Abstract
The serine threonine kinase checkpoint kinase 2 (CHK2) is a DNA damage checkpoint protein important for the ATM-p53 signaling pathway. In addition to its phosphorylation, CHK2 is also ubiquitylated, and both post-translational modifications are important for its function. However, although the mechanisms that regulate CHK2 phosphorylation are well established, those that control its ubiquitylation are not fully understood. In this study, we demonstrate that the ubiquitin E3 ligase PIRH2 (p53-induced protein with a RING (Really Interesting New Gene)-H2 domain) interacts with CHK2 and mediates its polyubiquitylation and proteasomal degradation. We show that the deubiquitylating enzyme USP28 forms a complex with PIRH2 and CHK2 and antagonizes PIRH2-mediated polyubiquitylation and proteasomal degradation of CHK2. We also provide evidence that CHK2 ubiquitylation by PIRH2 is dependent on its phosphorylation status. Cells deficient in Pirh2 displayed accumulation of Chk2 and enhanced hyperactivation of G1/S and G2/M cell-cycle checkpoints. This hyperactivation was, however, no longer observed in Pirh2−/−Chk2−/− cells, providing evidence for the importance of Chk2 regulation by Pirh2. These findings indicate that PIRH2 has central roles in the ubiquitylation of Chk2 and its turnover and in the regulation of its function.
Similar content being viewed by others
Main
PIRH2 (p53-induced protein with a RING (Really Interesting New Gene)-H2 domain), also known as RCHY1, encodes for an ubiquitin (Ub) ligase that is transcriptionally regulated by p53.1 PIRH2 interacts with the active tetrameric form of p53, mediates its ubiquitylation and regulates its turnover via Ub–proteasome mechanisms.1, 2 PIRH2 exerts its ubiquitylation function in cooperation with the E2 Ub-conjugating enzyme UbcH5b.1 A recent study indicated that PIRH2 negative regulation of p53 is enhanced in the presence of the ubiquitylation factor E4B.3 In addition to p53, other ubiquitylation substrates have been also reported for PIRH2, including c-Myc,4 p27kip1,5 ɛ-COP,6 the signal recognition particle receptor β subunit,7 the DNA polymerase eta8 and p73.9, 10
The serine threonine kinase CHK2 (checkpoint kinase 2) is a checkpoint effector important for the signaling of DNA double-strand breaks and the activation of cell-cycle checkpoints.11 In response to DNA damage, ataxia telangiectasia mutated (ATM) phosphorylates CHK2 on its threonine 68 (T68) and allows it to interact with the FHA domain of another CHK2.12, 13, 14 This dimerization of CHK2 leads to its autophosphorylation on T383 and T387 and promotes its full activation.15, 16, 17 CHK2 phosphorylates p53 on Serine 20 (S23 for mouse) in response to DNA damage and has also other phosphorylation substrates, including PML, E2F1, CDC25C and BRCA1, highlighting its importance in processes such as DNA repair, cell-cycle arrest, apoptosis and senescence.18 CHK2 is also involved in DNA repair via the phosphorylation of Forkhead Box M1.19 In addition, in collaboration with BRCA1, and independently of p53, CHK2 is required for the normal progression of mitosis and chromosomal stability.20, 21
Although Chk2 is phosphorylated and activated primarily by the kinase ATM, it is also ubiquitylated; however, the mechanism of this ubiquitylation remains poorly understood.11 In this study, we report that PIRH2 interacts with CHK2 and mediates its polyubiquitylation and proteasomal degradation. Our data indicate that phosphorylation status of CHK2 and Ub-specific-processing protease 28 (USP28) have important roles in balancing PIRH2-mediated CHK2 ubiquitylation turnover. Consistent with the role PIRH2 has in the regulation of CHK2 stability, its deficiency resulted in higher levels of Chk2 protein and increased DNA damage induced phosphorylation of its substrates. Thus, CHK2 ubiquitylation by PIRH2 is important for the regulation of its turnover and therefore represents an important regulatory mechanism for the function of this kinase.
Results
Pirh2 deficiency leads to accumulation of Chk2 protein
Immunohistochemistry analysis of the expression levels of Chk2 in the spleen and thymus of Pirh2−/− mice and their wild-type (WT) controls indicated elevated expression level of Chk2 in Pirh2-deficient cells (Figures 1a and b). We next examined the effect of Pirh2 deficiency on Chk2 expression under DNA-damaging conditions. WT and Pirh2−/− splenocytes and thymocytes were subjected to 6 Gy of γ-irradiation and Chk2 expression was examined by western immunoblotting at different time points post irradiation. Our data indicated higher accumulation of Chk2 protein in irradiated splenocytes and thymocytes from Pirh2−/− mice compared with WT littermates (Figures 1c and d). As the kinase ATM is central to the DNA damage response and functions upstream of Chk2 in the DNA double-strand break signaling cascade,22 we examined the effect of Pirh2 deficiency on its levels of expression and autophosphorylation (Ser1981). Western blot analysis indicated no significant changes in the expression or autophosphorylation levels of Atm both under untreated conditions or in response to γ-irradiation (Figure 1c). However, consistent with the elevated expression level of Chk2 in irradiated Pirh2-deficient cells, the phosphorylation levels of Chk2 substrates p53 (S23) and E2F1 were found higher post irradiation in Pirh2−/− thymocytes compared with WT controls (Figures 1e and f). These data support the importance of Pirh2 in the regulation of Chk2 turnover under normal conditions and in response to DNA damage.
Loss of Pirh2 leads to accumulation of Chk2. (a) Immunohiostochemistry analysis of the expression level of Chk2 in spleen of 10-week-old Pirh2−/− mice and WT littermates. (Bar=10 μm). (b) Immunohiostochemistry analysis of the expression level of Chk2 in thymus of 10-week-old Pirh2−/− mice and WT littermates. (Bar=10 μm). (c) Immunoblot (IB) analysis of the expression level of Chk2, Atm and pAtm (Ser1981) at different time points (h) post irradiation (6 Gy) of splenocytes from Pirh2−/− mice and WT littermates. (d) IB analysis of the expression level of Chk2 in Pirh2−/− and WT thymocytes at different time points (h) post irradiation (6 Gy). (e) IB analysis of the level of expression of Chk2 and its downstream substrate p53. WT and Pirh2−/− thymocytes were subjected to 6 Gy of irradiation, and the levels of Chk2, total p53 and Ser23 phosphorylated p53 were assessed at different time points post irradiation. (f) Serine phosphorylation level of E2F1, a Chk2 substrate, in Pirh2-deficient cells. WT and Pirh2−/− thymocytes were subjected to 6 Gy of irradiation, and cells were lysed 6 h later and immunoprecipitation (IP) performed using anti-E2F1 antibody. Immunoprecipitates were subjected to IB analysis with anti-phosphoserine antibody. Data shown is representative of at least three independent experiments. IgG, immunoglobulin G; WCL, whole-cell lysates
Pirh2-deficient cells display Chk2-dependent defects of G1/S and G2/M cell-cycle checkpoints
Chk2 has important roles in the activation of G1/S and G2/M cell-cycle checkpoints in response to DNA double-strand breaks.22 Therefore, we wanted to determine whether its accumulation in the absence of Pirh2 affects the activation of G1/S and G2/M checkpoints. Pirh2−/− mouse embryonic fibroblasts (MEFs) displayed elevated activation of the G1/S checkpoint as indicated by the significantly reduced fraction of these cells in S phase compared with WT controls in response to 10 Gy of γ-irradiation (14.9%±4 versus 40%±6.9; P=0.018; Figures 2a and b). This enforced activation of G1/S checkpoint in Pirh2−/− cells was no longer observed in irradiated Chk2−/−Pirh2−/− MEFs (47.7%±2.7; P=0.0015; Figures 2a and b). Consistent with the role Chk2 has in G2/M checkpoint activation, Pirh2−/− MEFs also displayed stronger γ-irradiation-induced activation of the G2/M cell-cycle checkpoint as evidenced by the significantly reduced fraction of these cells in M phase compared with WT controls (30.85%±3.1 versus 18.1%±3; % P=0.024; Figures 2c and d). Interestingly, Chk2 deficiency in Pirh2−/− MEFs restored their level of G2/M activation to a level similar to WT controls (Chk2−/−Pirh2−/− MEFs versus WT MEFs; P>0.05; Figures 2c and d). Based on these data, we conclude that increased Chk2 level in the absence of Pirh2 enforces the activation of G1/S and G2/M cell-cycle checkpoints.
Increased activation of G1/S and G2/M cell-cycle checkpoints in Pirh2−/− cells is abrogated by additional loss of Chk2. (a) Analysis of the effect of Pirh2 loss on G1/S checkpoint activation. Early passage primary Pirh2−/−, Chk2−/−Pirh2−/−, Chk2−/− and WT MEFs were left untreated (UT) or irradiated with 10 Gy, and 18 h later cells were pulsed with bromodeoxyuridine (BrdU) for 4 h. The percentage of cells in S-phase was determined by flow cytometry. A representative of at least three independent experiments is shown. (b) Quantification of the percentage of cells remaining in S phase after irradiation relative to untreated controls. Data are presented as the mean±S.D. of at least three independent experiments. Asterisk denotes statistical significance (P<0.05, Student’s t-test). (c) Analysis of the effect of Pirh2 loss on G2-M checkpoint activation. Pirh2−/−, Chk2−/−Pirh2−/−, Chk2−/− and WT primary MEFs were irradiated with 2 Gy or left untreated, then allowed to recover for 1 h. Cells in M phase were detected by staining with propidium iodide (PI) and phospho-H3 and flow cytometry. A representative of at least three independent experiments is shown. (d) Quantification of the percentage of cells in M phase following irradiation relative to untreated. Data are presented as the mean±S.D. of at least three independent experiments. Asterisk denotes statistical significance (P<0.05; Student’s t-test)
Pirh2 interacts with Chk2
To examine the mechanisms that lead to the increased level of Chk2 proteins in the absence of Pirh2, we first investigated the possible physical interaction of these two proteins. Therefore, using HEK293T cells, we expressed human PIRH2 alone or in the presence of hemagglutinin epitope (HA)-tagged-WT or mutated (kinase dead) Chk2. PIRH2 was readily co-immunoprecipitated by anti-HA antibody in the presence of either HA-Chk2-WT or HA-Chk2-kinase dead (Figure 3a). We also examined the interaction of endogeneous Pirh2 and Chk2 using HEK293T cells and thymocytes. Immunoprecipitation of human CHK2 brought down PIRH2 in HEK293T cells (Figure 3b). Murine Chk2 was also detected in the Pirh2 immunoprecipitates from WT but not Pirh2−/− thymocytes (Figure 3c).
Chk2 interacts with Pirh2. (a) Interaction between CHK2 and PIRH2 in HEK293T cells. HEK293T cells were transfected with expression plasmids encoding HA-tagged WT Chk2, kinase-dead (KD) Chk2 and PIRH2 as indicated. Cells were lysed and subjected to immunoprecipitation (IP) with an anti-PIRH2 antibody. The resulting precipitates were subjected to immunoblot (IB) analysis with anti-HA or anti-PIRH2 antibody. A portion of the whole-cell lysate (WCL) corresponding to 3% of the input for IP was also subjected to IB analysis. (b) Endogenous interaction between human CHK2 and PIRH2 in HEK293T cells. HEK293T cells were lysed and subjected to IP with anti-CHK2 antibody, and the resulting precipitates were subjected to IB analysis with anti-CHK2 or anti-PIRH2 antibody. (c) Interaction of endogenous murine Chk2 and Pirh2 in thymocytes. WT and Pirh2−/− thymocytes were lysed and subjected to IP with an anti-Pirh2 antibody. The resulting precipitates were subjected to IB analysis with anti-Chk2 or anti-Pirh2 antibodies. (d) Interaction between recombinant Chk2 and PIRH2 proteins. His6-tagged Chk2 and glutathione S-transferase (GST)-tagged PIRH2 were subjected to IP with anti-GST, and the resulting precipitates were subjected to IB analysis with anti-His or anti-GST antibody. A portion (3%) of the input for IP was also subjected to IB analysis. Data shown is representative of at least three independent experiments. IgG, immunoglobulin G
We next examined whether Chk2 and Pirh2 directly interact. Using in vitro GST pull-down assay, purified recombinant hexahistidine epitope (His6)-tagged Chk2 and GST-tagged PIRH2 proteins, we observed that Chk2 was readily pulled down with PIRH2 (Figure 3d).
Collectively our data support direct physical interaction of CHK2 and PIRH2 in mammalian cells.
Pirh2 polyubiquitylates Chk2 and mediates its proteasomal degradation
Despite the compelling evidence for the importance of ubiquitylation for the regulation of CHK2 stability,23, 24, 25 the mechanisms for this regulation remain to be identified. Based on Chk2 interaction with Pirh2 and its accumulation in the absence of this E3 ligase, we hypothesized that Pirh2 might ubiquitylate Chk2 and regulate its turnover. To test this hypothesis, we first used an intracellular ubiquitylation assay and HEK293T cells expressing PIRH2, HA-Chk2 and Myc-Ub. When HA-Chk2 proteins were expressed at similar levels, the additional overexpression of full-length PIRH2 significantly increased Chk2 polyubiquitylation (Figures 4a and b and Supplementary Figure S1). By contrast, Chk2 polyubiquitylation was reduced in the presence of the RING domain deleted PIRH2 (PIRH2ΔRing) compared with full-length PIRH2 (Figure 4c and Supplementary Figure S2a).
Pirh2 mediates polyubiquitylation of Chk2. (a) PIRH2 mediates Chk2 polyubiquitylation in vivo. HEK293T cells were transfected with expression plasmids encoding Myc-tagged Ub, HA-tagged Chk2 and PIRH2. Whole-cell lysate (WCL) of transfected HEK293T cells were subjected to immunoblot (IB) analysis with anti-HA to detect HA-tagged Chk2, anti-Ub and with anti-PIRH2 to assess the level of expression of transfected PIRH2. (b) WCL of transfected HEK293T cells as indicated in (a) were immunoprecipitated using anti-HA, and anti-Ub IB analysis was performed to detect ubiquitylated HA-tagged Chk2 proteins. (c) Role of the PIRH2 RING domain in the ubiquitylation of Chk2. HEK293T cells were transfected with expression plasmids encoding Myc-tagged Ub, HA-tagged Chk2 and WT PIRH2 or its RING finger domain deleted form (ΔR). Immunoprecipitation (IP) performed with anti-HA were subjected to anti-HA IB analysis to detect ubiquitylated HA-tagged Chk2. (d) In vivo ubiquitylation assay. Thymocytes from WT and Pirh2−/− mice were cultured in the presence or absence of MG-132 for 3 h. Cell lysates were adjusted to contain equivalent amounts of Chk2 protein between WT and Pirh2−/− samples. IP performed with anti-Chk2 antibody was subjected to IB analysis with anti-Ub. (e and f) In vitro ubiquitylation of CHK2 by PIRH2. PIRH2-mediated CHK2 in vitro ubiquitylation assay. In all, 6 × His tagged-Ub, human Ub activating enzyme E1, UBE2D2/UbcH5b, recombinant human PIRH2 (0.5–0.05 μg) and CHK2 were used to assess PIRH2-mediated CHK2 ubiquitylation in vitro. Ubiquitylated CHK2 proteins were visualized by western blot using anti-CHK2 antibody (e) or anti-Ub antibody (f). CHK2 aggregates are indicated with an asterisk. Data shown is representative of at least three independent experiments. Ubn, polyubiquitylation
We next examined the effect of Pirh2 loss on the in vivo level of Chk2 ubiquitylation. Thymocyte lysates were prepared from WT and Pirh2−/− mice and were adjusted to contain equivalent amounts of Chk2 protein to balance for the higher Chk2 level in the absence of Pirh2. When the Chk2 proteins immunoprecipitated from WT thymocytes were immunoblotted with anti-Ub antibody, we observed smears with slower mobility (Figure 4d). These smears became more pronounced when Chk2 was immunoprecipitated from WT thymocytes treated with the proteasome inhibitor MG-132 (Figure 4d and Supplementary Figure S2b). By contrast, anti-Ub antibody immunoblot analysis of immunoprecipitated Chk2 from Pirh2−/− thymocytes indicated significantly reduced smear intensity, even in the presence of MG-132 (Figure 4d and Supplementary Figure S2b).
To examine the ability of Pirh2 to directly ubiquitylate Chk2, we performed in vitro ubiquitylation assays and assessed the E3 ligase activity of GST-PIRH2 against His6-CHK2. Polyubiquitylation of His6-CHK2 was observed only with the additional presence of the GST-PIRH2 (Figures 4e and f and Supplementary Figures S3a and b). This polyubiquitylation was stronger in the presence of increasing concentration of either PIRH2 or CHK2 (Figures 4e and f and Supplementary Figure S3c).
Collectively, these data indicate that Chk2 is a substrate for Pirh2-mediated ubiquitylation and that this ubiquitylation leads to its proteasomal degradation.
Phosphorylation of Chk2 is critical for its ubiquitylation by Pirh2
Chk2 stability is regulated by both ubiquitylation and phosphorylation.23, 26 Phosphorylation of human CHK2 on its S456 residue (S460 of mouse Chk2) regulates its stability in response to DNA damage.26 We therefore examined whether this Chk2 phosphorylation affects its ubiquitylation by Pirh2. Mutation of S460 residue of mouse Chk2 to alanine (SA mutant), to prevent phosphorylation of this site, resulted in hyper-ubiquitylation of Chk2 (Figure 5). In addition, mouse Chk2 carrying a substitution of S460 residue to aspartic acid (SD mutant), to mimic the phosphorylation of this site, displayed a significantly lower level of ubiquitylation by Pirh2 compared with the SA Chk2 mutant (Figure 5).
Chk2 ubiquitylation by PIRH2 is controlled by its phosphorylation status. Phosphorylation of Chk2 S460 enhances its ubiquitylation by PIRH2. HEK293T cells were transfected with expression plasmids encoding PIRH2, Myc-tagged Ub and HA-tagged Chk2 derivatives (WT, S460A ‘SA’ and S460D ‘SD’) as indicated. (Left panel) Immunoprecipitation (IP) performed with anti-HA were subjected to anti-HA immunoblot (IB) analysis to detect ubiquitylation of HA-tagged Chk2 proteins. (Right panel) Whole cell lysates (WCL) of transfected cells were subjected to IB analysis with anti-HA and anti-PIRH2. Data shown is representative of at least three independent experiments. Ubn, polyubiquitylation
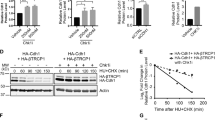
We next performed pulse-chase analyses and examined the effect of Pirh2 on the in vivo stability of Chk2. These analyses indicated that expression in HEK293T cells of the full-length PIRH2, but not its mutated form PIRH2ΔRING, leads to decrease stability of HA-Chk2 proteins (Figures 6a and b). In addition, our data indicated that S460A substitution of Chk2 promoted its degradation by PIRH2, whereas its S460D substitution attenuated this degradation (Figures 6c and d).
Regulation of CHK2 turnover by PIRH2 is dependent on its phosphorylation status. (a) PIRH2 mediates proteasomal degradation of Chk2. HEK293T cells were transfected with empty vector or expression plasmids as indicated. Twenty-four hours post transfection, cells were cultured in the presence of cycloheximide (50 μg/ml) for the indicated time. Cell lysates were then subjected to immunoblot (IB) analysis with anti-HA, anti-PIRH2 and anti-Actin antibodies. (b) Intensity of the HA-Chk2 bands was normalized by that of the corresponding Actin bands and then expressed as a percentage of the normalized value for time 0. Data shown are representative of at least three independent experiments. (c) PIRH2 mediates proteasomal degradation of S460 unphosphorylated Chk2. HEK293T cells were transfected with expression plasmids encoding HA-tagged Chk2 derivatives (WT, S460A and S460D) and either PIRH2 or empty vector (mock), and pulse-chase assay was performed as described above. (d) Intensity of the HA-Chk2 bands was normalized by that of the corresponding Actin bands and then expressed as a percentage of the normalized value for time 0. Data shown are representative of at least three independent experiments
Together, these data indicate that the phosphorylation status of Chk2 on S460 residue is critical for the regulation of its turnover mediated by the E3 ligase Pirh2.
Cooperation of PIRH2 and USP28 in the control of CHK2 ubiquitylation and stability
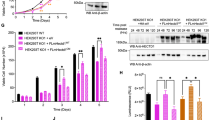
It has been previously demonstrated that Chk2 stability is regulated by the USP28.26, 27 We therefore hypothesized that PIRH2, CHK2 and USP28 may form a complex and that through its deubiquitylase activities, USP28 may restrain CHK2 ubiquitylation by PIRH2. To test these hypotheses, we first expressed PIRH2, HA-Chk2 and FLAG-USP28 in HEK293T cells and examined the ability of these proteins to interact. Immunoprecipitation and western blotting analyses indicated interactions of PIRH2, Chk2 and USP28 (Figure 7a and Supplementary Figure S4). We next examined the effect of USP28 on PIRH2-mediated ubiquitylation of Chk2. Intracellular ubiquitylation assays using HEK293T overexpressing PIRH2, HA-Chk2 and FLAG-tagged USP28 indicated that USP28 decreased the level of PIRH2-mediated ubiquitylation of Chk2 (Figure 7b). To further confirm the importance of USP28 in counteracting Chk2 ubiquitylation by PIRH2, we performed similar intracellular ubiquitylation assays in the presence of the catalytic-dead mutant USP28-C171A.27, 28 In contrast to WT USP28, its catalytic-dead form failed to significantly restrain Chk2 ubiquitylation by PIRH2 (Figure 7c).
USP28 balances Chk2 ubiquitylation and proteasomal degradation mediated by PIRH2. (a) PIRH2 interacts with the deubiquitylase USP28. HEK293T cells were transfected with expression plasmids as indicated. Cells were lysed and subjected to immunoprecipitation (IP) with anti-PIRH2 antibody, and the resulting precipitates were subjected to immunoblot (IB) analysis with anti-PIRH2, anti-HA and anti-FLAG-M2 antibody. Data shown are representative of at least three independent experiments. (b) USP28 restrains PIRH2 ubiquitylation of Chk2. HEK293T cells were transfected with expression plasmids encoding Myc-tagged Ub, HA-tagged Chk2, PIRH2 and FLAG-tagged USP28. IP performed with anti-HA were subjected to anti-Ub IB analysis to detect ubiquitylated HA-tagged Chk2. Data shown are representative of at least three independent experiments. (c) Effect of the catalytic-dead USP28 on PIRH2 ubiquitylation of Chk2. Experiment were performed as in (b) with the addition of catalytic-dead USP28 (C171A) as indicated. Data shown are representative of at least three independent experiments. (d) PIRH2-mediated proteasomal degradation of Chk2 is suppressed by USP28. HEK293T cells were transfected as indicated with expression plasmids encoding HA-tagged Chk2, FLAG-tagged USP28 and PIRH2. Twenty-four hours post transfection, cells were cultured in the presence of cycloheximide (50 μg/ml) for the indicated time. Cell lysates were then subjected to IB analysis with anti-HA, anti-PIRH2, anti-Flag and anti-Actin antibodies. Pulse-chase analysis was performed as described in Figure 6a. (e) Intensity of the HA-Chk2 bands was normalized by that of the corresponding Actin bands and then expressed as a percentage of the normalized value for time 0. Data shown are representative of at least three independent experiments. WCL, whole-cell lysates; Ubn, polyubiquitylation; *, non-specific
Consistent with these data, elevated instability of Chk2 protein in the presence of PIRH2 was significantly rescued by co-transfection of USP28 (Figures 7d and e). Collectively, these data support a role for PIRH2 and USP28 in the regulation of Chk2 ubiquitylation and turnover.
Discussion
The Ub–proteasome pathway is required for various cellular processes, including DNA transcription, immune response, cell cycle, cell death, DNA damage signaling and DNA damage repair.29, 30 The E3 ligase Pirh2 has been initially reported to regulate the expression level of activated p53 via its polyubiquitylation and proteasomal degradation.1, 2 However, Pirh2 is not required for maintaining steady-state levels of p53 under unstressed conditions.4, 5, 31
Similar to p53, the serine threonine kinase CHK2 has important roles in response to stimuli that include DNA damage and oncogenic activation and is an important component of the ATM-p53 signaling pathway.18, 32 Although CHK2 is a stable protein with a half-time over 6 h,33, 34 this stability is reduced by a number of CHK2 mutations associated with human malignancies, including testicular tumors, lung tumors and tumors from Li–Fraumeni Syndrome patients.35, 36, 37, 38 Several lines of evidence suggest that in addition to CHK2 phosphorylation its ubiquitylation also has important role in the regulation of its stability.23, 26, 27 Both the Ub E3 ligase Mdm2 and the acetyltransferase PCAF, which also possesses an intrinsic E3 ligase activity, have been reported to regulate CHK2 ubiquitylation and degradation.23 However, ubiquitylation of CHK2 and the regulation of its turnover were independent of Mdm2 and PCAF E3 ligase activities.23 Similarly, EDD was also reported to have a role in the activation of CHK2; however, the ability of this Ub E3 ligase to mediate CHK2 ubiquitylation remains to be demonstrated.39, 40 A recent study has indicated the ability of the E3 ligase Rnf8 to ubiquitylate Chk2 and regulate its turnover.25
In this study, we provide evidence to support the importance of the E3 ligase PIRH2 in mediating CHK2 ubiquitylation. PIRH2 physically associated with CHK2 and mediated its polyubiquitylation in vivo and in vitro. We also report that CHK2 ubiquitylation by PIRH2 is important for the regulation of its turnover. Expression of full-length PIRH2 protein, but not its mutant form lacking E3 ligase activity, resulted in reduced stability of CHK2 protein. In addition, and consistent with the importance of Chk2 ubiquitylation by Pirh2 for its proteasomal degradation, Pirh2-deficient cells displayed elevated expression level of Chk2 protein.
Ubiquitylation and phosphorylation processes are highly interdependent, with many cases where phosphorylation of proteins triggers their ubiquitylation41 (Figure 8). In response to DNA double-strand breaks, the kinase activity of CHK2 is upregulated following its phosphorylation by ATM and its autophosphorylation.18 Phosphorylation of CHK2 is also important for its protein stability. For instance, phosphorylation of human CHK2 protein on S456 (equivalent to mouse S460), located in its kinase domain, is critical for the regulation of its stability.23, 26 Furthermore, in response to genotoxic stress, CHK2 autophosphorylation on S379 (S383 for mouse Chk2) is required for its ubiquitylation by Cullin 1 containing E3 ligase complex and for its function in inducing apoptosis.24 Data presented in this study provide evidence to support the importance of phosphorylation of CHK2 for its ubiquitylation and proteasomal degradation mediated by PIRH2 (Figure 8). Although PIRH2 efficiently polyubiquitylated WT Chk2 and mediated its proteasomal degradation, the phospho-mimic S460D mutant of Chk2 was resistant to PIRH2-mediated polyubiquitylation and destabilization. Conversely, the inability of Chk2 to be phosphorylated on its S460 residue (S460A mutation) resulted in its hyper-ubiquitylation and increased proteasomal degradation by PIRH2.
A model for CHK2 ubiquitylation during DNA damage response (DDR). A model illustrating CHK2 phosphorylation and polyubiquitylation status during DDR (left; Antoni et al.18 and Kass et al.23, 26) and proposed role for PIRH2 in the termination of DDR through its polyubiquitylation and degradation of CHK2 (right). Left: In response to genotoxic stress, CHK2 is phosphorylated on T68 (mouse T77) by ATM, followed by CHK2 autophosphorylation on S379 (mouse S383).18, 24 Ubiquitin E3 ligase Cul-1 complex polyubiquitylates CHK2 proteins phosphorylated on T68 and S379 and leads to full activation of CHK2.24 CHK2 is also phosphorylated on S456 (mouse S460), by a yet to be identified kinase (?), and this phosphorylation was reported to increase the stability of CHK2 protein.26 Right: The Ub E3 ligase PIRH2 mediates polyubiquitylation of CHK2 proteins dephosphorylated on S456, by an unknown phosphatase, and leads to CHK2 proteasomal degradation. This PIRH2 action on S456-dephosphorylated CHK2 contributes to the termination of DDR signaling. RNF8 has also been recently shown to ubiquitylate CHK2 and regulate its turnover.25 P, phosphorylation; ?, unknown; H, human; m, mouse
The kinase Chk2 is important for the activation of the G1/S and G2/M cell-cycle checkpoints in response to DNA double-strand breaks.18 Our observation that Chk2 ubiquitylation was defective in the absence of PIRH2, leading to its increased expression level, suggested the possibility that the accumulated Chk2 in Pirh2-deficient cells may lead to enhanced activation of G1/S and G2/M cell-cycle checkpoints. Indeed, our data indicate higher activation of these cell-cycle checkpoints in the absence of Pirh2. Interestingly, loss of Chk2 in Pirh2−/− cells prevented the hyperactivation of these checkpoints associated with Pirh2 deficiency. These data suggest that the pronounced activation of G1/S and G2/M cell checkpoints in Pirh2-deficient cells is associated with their accumulated Chk2. Therefore, through its ubiquitylation of Chk2 and the regulation of its turnover, Pirh2 emerges as an important regulator of Chk2 physiological functions, including in the activation of G1/S and G2/M checkpoints.
Collectively, our data indicate that PIRH2 regulates the stability of CHK2 protein under unstressed conditions as well as following CHK2 dephosphorylation post-genotoxic stress (Figure 8). It is interesting to note that, in addition to PIRH2, the E3 ligase RNF8 has also been shown to polyubiquitylate CHK2 and mediate its proteasomal degradation in response to DNA damage.25 The involvement of different E3 ligases in the downregulation of CHK2 highlights the importance of the regulation of the turnover of this checkpoint protein. We propose that by ubiquitylating dephosphorylated CHK2, and targeting it for proteasomal degradation, PIRH2 might contribute to the shutdown of the DNA damage response. We also conclude that, by regulating the turnover of CHK2 and a number of other key proteins, including p53, p73 and c-MYC, PIRH2 emerges as an E3 ligase that has important roles, including in the DNA damage response and cancer.
Materials and Methods
Mice and MEFs
Pirh2−/− mice lacking exon 2 and 3 were previously described4 and were in a C57BL/6 genetic background. Mice were genotyped by PCR (primer sequences and PCR conditions available upon request). Pirh2−/− mice were crossed to Chk2−/− mice42 to generate Chk2+/−Pirh2+/− mutants. These double heterozygotes were subsequently intercrossed to generate Chk2−/−Pirh2−/−, Pirh2−/−, Chk2−/− and WT MEFs from E13.5 embryos. All experiments were performed in compliance with the Ontario Cancer Institute animal care committee guidelines.
Cell culture
MEFs and HEK293T cell lines were cultured in Dulbecco’s modified Eagle’s medium (Gibco; Invitrogen, Burlington, ON, Canada) supplemented with 10% fetal bovine serum (FBS; Wisent, St-Jean-Baptiste, QC, Canada). Thymocytes were cultured in RPMI1640 (Gibco) with 10% FBS.
Western blot and immunoprecipitation analysis
Proteins were separated on 7.5, 10 or 12% Tris-Glysine SDS-PAGE home-made gels. For detection of Atm, proteins were separated on a 3–8% Tris-Acetate gradient polyacrylamide gel (Invitrogen, Burlington, ON, Canada). The following antibodies were used for Western blotting in 5% powdered milk in TBS-T: rabbit polyclonal anti-p53 (FL393, Santa Cruz Biotechnology, Dallas, TX, USA), anti-phospho-p53 (Ser23; #9287, Cell Signaling), affinity purified anti-Chk2 (raised against murine Chk230–47), anti-Atm (Cell Signaling, Danvers, MA, USA), anti-phospho-Atm (Ser1981; Cell Signaling), anti-phospho-serines (Cell Signaling Technology), anti-E2F1 (sc-193, Santa Cruz), anti-Pirh2 (BETHYL Laboratories, Montgomery, TX, USA) and anti-β actin (Sigma, Oakville, ON, Canada).
HEK293T cells were transfected with expression plasmids encoding human PIRH2 (pcDNA3-hPIRH2) and HA-tagged murine Chk2 (pcDNA3-HA-Chk2) using the calcium phosphate method. Cells were lysed 48 h later in a solution containing 50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1% Triton-X, 1 mM PMSF, 400 μM Na3VO4 and protease inhibitor cocktail tablet (Roche, Laval, QC, Canada). Lysates of transfected HEK293T cells were centrifuged at 14 000 × g for 10 min at 4 °C, and the resulting supernatant was incubated with anti-HA antibody for 2 h at 4 °C. Protein A-Sepharose (GE Healthcare, Mississauga, ON, Canada) that had been equilibrated with the same solution was added to the mixture and the sample rotated overnight at 4 °C. The resin was separated by centrifugation, washed four times with ice-cold lysis buffer and then boiled in SDS sample buffer. Immunoblot analysis was performed with anti-human PIRH2, anti-HA antibody (Y-11; Santa Cruz), anti-Ub antibody (FK2; mouse monoclonal antibody; BIOMOL International (Plymouth Meeting, PA, USA) and Covance, Princeton, NJ, USA), anti-FLAG M2 antibody (Sigma), HRP-conjugated antibodies to mouse or rabbit immunoglobulin G (1 : 10 000 dilutions, Cell Signaling) and an enhanced chemiluminescence system (GE Healthcare).
Intracellular ubiquitination assay
HEK293T cells were transfected using the calcium phosphate method. Expression plasmids encoding the full-length human PIRH2 (WT) or a mutant lacking the ring finger domain (ΔR), HA-tagged WT or mutant Chk2, Flag-tagged WT USP28 or USP28-(C171A) (gifts from Dr. Elledge and Dr. Eilers) and Myc-tagged Ub were used for these transfections. After 48 h, cells were lysed in a modified RIPA buffer ((2 mM Tris-HCl (pH 7.5), 5 mM EDTA, 150 mM NaCl, 1% NP-40, 1% deoxycholate, 0.025% SDS, 1 mM PMSF and protease inhibitor cocktail tablets (Roche)). Cell lysates were centrifuged at 14 000 × g for 10 min at 4 °C, and the resulting supernatant was incubated with an affinity-purified rabbit polyclonal anti-Chk2 antibody for 2 h at 4 °C. Protein A-Sepharose (GE Healthcare) that had been equilibrated with the same solution was added to the mixture, which was then rotated overnight at 4 °C. The resin was separated by centrifugation, washed four times with ice-cold lysis buffer and then boiled in SDS sample buffer. Immunoblot analysis was performed with anti-Ub (FK2; anti-mouse monoclonal antibody, BIOMOL International and Covance) or anti-HA antibody (Y11; Santa Cruz), HRP-conjugated antibodies to mouse or rabbit immunoglobulin G and enhanced chemiluminescence system.
In vivo ubiquitination assay
Thymocytes from 8-week-old WT and Pirh2−/− mice were lysed in a modified RIPA buffer as described earlier. Cell lysates were immunoprecipitated with affinity-purified rabbit polyclonal anti-Chk2 antibody (first IP). The immunocomplexes were denatured in Laemmli’s sample buffer (25 mM Tris pH8.3, 192 mM glycine, 0.1% SDS) to dissociate contaminant proteins associated with Chk2 and reimmunoprecipitation (second IP) was performed using anti-Chk2 antibody. The resin was separated by centrifugation, washed four times with ice-cold lysis buffer and then boiled in SDS sample buffer. Immunoblot analysis was performed with anti-Ub or anti-Chk2 antibodies, HRP-conjugated antibodies to mouse or rabbit immunoglobulin G (1 : 5000 dilutions, Cell Signaling) and an enhanced chemiluminescence system (Amersham Pharmacia, Baie d'Urfe, QC, Canada).
In vitro ubiquitylation assay
The ubiquitylation reaction was performed in a volume of 20 μl in a buffer of 50 mM Tris pH 7.6, 5 mM MgCl2, 2 mM ATP and 2 mM DTT. The reaction mixture contained recombinant human Ub-activating enzyme E1 (50 ng), UBE2D2/UbcH5b (100 ng), Ub (5 μg), GST-PIRH2 (0.05–0.5 μg) and His6-tagged CHK2 (0.05–0.5 μg). After incubation at 30 °C for 90 min, the reactions were stopped by addition of SDS-PAGE sample buffer and resolved on 7.5% SDS-PAGE gels. Ubiquitylated proteins were detected by western blot using anti Chk2 and anti-Ub antibodies.
In vitro protein-binding assay
Recombinant proteins (1 μg of GST-PIRH2 and 1 μg of His6-Chk2) were mixed at 4 °C in 100 μl of protein-binding buffer (25 mM HEPES at pH 7.4, 150 mM KCl, 2 mM MgCl2, 0.5% Triton X-100, 1 mM EGTA and 1 mg/ml BSA). Immunoprecipitation was then performed using anti-Chk2 antibody, and the resulting precipitates were subjected to SDS-PAGE and immunoblot analysis with antibodies to GST or His.
Pulse-chase analysis with cycloheximide
Cells were cultured with cycloheximide at the concentration of 50 μg/ml and incubated for various times. Cell lysates were then prepared and subjected to SDS-PAGE. Immunoblot analyses were performed using anti-HA and anti-PIRH2 antibodies.
Immunohistochemistry
Tissues fixed in 10% buffered formalin were processed for paraffin-embedded sectioning at 5 μm and stained with hematoxylin and eosin. Immunohistochemistry was performed using an anti-Chk2 antibody raised against murine Chk2.30–47 Images were captured with HCX lenses on a Leica DM4000B microscope (Wetzlar, Germany) equipped with digital camera (Leica DC 300RF). Images were acquired using Leica Image Manager software. All digital images were imported into Photoshop (Adobe, San Jose, CA, USA) and adjusted for gain, contrast and gamma settings.
Recombinant proteins
His-tagged Chk2 was expressed in Escherichia coli strain BL21 (DE3; Invitrogen) and purified using ProBond metal affinity beads (Invitrogen). GST-tagged PIRH2 expressed in BL21 cells was purified by glutathione-Sepharose beads (Roche).
G1/S checkpoint assay
Primary MEFs (passages 1–3) were either untreated or irradiated (10 Gy) and left to recover for 16 h before pulsing with 10 μM BrDU for 4 h. Cells were then fixed overnight with 70% ethanol and stained with FITC-conjugated anti-BrDU antibody (e-Bioscience, San Diego, CA, USA) and propidium iodide as previously described.43
G2/M checkpoint assay
Early passage primary MEFs (passages 1–3) were left untreated or irradiated (2 Gy). One hour post irradiation, cells were fixed with 70% ethanol overnight and then stained with FITC-conjugated anti-phospho histone H3 (Ser10) antibody (Cell Signaling) and propidium iodide. Analysis of the cells was performed using flow cytometry as previously described.43
Statistical analysis
The statistical significance of experimental data (P-values ⩽0.05) was determined using Graph Prism (Graph pad software, Inc La Jolla, CA, USA) and ANOVA with or without Tukey–Kramer test.
Abbreviations
- ATM:
-
ataxia telangiectasia mutated
- CHK2:
-
checkpoint kinase 2
- HA:
-
hemagglutinin
- ΔR:
-
mutant lacking the ring finger domain
- MEF:
-
mouse embryonic fibroblast
- PIRH2:
-
p53-induced protein with a RING (Really Interesting New Gene)-H2
- Ub:
-
ubiquitin
- USP28:
-
ubiquitin-specific-processing protease 28
- WT:
-
wild type
References
Leng RP, Lin Y, Ma W, Wu H, Lemmers B, Chung S et al. Pirh2, a p53-induced ubiquitin-protein ligase, promotes p53 degradation. Cell 2003; 112: 779–791.
Sheng Y, Laister RC, Lemak A, Wu B, Tai E, Duan S et al. Molecular basis of Pirh2-mediated p53 ubiquitylation. Nat Struct Mol Biol 2008; 15: 1334–1342.
Wu H, Pomeroy SL, Ferreira M, Teider N, Mariani J, Nakayama KI et al. UBE4B promotes Hdm2-mediated degradation of the tumor suppressor p53. Nat Med 2011; 17: 347–355.
Hakem A, Bohgaki M, Lemmers B, Tai E, Salmena L, Matysiak-Zablocki E et al. Role of Pirh2 in mediating the regulation of p53 and c-Myc. PLoS Genet 2011; 7: e1002360.
Hattori T, Isobe T, Abe K, Kikuchi H, Kitagawa K, Oda T et al. Pirh2 promotes ubiquitin-dependent degradation of the cyclin-dependent kinase inhibitor p27Kip1. Cancer Res 2007; 67: 10789–10795.
Maruyama S, Miyajima N, Bohgaki M, Tsukiyama T, Shigemura M, Nonomura K et al. Ubiquitylation of epsilon-COP by PIRH2 and regulation of the secretion of PSA. Mol Cell Biochem 2008; 307: 73–82.
Abe K, Hattori T, Isobe T, Kitagawa K, Oda T, Uchida C et al. Pirh2 interacts with and ubiquitylates signal recognition particle receptor beta subunit. Biomed Res 2008; 29: 53–60.
Jung YS, Liu G, Chen X . Pirh2 E3 ubiquitin ligase targets DNA polymerase eta for 20S proteasomal degradation. Mol Cell Biol 2010; 30: 1041–1048.
Wu H, Zeinab RA, Flores ER, Leng RP . Pirh2, a ubiquitin E3 ligase, inhibits p73 transcriptional activity by promoting its ubiquitination. Mol Cancer Res 2011; 9: 1780–1790.
Jung YS, Qian Y, Chen X . The p73 tumor suppressor is targeted by Pirh2 RING finger E3 ubiquitin ligase for the proteasome-dependent degradation. J Biol Chem 2011; 286: 35388–35395.
Stracker TH, Usui T, Petrini JH . Taking the time to make important decisions: the checkpoint effector kinases Chk1 and Chk2 and the DNA damage response. DNA Repair (Amst) 2009; 8: 1047–1054.
Matsuoka S, Rotman G, Ogawa A, Shiloh Y, Tamai K, Elledge SJ . Ataxia telangiectasia-mutated phosphorylates Chk2 in vivo and in vitro. Proc Natl Acad Sci USA 2000; 97: 10389–10394.
Ahn JY, Schwarz JK, Piwnica-Worms H, Canman CE . Threonine 68 phosphorylation by ataxia telangiectasia mutated is required for efficient activation of Chk2 in response to ionizing radiation. Cancer Res 2000; 60: 5934–5936.
Melchionna R, Chen XB, Blasina A, McGowan CH . Threonine 68 is required for radiation-induced phosphorylation and activation of Cds1. Nat Cell Biol 2000; 2: 762–765.
Ahn JY, Li X, Davis HL, Canman CE . Phosphorylation of threonine 68 promotes oligomerization and autophosphorylation of the Chk2 protein kinase via the forkhead-associated domain. J Biol Chem 2002; 277: 19389–19395.
Lee CH, Chung JH . The hCds1 (Chk2)-FHA domain is essential for a chain of phosphorylation events on hCds1 that is induced by ionizing radiation. J Biol Chem 2001; 276: 30537–30541.
Xu X, Tsvetkov LM, Stern DF . Chk2 activation and phosphorylation-dependent oligomerization. Mol Cell Biol 2002; 22: 4419–4432.
Antoni L, Sodha N, Collins I, Garrett MD . CHK2 kinase: cancer susceptibility and cancer therapy – two sides of the same coin? Nat Rev Cancer 2007; 7: 925–936.
Tan Y, Raychaudhuri P, Costa RH . Chk2 mediates stabilization of the FoxM1 transcription factor to stimulate expression of DNA repair genes. Mol Cell Biol 2007; 27: 1007–1016.
Stolz A, Ertych N, Kienitz A, Vogel C, Schneider V, Fritz B et al. The CHK2-BRCA1 tumour suppressor pathway ensures chromosomal stability in human somatic cells. Nat Cell Biol 2010; 12: 492–499.
Castedo M, Perfettini JL, Roumier T, Yakushijin K, Horne D, Medema R et al. The cell cycle checkpoint kinase Chk2 is a negative regulator of mitotic catastrophe. Oncogene 2004; 23: 4353–4361.
Lavin MF . Ataxia-telangiectasia: from a rare disorder to a paradigm for cell signalling and cancer. Nat Rev Mol Cell Biol 2008; 9: 759–769.
Kass EM, Poyurovsky MV, Zhu Y, Prives C . Mdm2 and PCAF increase Chk2 ubiquitination and degradation independently of their intrinsic E3 ligase activities. Cell Cycle 2009; 8: 430–437.
Lovly CM, Yan L, Ryan CE, Takada S, Piwnica-Worms H . Regulation of Chk2 ubiquitination and signaling through autophosphorylation of serine 379. Mol Cell Biol 2008; 28: 5874–5885.
Feng L, Chen J . The E3 ligase RNF8 regulates KU80 removal and NHEJ repair. Nat Struct Mol Biol 2012; 19: 201–206.
Kass EM, Ahn J, Tanaka T, Freed-Pastor WA, Keezer S, Prives C . Stability of checkpoint kinase 2 is regulated via phosphorylation at serine 456. J Biol Chem 2007; 282: 30311–30321.
Zhang D, Zaugg K, Mak TW, Elledge SJ . A role for the deubiquitinating enzyme USP28 in control of the DNA-damage response. Cell 2006; 126: 529–542.
Popov N, Wanzel M, Madiredjo M, Zhang D, Beijersbergen R, Bernards R et al. The ubiquitin-specific protease USP28 is required for MYC stability. Nat Cell Biol 2007; 9: 765–774.
Hershko A, Ciechanover A . The ubiquitin system. Annu Rev Biochem 1998; 67: 425–479.
Grabbe C, Husnjak K, Dikic I . The spatial and temporal organization of ubiquitin networks. Nat Rev Mol Cell Biol 2011; 12: 295–307.
Li Q, Lin S, Wang X, Lian G, Lu Z, Guo H et al. Axin determines cell fate by controlling the p53 activation threshold after DNA damage. Nat Cell Biol 2009; 11: 1128–1134.
Di Micco R, Fumagalli M, Cicalese A, Piccinin S, Gasparini P, Luise C et al. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature 2006; 444: 638–642.
Lukas C, Bartkova J, Latella L, Falck J, Mailand N, Schroeder T et al. DNA damage-activated kinase Chk2 is independent of proliferation or differentiation yet correlates with tissue biology. Cancer Res 2001; 61: 4990–4993.
Yu X, Chini CC, He M, Mer G, Chen J . The BRCT domain is a phospho-protein binding domain. Science 2003; 302: 639–642.
Lee SB, Kim SH, Bell DW, Wahrer DC, Schiripo TA, Jorczak MM et al. Destabilization of CHK2 by a missense mutation associated with Li-Fraumeni Syndrome. Cancer Res 2001; 61: 8062–8067.
Bartkova J, Falck J, Rajpert-De Meyts E, Skakkebaek NE, Lukas J, Bartek J . Chk2 tumour suppressor protein in human spermatogenesis and testicular germ-cell tumours. Oncogene 2001; 20: 5897–5902.
Wu X, Webster SR, Chen J . Characterization of tumor-associated Chk2 mutations. J Biol Chem 2001; 276: 2971–2974.
Matsuoka S, Nakagawa T, Masuda A, Haruki N, Elledge SJ, Takahashi T . Reduced expression and impaired kinase activity of a Chk2 mutant identified in human lung cancer. Cancer Res 2001; 61: 5362–5365.
Henderson MJ, Munoz MA, Saunders DN, Clancy JL, Russell AJ, Williams B et al. EDD mediates DNA damage-induced activation of CHK2. J Biol Chem 2006; 281: 39990–40000.
Munoz MA, Saunders DN, Henderson MJ, Clancy JL, Russell AJ, Lehrbach G et al. The E3 ubiquitin ligase EDD regulates S-phase and G(2)/M DNA damage checkpoints. Cell Cycle 2007; 6: 3070–3077.
Hunter T . The age of crosstalk: phosphorylation, ubiquitination, and beyond. Mol Cell 2007; 28: 730–738.
Hirao A, Kong YY, Matsuoka S, Wakeham A, Ruland J, Yoshida H et al. DNA damage-induced activation of p53 by the checkpoint kinase Chk2. Science 2000; 287: 1824–1827.
Theunissen JW, Petrini JH . Methods for studying the cellular response to DNA damage: influence of the Mre11 complex on chromosome metabolism. Methods Enzymol 2006; 409: 251–284.
Acknowledgements
We thank V Stambolic, S Benchimol, C Arrowsmith, L Salmena, B Raught and the Hakem laboratory members for helpful discussions. We also thank Dr. T. Mak, Dr. S. Benchimol, Dr. S. Elledge and Dr. M. Eilers for providing reagents. This work was supported by the Canadian Institute of Health Research. RH was supported by a salary award from the Canadian Institute of Health Research.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by V Dixit
Supplementary Information accompanies the paper on Cell Death and Differentiation website
Rights and permissions
About this article
Cite this article
Bohgaki, M., Hakem, A., Halaby, M. et al. The E3 ligase PIRH2 polyubiquitylates CHK2 and regulates its turnover. Cell Death Differ 20, 812–822 (2013). https://doi.org/10.1038/cdd.2013.7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cdd.2013.7
Keywords
This article is cited by
-
The RNA-binding protein HuR is a novel target of Pirh2 E3 ubiquitin ligase
Cell Death & Disease (2021)
-
SIRT1 modulates cell cycle progression by regulating CHK2 acetylation−phosphorylation
Cell Death & Differentiation (2020)
-
Targeting deubiquitinase USP28 for cancer therapy
Cell Death & Disease (2018)
-
High Expression of Pirh2 is Associated with Poor Prognosis in Glioma
Cellular and Molecular Neurobiology (2017)
-
The emerging role of deubiquitinating enzymes in genomic integrity, diseases, and therapeutics
Cell & Bioscience (2016)