which recognizes the N-terminus of SQSTM1 (a) or using an anti-C-terminal SQSTM1 antibody (b), NBR1 using anti-N-terminal NBR1 antibody and β-actin (loading control). (c and d) Effects of overexpression or knockdown of NBR1 on SQSTM1 protein expression. HeLa cells were transiently transfected with HA-NBR1 construct (c) or NBR1 siRNA (si-NBR1, Dharmacon, 100 nM) (d) for 24 h, following by CVB3 infection for 7 h. Western blotting was performed to examine the expression of NBR1 using an anti-HA antibody, which recognizes the N-terminus of NBR1 (c) or using an anti-N-terminal NBR1 antibody (d), and SQSTM1 using either anti-C-terminal or anti-N-terminal SQSTM1 antibody as indicated. Protein expression of β-actin was examined as a loading control. Si-control, scramble siRNA control. (e and f) Effects of overexpression or knockdown of SQSTM1 on NBR1 gene expression. HeLa cells were transiently transfected with Flag-SQSTM1 construct (e) or si-SQSTM1 (f) for 24 h, followed by CVB3 infection for 7 h. qRT-PCR was performed to examine mRNA levels of NBR1 and GAPDH (internal control). Results are presented as mean±S.D. (n=4). (g and h) Effects of overexpression or knockdown of NBR1 on SQSTM1 gene expression. HeLa cells were transiently transfected with HA-NBR1 construct (g) or si-NBR1 (h) for 24 h, followed by CVB3 infection for 7 h. qRT-PCR was performed to examine mRNA levels of SQSTM1 and GAPDH (internal control). Results are presented as mean±S.D. (n=4)
Discussion
Accumulating evidence has revealed that disruption of protein homeostasis is a key contributor to the development and pathogenesis of many human diseases, including those related to coxsackievirus infection.8, 17, 18 Protein homeostasis is achieved through the function of protein quality control system. Autophagy-mediated selective recycling of the terminally misfolded proteins/aggregates serves as a key component of protein quality control and defects in this pathway have been linked to protein conformation diseases, such as bone, liver, heart and neurodegenerative diseases.3, 19, 20, 21, 22, 23, 24, 25, 26
Misfolded and ubiquitinated proteins are commonly detected in CVB3-infected cells and tissues, suggesting that dysfunction of the protein degradation pathway may have a role in viral pathogenesis.7, 9 We have previously demonstrated that autophagy adapter protein SQSTM1 is cleaved following CVB3 infection, resulting in the loss-of-function of SQSTM1 in selective autophagy.11 The present research extends our previous study to investigate the interaction between NBR1 and coxsackievirus infection. NBR1 is a SQSTM1-like cargo receptor and has been suggested to have a compensatory effect when SQSTM1 is malfunctioning.3 In this study, we showed that NBR1 is also cleaved after viral infection and the cleavage is catalyzed by both viral protease 2Apro and 3Cpro. More interestingly, we demonstrated that, in addition to loss-of-function, the C-terminal truncated mutants of SQSTM1 and NBR1 also exhibit a dominant-negative regulatory effect against the function of native proteins in the clearance of ubiquitin conjugates. First, we showed that the SQSTM1-C mutant inhibits full-length SQSTM1-dependent protein aggregate formation, a prerequisite step for SQSTM1-mediated selective degradation by autophagosome.27 Second, we demonstrated that overexpression of SQSTM1-C results in elevated accumulation of both SQSTM1 and NBR1, two known substrate targets of selective autophagy. The mechanism of such action of SQSTM1-C remains unclear. We speculate that SQSTM1-C stabilizes full-length SQSTM1 and NBR1 by blocking their degradation through competing for the binding of ubiquitin chain and/or LC3.
Unlike SQSTM1, we found NBR1 by itself does not facilitate the formation of protein aggregates and promote the accumulation of insoluble ubiquitin conjugates (data not shown). This is presumably due to the difference in the function of the PB1 domains of these two proteins. The PB1 domain enables SQSTM1 to form self-aggregates, which is a crucial step for SQSTM1-mediated selective autophagy;28 however, Nbr1 requires the coiled-coil (cc) domain, but not the PB1 domain, for its dimerization.4 Our hypothesis is that cc domain-mediated NBR1 dimerization is not sufficient to induce the formation of large protein aggregates. In other words, the step of protein aggregate formation is dispensable for NBR1-mediated selective autophagy. Interestingly, we found that coexpression of SQSTM1 and NBR1 facilitates the formation of the punctate structures of NBR1 (data not shown). This is likely a result of the interaction between NBR1 and SQSTM1 and the ability of SQSTM1 to polymerize.
Another interesting observation of this study is that the cleavage product 3C-NBR1-C, but not 2A-NBR1-C, induces the accumulation of endogenous SQSTM1. As shown in the schematic diagram (Figure 3c), as compared with 3C-NBR1-C, 2A-NBR1-C contains extra 280 amino acids at its N-terminus, which include a second LIR and probably other undefined functional domains. It is postulated that the presence of additional domains may cause the failure to interfere with the function of native proteins. Future investigation is required to dissect the molecular basis of this assumption.
In this study, we reported a positive regulatory relationship between SQSTM1 and NBR1. The observation that knockdown of SQSTM1 results in a downregulation, rather than an upregulation of NBR1, implies that NBR1 may not have a compensatory role for the absence of SQSTM1. What are the possible mechanisms of the mutual regulation of SQSTM1 and NBR1? Our results showed that overexpression or knockdown of NBR1 does not affect mRNA expression of SQSTM1, and vice versa, indicating that mutual regulation between SQSTM1 and NBR1 takes place at the post-transcriptional level. It has been previously demonstrated that NBR1 and SQSTM1 interact directly to form heterodimer through respective PB1 domain.29 GST pull-down assay also showed that the UBA domain of NBR1 can bind directly with SQSTM1.30 These findings raise the possibility that interaction between these two proteins prevents them from being recognized and degraded through autophagy. We speculate that overexpressed SQSTM1 interacts and stabilizes its binding partner, NBR1, whereas depletion of SQSTM1 results in the release of NBR1 from the binding complex and subsequent degradation, and vice versa. In line with this hypothesis, it was recently reported that the N-terminal truncated form of NBR1 (aa1-135) that contains an intact PB1 domain stabilizes SQSTM1 and reduce its turnover through autophagy pathway.30 The other possible explanation for the reciprocal regulation of NBR1 and SQSTM1 is that they compete for degradation through the shared autophagic pathway. For example, the decrease in NBR1 protein levels when SQSTM1 is silenced could be a result of accelerated disposal of NBR1 when there is less competition from SQSTM1. It was previously reported that SQSTM1 can also be degraded through the proteasome pathway;31 however, in the presence of proteasome inhibitors, we found a minimal increase in SQSTM1 and NBR1 protein levels compared with the treatment with lysosome inhibitors (data not shown), suggesting that proteasome pathway is not a major route for the degradation of SQSTM1 and NBR1.
In addition to NBR1 and SQSTM1, CVB3 proteases were reported to target a number of other host proteins, such as eukaryotic translation initiation factor 4γ,32 poly(A)-binding protein,33 serum response factor,34 dystrophin,35 adenosine-uridine-rich element RNA binding factor 1,36 GTPase activating protein (SH3 domain) binding protein 1,37 melanoma differentiation-associated protein 5,38 mitochondrial antiviral-signaling protein38 and retinoic acid-inducible gene 1.38 Through modulating cellular processes and disrupting host innate immune response, viral proteases have a crucial role in viral infection. SQSTM1 has been shown to have an antiviral effect against Sindbis viral infection by directing viral capsid protein for autophagic degradation.39 However, in this study and our previous report,11 we found that overexpression or knockdown of NBR1 and SQSTM1 had little impact on viral replication (data not shown), suggesting that loss of functional SQSTM1/NBR1 alone is not sufficient enough to benefit viral replication. We postulate that other cellular machineries manipulated by viral proteases are also needed to work in concert with the cleavage of SQSTM1/NBR1 to ensure successful viral replication.
In conclusion, this study provides the first proof demonstrating the toxic gain-of-function or dominant-negative effects of the cleavage mutants of SQSTM1 and NBR1. Our findings that CVB3 induces the cleavage of the two functionally similar autophagy adapter proteins and subsequent impairment of selective clearance of ubiquitin aggregates suggest a novel mechanism contributing to the pathogenesis of CVB3 infection.
Materials and Methods
Reagents
The antibodies used in this study include: anti-N-terminal NBR1 (Santa Cruz Biotechnology, Dallas, TX, USA; sc-130380), anti-C-terminal SQSTM1 (PROGEN Biotechnik GmbH, Heidelberg, Germany; GSQSTM1-C) and anti-N-terminal SQSTM1 (PROGEN Biotechnik GmbH, GSQSTM1-N), anti-FLAG (Santa Cruz Biotechnology, sc-807), anti-cleaved caspase-3 (Asp175) (Cell Signaling, Danvers, MA, USA; 9661S), anti-ubiquitin (Sigma, St. Louis, MO, USA; U5379), anti-ACTN/α-actinin (Sigma, A5044), anti-ACTB/β-actin (Sigma, A5316), anti-VP1 (DakoCytomation, Glostrup, Denmark; M706401-1) and anti-HA (Roche, Indianapolis, IN, USA; 11867423001). General caspase inhibitor benzyloxycarbonyl-Val-Ala-Asp-fluoromethylketone (Z-VAD-FMK) was purchased from EMD Millipore (Billerica, MA, USA; #219007). VECTASHIELD Mounting Medium with DAPI (4′, 6-diamidino-2-phenylindole) was from Vector Laboratories (Burlingame, CA, USA; H-1200). Fluorophore-labeled secondary antibodies (goat anti-rat Alexa 488, goat anti-rat Alexa 594, goat anti-rabbit Oregon green and goat anti-rabbit Alexa 594), and Lipofectamine 2000 (#11668-019) were all from Life Technologies (Grand Island, NY, USA). Complete protease inhibitor tablets were from Roche (#11836170001).
Cell culture
HeLa cells (American Type Culture Collection, Manassas, VA, USA; ATCC CCL-2) were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 100 μg/ml penicillin and 100 μg/ml streptomycin at 37 °C incubator with 5% CO2.
Viral infection
HeLa cells were infected with CVB3 (Kandolf strain) at multiplicity of infections of 10 for 1 h in DMEM without FBS, and then washed in Dulbecco’s PBS (DPBS) and maintained in the culture medium containing DMEM and 10% FBS for a variety of time points as indicated.
Plasmids, siRNAs and transient transfection
Plasmids expressing SQSTM1 cleavage fragments (SQSTM1-C and SQSTM1-N) were generated from the Flag-tagged, full-length SQSTM1 construct (a generous gift from Dr. Brett Finlay, University of British Columbia, Vancouver, BC, Canada). Plasmids expressing C-terminal NBR1 fragments (Flag-2A-NBR1-C and Flag-3C-NBR1-C) were established from an HA-NBR1 template provided generously by Dr. Caroline Whitehouse, King’s College London, London, UK. The small interfering RNAs (siRNAs) against SQSTM1 (L-010230-00-0005) and NBR1 (L-010522-00-0005) were purchased from Dharmacon (Lafayette, CO, USA). Cells were transfected with plasmids or siRNAs using Lipofectamine 2000 for 24–48 h as per the manufacturer’s instructions.
Subcellular fractionation
Triton X-100-soluble and -insoluble fractions were isolated as previously described.11 Briefly, cells were initially lysed in DPBS containing 1% Triton X-100 and protease inhibitors; the supernatants were collected as Triton X-100-soluble fractions. The pellets were further lysed in DPBS containing 2% SDS and complete protease inhibitors; the resulting solubilized supernatants were harvested as Triton X-100-insoluble fractions.
Western blotting
Cells were washed in cold DPBS and lysed in Modified Oncogene Science lysis buffer (50 mM NaPyrophosphate, 50 mM NaF, 50 mM NaCl, 5 mM EDTA, 5 mM EGTA, 100 μM Na3VO4, 10 mM HEPES and 0.1% Triton X-100). Either Bradford or BCA assay was performed to measure the protein concentration. A total of 20–40 μg of protein per sample was loaded for SDS-PAGE. Western blotting was performed according to the protocol as previous described.40
Real-time quantitative reverse transcriptase PCR (qRT-PCR)
Total cellular RNA was extracted using the RNeasy Mini kit (Qiagen, Venlo, Netherlands; #74104). cDNA was synthesized using the SuperScript III First-Strand Synthesis kit (Life Technologies, #18080-051) as per the manufacturer’s instructions. NBR1 or SQSTM1 was amplified in a 20 μl PCR reaction system composed of 100 ng of cDNA template, 1 μl of 20 × TaqMan probe (Life Technologies, #4331182, Hs01061917_g1 (SQSTM1), Hs00245918_m1(NBR1), Hs02758991_g1 (GAPDH)), and 2 × TaqMan Universal Master Mix II with UNG (Life Technologies, #4440038). The PCR protocol performed on a ViiA 7 Real-Time PCR System (Applied Biosystems, Foster City, CA, USA) was: 50 °C for 2 min; 95 °C for 10 min; 40 cycles of 95 °C for 15 s and finally 60 °C for 1 min.
CVB3 proteases 2Apro and 3Cpro in vitro cleavage assay
Coxsackieviral proteases 2Apro, 2Apro mutant and 3Cpro were purified as previously described.41 HeLa cells were collected into the cleavage reaction buffer (20 mM Hepes, pH 7.4, 150 mM KOAc, 1 mM DTT) and grinded with a dounce homogenizer for 30–50 strokes on ice. Cell lysates were centrifuged to remove the debris. Purified 2Apro, 2Apro mutant or 3Cpro was incubated with 50 μg of HeLa cell extracts in the cleavage reaction buffer at 37 °C for increasing periods of time as indicated. The reaction was stopped by addition of SDS-PAGE sample buffer.
Immunocytochemical staining and confocal laser-scanning microscopy
Cells were fixed with 4% paraformaldehyde diluted in DPBS for 15 min at room temperature, permeabilized with 0.25% Triton X-100 in DPBS for 10 min and blocked with 1% bovine serum album in DPBS plus Tween 20 for 30 min. Coverslips were incubated with primary antibodies at 4 °C overnight. After washing for 5 min × 3 times, secondary antibodies were added and incubated for 1 h at room temperature in the dark. The coverslips were washed for 5 min × 3 times after decanting secondary antibodies and cells were counterstained with DAPI. Images were taken with a Leica SP2 AOBS inverted confocal laser-scanning microscope (Wetzlar, Germany). The number of cells with GFP-SQSTM1 puncta relative to the number of Flag-SQSTM1-WT, -N or-C co-expressing cells was counted (a minimum of 200 co-expressing cells were counted for each group) using Image-Pro Plus 5.1 software (Rockville, MD, USA). The quantification was performed and presented as a percentage.
Statistical analysis
All data presented are representative of at least three independent experiments. Results are presented as mean±S.D. Unpaired Student’s t-test was performed. P<0.05 was considered to be statistically significant.
Abbreviations
- SQSTM1/p62:
-
sequestosome 1
- NBR1:
-
neighbor of BRCA1 gene 1 protein
- CVB3:
-
coxsackievirus type B3
- PB1:
-
Phox/Bem1p domain
- LIR:
-
microtubule-associated protein light chain (LC3)-interacting region
- UBA:
-
ubiquitin association domain
- ZZ:
-
zinc-finger domain
- CC:
-
coiled-coil domain
- TB:
-
tumor necrosis-associated factor 6 binding domain
References
Klionsky DJ, Emr SD . Autophagy as a regulated pathway of cellular degradation. Science 2000; 290: 1717–1721.
Ichimura Y, Kominami E, Tanaka K, Komatsu M . Selective turnover of p62/A170/SQSTM1 by autophagy. Autophagy 2008; 4: 1063–1066.
Kirkin V, Lamark T, Sou YS, Bjorkoy G, Nunn JL, Bruun JA et al. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol Cell 2009; 33: 505–516.
Johansen T, Lamark T . Selective autophagy mediated by autophagic adapter proteins. Autophagy 2011; 7: 279–296.
Fujita N, Yoshimori T . Ubiquitination-mediated autophagy against invading bacteria. Curr Opin Cell Biol 2011; 23: 492–497.
Komatsu M, Ichimura Y . Selective autophagy regulates various cellular functions. Genes Cells 2010; 15: 923–933.
Gao G, Zhang J, Si X, Wong J, Cheung C, McManus B et al. Proteasome inhibition attenuates coxsackievirus-induced myocardial damage in mice. Am J Physiol Heart Circ Physiol 2008; 295: H401–H408.
Luo H, Wong J, Wong B . Protein degradation systems in viral myocarditis leading to dilated cardiomyopathy. Cardiovasc Res 2010; 85: 347–356.
Si X, Gao G, Wong J, Wang Y, Zhang J, Luo H . Ubiquitination is required for effective replication of coxsackievirus B3. PLoS One 2008; 3: e2585.
Klingel K, Sauter M, Bock CT, Szalay G, Schnorr JJ, Kandolf R . Molecular pathology of inflammatory cardiomyopathy. Med Microbiol Immunol 2004; 193: 101–107.
Shi J, Wong J, Piesik P, Fung G, Zhang J, Jagdeo J et al. Cleavage of sequestosome 1/p62 by an enteroviral protease results in disrupted selective autophagy and impaired NFKB signaling. Autophagy 2013; 9: 1591–1603.
Lamark T, Kirkin V, Dikic I, Johansen T . NBR1 and p62 as cargo receptors for selective autophagy of ubiquitinated targets. Cell Cycle 2009; 8: 1986–1990.
Cemma M, Kim PK, Brumell JH . The ubiquitin-binding adaptor proteins p62/SQSTM1 and NDP52 are recruited independently to bacteria-associated microdomains to target Salmonella to the autophagy pathway. Autophagy 2011; 7: 341–345.
Deosaran E, Larsen KB, Hua R, Sargent G, Wang Y, Kim S et al. NBR1 acts as an autophagy receptor for peroxisomes. J Cell Sci 2013; 126 (Pt 4): 939–952.
Komatsu M, Waguri S, Koike M, Sou YS, Ueno T, Hara T et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell 2007; 131: 1149–1163.
Carthy CM, Yanagawa B, Luo H, Granville DJ, Yang D, Cheung P et al. Bcl-2 and Bcl-xL overexpression inhibits cytochrome c release, activation of multiple caspases, and virus release following coxsackievirus B3 infection. Virology 2003; 313: 147–157.
Willis MS, Patterson C . Proteotoxicity and cardiac dysfunction—Alzheimer’s disease of the heart? N Engl J Med 2013; 368: 455–464.
Toyama BH, Hetzer MW . Protein homeostasis: live long, won’t prosper. Nat Rev Mol Cell Biol 2013; 14: 55–61.
Ding WX . Role of autophagy in liver physiology and pathophysiology. World J Biol Chem 2010; 1: 3–12.
Rue L, Lopez-Soop G, Gelpi E, Martinez-Vicente M, Alberch J, Perez-Navarro E . Brain region- and age-dependent dysregulation of p62 and NBR1 in a mouse model of Huntington’s disease. Neurobiol Dis 2013; 52: 219–228.
Mori F, Tanji K, Odagiri S, Toyoshima Y, Yoshida M, Kakita A et al. Autophagy-related proteins (p62, NBR1 and LC3) in intranuclear inclusions in neurodegenerative diseases. Neurosci Lett 2012; 522: 134–138.
Mori F, Tanji K, Odagiri S, Toyoshima Y, Yoshida M, Ikeda T et al. Ubiquilin immunoreactivity in cytoplasmic and nuclear inclusions in synucleinopathies, polyglutamine diseases and intranuclear inclusion body disease. Acta Neuropathol 2012; 124: 149–151.
Odagiri S, Tanji K, Mori F, Kakita A, Takahashi H, Wakabayashi K . Autophagic adapter protein NBR1 is localized in Lewy bodies and glial cytoplasmic inclusions and is involved in aggregate formation in alpha-synucleinopathy. Acta Neuropathol 2012; 124: 173–186.
Willis MS, Townley-Tilson WH, Kang EY, Homeister JW, Patterson C . Sent to destroy: the ubiquitin proteasome system regulates cell signaling and protein quality control in cardiovascular development and disease. Circ Res 2010; 106: 463–478.
Zheng Q, Wang X . Autophagy and the ubiquitin-proteasome system in cardiac dysfunction. Panminerva Med 2010; 52: 9–25.
Zheng Q, Su H, Ranek MJ, Wang X . Autophagy and p62 in cardiac proteinopathy. Circ Res 2011; 109: 296–308.
Kirkin V, McEwan DG, Novak I, Dikic I . A role for ubiquitin in selective autophagy. Mol Cell 2009; 34: 259–269.
Itakura E, Mizushima N . p62 Targeting to the autophagosome formation site requires self-oligomerization but not LC3 binding. J Cell Biol 2011; 192: 17–27.
Lamark T, Perander M, Outzen H, Kristiansen K, Overvatn A, Michaelsen E et al. Interaction codes within the family of mammalian Phox and Bem1p domain-containing proteins. J Biol Chem 2003; 278: 34568–34581.
Whitehouse CA, Waters S, Marchbank K, Horner A, McGowan NW, Jovanovic JV et al. Neighbor of Brca1 gene (Nbr1) functions as a negative regulator of postnatal osteoblastic bone formation and p38 MAPK activity. Proc Natl Acad Sci USA 2010; 107: 12913–12918.
Seibenhener ML, Babu JR, Geetha T, Wong HC, Krishna NR, Wooten MW . Sequestosome 1/p62 is a polyubiquitin chain binding protein involved in ubiquitin proteasome degradation. Mol Cell Biol 2004; 24: 8055–8068.
Lamphear BJ, Yan R, Yang F, Waters D, Liebig HD, Klump H et al. Mapping the cleavage site in protein synthesis initiation factor eIF-4 gamma of the 2A proteases from human coxsackievirus and rhinovirus. J Biol Chem 1993; 268: 19200–19203.
Kerekatte V, Keiper BD, Badorff C, Cai A, Knowlton KU, Rhoads RE . Cleavage of Poly(A)-binding protein by coxsackievirus 2A protease in vitro and in vivo: another mechanism for host protein synthesis shutoff? J Virol 1999; 73: 709–717.
Wong J, Zhang J, Yanagawa B, Luo Z, Yang X, Chang J et al. Cleavage of serum response factor mediated by enteroviral protease 2A contributes to impaired cardiac function. Cell Res 2012; 22: 360–371.
Badorff C, Berkely N, Mehrotra S, Talhouk JW, Rhoads RE, Knowlton KU . Enteroviral protease 2A directly cleaves dystrophin and is inhibited by a dystrophin-based substrate analogue. J Biol Chem 2000; 275: 11191–11197.
Wong J, Si X, Angeles A, Zhang J, Shi J, Fung G et al. Cytoplasmic redistribution and cleavage of AUF1 during coxsackievirus infection enhance the stability of its viral genome. FASEB J 2013; 27: 2777–2787.
Fung G, Ng CS, Zhang J, Shi J, Wong J, Piesik P et al. Production of a dominant-negative fragment due to G3BP1 cleavage contributes to the disruption of mitochondria-associated protective stress granules during CVB3 infection. PLoS One 2013; 8: e79546.
Feng Q, Langereis MA, Lork M, Nguyen M, Hato SV, Lanke K et al. Enteroviruses 2Apro targets MDA5 and MAVS in infected cells. J Virol 2014; 88: 3369–3378.
Orvedahl A, MacPherson S, Sumpter R Jr, Talloczy Z, Zou Z, Levine B . Autophagy protects against Sindbis virus infection of the central nervous system. Cell Host Microbe 2010; 7: 115–127.
Zhang J, Wong J, Gao G, Luo H . Tripeptidyl peptidase II serves as an alternative to impaired proteasome to maintain viral growth in the host cells. FEBS Lett 2011; 585: 261–265.
Joachims M, Van Breugel PC, Lloyd RE . Cleavage of poly(A)-binding protein by enterovirus proteases concurrent with inhibition of translation in vitro. J Virol 1999; 73: 718–727.
Acknowledgements
We thank Dr. Eric Jan at the University of British Columbia for his generous gifts of the purified proteins of coxsackieviral protease 2Apro, mutant 2Apro and 3Cpro. This work was supported by the Canadian Institutes of Health Research (CIHR) (HL). JS is supported by a Doctoral Fellowship from the China Scholarship Council.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by RA Knight
Rights and permissions
About this article
Cite this article
Shi, J., Fung, G., Piesik, P. et al. Dominant-negative function of the C-terminal fragments of NBR1 and SQSTM1 generated during enteroviral infection. Cell Death Differ 21, 1432–1441 (2014). https://doi.org/10.1038/cdd.2014.58
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cdd.2014.58
This article is cited by
-
Autophagy receptors as viral targets
Cellular & Molecular Biology Letters (2021)
-
SG formation relies on eIF4GI-G3BP interaction which is targeted by picornavirus stress antagonists
Cell Discovery (2019)
-
CALCOCO2/NDP52 and SQSTM1/p62 differentially regulate coxsackievirus B3 propagation
Cell Death & Differentiation (2019)
-
VPS34 stimulation of p62 phosphorylation for cancer progression
Oncogene (2017)
-
NBR1 is dispensable for PARK2-mediated mitophagy regardless of the presence or absence of SQSTM1
Cell Death & Disease (2015)