Abstract
Although Bcl-2 family members control caspase activity by regulating mitochondrial permeability, caspases can, in turn, amplify the apoptotic process upstream of mitochondria by ill-characterized mechanisms. We herein show that treatment with a potent inhibitor of Bcl-2 and Bcl-xL, ABT-737, triggers caspase-dependent induction of the BH3-only protein, Mcl-1 inhibitor, Noxa. RNA interference experiments reveal that induction of Noxa, and subsequent cell death, rely not only on the transcription factor E2F-1 but also on its regulator pRb. In response to ABT-737, pRb is cleaved by caspases into a p68Rb form that still interacts with E2F-1. Moreover, pRb occupies the noxa promoter together with E2F-1, in a caspase-dependent manner upon ABT-737 treatment. Thus, caspases contribute to trigger the mitochondrial apoptotic pathway by coupling Bcl-2/Bcl-xL inhibition to that of Mcl-1, via the pRb/E2F-1-dependent induction of Noxa.
Similar content being viewed by others
Main
The Bcl-2 family, composed of anti- and pro-apoptotic proteins, are major regulators of apoptosis1, 2, 3, 4 upstream of mitochondrial permeability and caspase activity. Pro-apoptotic proteins include multi-domain proteins, such as Bax and Bak, and their upstream effectors the Bcl-2 homology 3 (BH3)-only proteins, such as Bid, Bim, Puma, Bad, Noxa and so on. The anti-apoptotic members are frequently overexpressed in cancer cells and they promote survival, in great part, by physically interacting with the BH3 domain of their pro-apoptotic counterparts. Small molecules that bind to the BH3-binding grove of Bcl-2 homologs (the so-called ‘BH3 mimetics’) have been developed as pro-apoptotic inhibitors of these proteins.5 There are subtle yet significant differences in the BH3-binding interfaces of Bcl-2 homologs: Bim or Puma interact with all known Bcl-2 homologs, whereas Bad interacts preferentially with Bcl-2 and Bcl-xL and Noxa with Mcl-1.6, 7 These differences explain why currently known BH3-mimetics only inhibit subsets of anti-apoptotic proteins.8 One of these compounds is ABT-737, which occasionally presents in vitro monotherapy toxicity.5 It potently inhibits the BH3-binding activity of Bcl-2, Bcl-xL and Bcl-w but not that of Mcl-1 and Bfl-1.9 ABT-737 promotes cell death by displacing, from its targets, ‘BH3 activators’ such as Bim or Puma (BH3-only proteins that can directly activate multi-domain proteins when free from anti-apoptotic proteins)10, 11 and/or active Bax.12 Efficient induction of apoptosis by ABT-737 requires that pro-apoptotic proteins are not sequestered by an excess of empty Mcl-1 or Bfl-1 that are not efficiently inhibited by ABT-737. Thus, sensitivity to ABT-737 is enhanced by combined treatments that decrease Mcl-1 expression and/or induce Noxa, a BH3-only protein that essentially functions as an inhibitor of Mcl-1.13, 14, 15, 16, 17, 18
ABT-737 is a powerful tool to investigate how death signals induced by direct inhibition of subsets of anti-apoptotic Bcl-2 family members lead to cell demise. Caspase activity contributes to the final stages of cell death induced by inhibition of Bcl-2 homologs. However, executioner caspases were found to be required for full-blown Bax activation and mitochondrial permeabilisation in response to diverse stimuli.19 In addition, when caspase activity is blocked, subsets of mitochondria remain refractory to permeabilisation and allow cells to survive to death stimuli.20, 21 Thus, caspase activity might also amplify the apoptotic process upstream of mitochondria, and fuel signals initiated by inhibition of some Bcl-2 homologs by ill-characterized mechanisms.
It is notable that, whereas the pro-apoptotic activity of ABT-737 relies on the nearly immediate ability of this compound to disrupt pre-existing complexes,8, 22 its effects on whole cells sometimes take numerous days to be manifest, implying that de novo synthesis of key actors might intervene. ABT-737 treatment was shown to induce the transcription of death receptor 523 and to induce a twofold change in the transcription of nearly 430 genes when added to renal carcinoma cells.24 Most relevantly here, low-level activation of the caspase cascade was incriminated in some of these transcriptomic effects.24 Thus, caspases may contribute to the long-term biological effects of Bcl-2/Bcl-xL inhibition. Whether and how this could actually amplify cell death induced by such inhibition remains to be determined.
In this manuscript, we show that caspase activity contributes to the response of cancer cells to ABT-737 by promoting the transcriptional induction of Noxa. Transcriptional pathways known to modulate levels of the Bcl-2 protein family (including that of Noxa) involve p53 or E2F-1.25, 26 E2F-1 differs from that of other E2F family members due to its ability to regulate not only cell-cycle progression but also apoptosis as it directly induces the expression of p73, of caspase 3 and 7 and of some pro-apoptotic Bcl-2 family members.27, 28, 29, 30, 31, 32, 33, 34 We show here that E2F-1 is a major contributor of caspase-dependent induction of Noxa in response to ABT-737 treatment. Caspases cleave the E2F-1 regulator pRb in ABT-737-treated cells, giving rise to a p68Rb truncated form, which has a direct role in Noxa and cell death inductions together with E2F-1. Thus, caspase activity provides a feed-forward mechanism that amplifies the mitochondrial apoptotic pathway by coupling inhibition of Bcl-2/Bcl-xL to that of Mcl-1, via the induction of E2F-1 transcription of Noxa by a pRb-dependent mechanism.
Results
ABT-737 induces late but specific Bax and caspase-dependent apoptosis in the glioma U251 cells
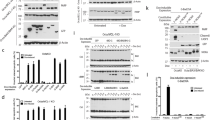
The effects of ABT-737 were investigated on glioma U251 cells in which significant cell death rates were measured only after 48 h of treatment with 2 μM of ABT-737, whereas no cell death was detected at earlier time points (Figure 1a). These delayed effects of ABT-737 were confirmed to be on-target effects, as simultaneous silencing of Bcl-2 and Bcl-xL, but not that of either one alone, induced cell death at a similar rate to that measured upon ABT-737 treatment (Figures 1a and b). This indicates that the viability of U251 requires the sustained and combined activities of Bcl-2 and Bcl-xL. Consistent with a role for the canonical mitochondrial apoptotic pathway in delayed cell death that results from Bcl-2/Bcl-xL inhibition, knock down of Bax expression by siRNA significantly reduced cell death rates induced by ABT-737 treatment (Figure 1c). Finally, pre-treatment of U251 cells with a pan-caspase inhibitor before addition of ABT-737 inhibited cell death (Figure 1a), showing that the late effects of ABT-737 on cell viability rely on caspase activation.
ABT-737 induces caspases and Bax-dependent cell death in U251. (a) U251 cells were treated for 24 or 48 h by ABT-737 at 2 μM and/or Z-VAD-FMK at 50 μM (zVAD). (b and c) U251 cells were transfected with the indicated siRNA. Forty-eight hours later, cells were harvested (b) or treated by ABT-737 at 2 μM for an additional 48 h (c). Western blot analysis of Bcl-2, Bcl-xL (b) and Bax (c) was performed. Cell death in a–c was assessed by a trypan blue staining procedure. Data presented in a–c are mean±S.E.M. of three independent experiments
E2F-1-dependent induction of the BH3-only Noxa has a key role in the induction of U251 cell death by ABT-737
The delayed effects on cell death of ABT-737 suggest that the mounting of a death signal, possibly via the de novo synthesis of a key factor, might contribute to the response. Kraft and colleagues recently identified the noxa gene (PMAIP1) as one gene whose expression is strongly induced by ABT-737.24 Consistently, we observed a strong increase of Noxa expression in U251 cells upon ABT-737 treatment at both protein and mRNA levels (Figures 2a and b). Importantly, silencing of Noxa significantly decreased cell death rates induced by ABT-737 treatment (Figure 2c), indicating that such induction has a role in cell death induction.
The BH3-only Noxa is involved in ABT-737 induction of cell death. (a) U251 cells were treated by ABT-737 at 2 μM for 48 h and western blot analysis of Noxa was performed. (b) noxa mRNA levels were measured in U251 cells treated or not by ABT-737 at 2 μM and/or QVD-OPh at 10 μM for 48 h by real-time RT-PCR analysis and normalized to rplpO and β2-microglobulin. (c) U251 cells were transfected with control (siScr) or Noxa siRNAs. Forty-eight hours later, cells were treated by ABT-737 at 2 μM for an additional 48 h. Western blot analysis of Noxa was performed and cell death was assessed by a trypan blue staining. Data presented in b and c are mean±S.E.M. of three independent experiments
Transcriptional factors such as p53 and E2F-1 are known to regulate Noxa expression. As U251 cells express a mutated p53 (R273H), we focused on E2F-1, as the mRNA levels of another E2F-1 target p73 were also found to be upregulated by ABT-737 (Supplementary Figure S1A). Silencing of E2F-1 by RNA interference prevented Noxa induction by ABT-737 (Figure 3a) and decreased cell death rates (Figure 3b). Conversely, transient overexpression of E2F-1 in U251 cells increased Noxa expression (Figure 3c), affected cell viability by itself and enhanced cell sensitivity to ABT-737 treatment (Figure 3d). By contrast, overexpression of a transcriptionally inactive mutant of E2F-1 (E132)30 had no effect (Figure 3d). Notably, upregulation of Noxa and sensitization to ABT-737 induction of cell death by overexpression of E2F-1 were also observed in the colorectal cancer p53-null HCT116 cells (Supplementary Figure S2), establishing that p53 is dispensable for E2F-1 -dependent induction of Noxa.
E2F-1 and pRb are involved in the induction of cell death and Noxa by ABT-737. (a and b) U251 cells were transfected with the indicated siRNA. Forty-eight hours later, cells were treated by ABT-737 at 2 μM for an additional 48 h. Western blot analysis of Noxa (a), pRb and E2F-1 (b) was performed, and cell death was assessed by a trypan blue staining (b). (c and d) U251 cells were transfected with pcDNA3Flag (vector control), pE2F1Flag (E2F1) or pE2F1E132 (E132) vector. Twenty-four hours later, cells were treated by ABT-737 at 2 μM for an additional 48 h. (c) Western blot analysis of Noxa and E2F-1 was performed and (d) cell death was assessed as in b. Data presented in b and d are mean±S.E.M. of three independent experiments
Taken together, these results indicate that ABT-737 treatment induces E2F-1-dependent Noxa upregulation, thereby contributing to cell death induced by the inhibition of Bcl-2 and Bcl-xL.
The E2F-1 regulatory protein pRb is cleaved by caspases upon ABT-737 treatment
To understand how ABT-737 treatment might influence E2F-1 activity, we investigated its influence on the expression of E2F-1 and of its regulator pRb. We observed no detectable modification in the expression levels of E2F-1 upon ABT-737 treatment (Figure 4a). We did not detect any modification of the phosphorylation of full-length pRb at Ser 807–811, a phosphorylation that relieves E2F-1 effects on cell-cycle progression from the negative regulation of pRb (Figure 4a). However, ABT-737 treatment modified the size of pRb polypeptides. Three bands of 100, 68 and 48 kDa, respectively, in addition to the 110 kDa band corresponding to full-length pRb, were immunodetected in lysates from ABT-737-treated U251 cells (Figures 4b and c). These bands were not detected in ABT-737-treated cells in which pRb was silenced, confirming that they all correspond to pRb polypeptides (Figure 4d). Notably, the truncated pRb polypeptides were also observed in cells in which Bcl-2 and Bcl-xL were silenced simultaneously, but not when either one of them was downregulated alone, indicating that their appearance results from an on-target effect of ABT-737 (Figure 4e).
pRb is truncated upon ABT-737 treatment in a Bax-dependent manner. (a–c) U251 cells were treated by ABT-737 at 2 μM for 48 h and western blot analysis of proteins expression were performed. (d and e) U251 cells were transfected with the indicated siRNA. Forty-eight hours later, cells were harvested (e) or treated by ABT-737 at 2 μM or not for an additional 48 h (d and e). Western blot analysis of protein expression was performed. Antibody (Ab) XZ55 directed against the full-length pRb, p100Rb and pRb68 forms was used in a, b, d and e panels while Ab G3-245 directed against the full-length pRb, p100Rb and pRb48 forms was used in c and d panels
Kinetic analysis of ABT-737-treated U251 cells indicated that the appearance of the truncated forms of pRb was concomitant with increases in cell death rates, suggesting that it may associate with a pro-death mechanism (Figure 5a, see also below). It is documented that cleavage of pRb by caspases 3 and 7 at the DEAD site truncates pRb at the C-terminal end, giving rise to a band of 100 kDa, whereas two bands of 48 and 68 kDa, respectively, are obtained after cleavage at the DSID site (Figure 5b).35, 36, 37 In U251 cells pre-treated with a pan-caspase inhibitor before ABT-737, the p68Rb form was no longer detected (Figure 5c). Moreover, neither the p68Rb nor the p48Rb forms were detected after treatment of cells with ABT-737 in which Bax expression had been silenced (Figure 4d).
pRb truncation and Noxa induction are caspase dependent. (a) U251 cells were treated by ABT-737 at 2 μM for indicated time. Western blot analysis of pRb expression with antibody (Ab) XZ55 was performed and cell death was assessed by a trypan blue staining. (b) Scheme of pRb with caspase cleavage sites, E2F interaction sites, antibody fixation sites and pRb cleaved forms p48Rb, p68Rb and p100Rb. (c) U251 cells were treated by ABT-737 at 2 μM for 48 h (c), or for indicated time (d and e), and/or the pan caspase inhibitor QVD-OPh at 10 μM. Western blot analysis of pRb (Ab XZ55) (c) or Noxa (d) was performed and caspase activities were measured (e). Data presented in e are mean±S.E.M. of three independent experiments
These data support the notion that pRb is cleaved by caspases that are activated as a result of Bax-dependent mitochondrial permeabilisation in response to ABT-737.
pRb contributes to caspase-dependent induction of Noxa
As pRb cleavages occurred concomitantly to cell death and as Noxa is an actor of this cell death, we wondered if pRb might have a role in Noxa induction upon ABT-737 treatment. Consistent with a positive role for pRb in these events, silencing of pRb by RNA interference prevented the upregulation of Noxa by ABT-737, as observed when E2F-1 was silenced (Figure 3a). Moreover, it had no effect on the viability of U251 cells by itself but it significantly decreased cell death rates induced by ABT-737 treatment (Figure 3b).
Involvement of pRb in Noxa induction prompted us to investigate whether caspases contribute to Noxa induction upon ABT-737 treatment. Pretreatment of U251 cells with a pan-caspase inhibitor impeded the induction of Noxa (at both protein and mRNA levels) by ABT-737 (Figures 2b and 5d). Importantly, kinetics of pRb cleavages and Noxa induction followed the kinetics of caspase activation. Indeed, induction of Noxa was detected at 40 h of ABT-737 treatment (Figure 5d), a time point that coincided with detectable appearance of pRb cleavage products (Figure 5a), detectable enhancement of specific caspase activity (Figure 5e) and detectable enhancement of cell death rates (Figure 5a). This puts forth a temporal link between caspase activation and Noxa induction by ABT-737.
Taken altogether, these data argue that pRb and caspases participate to Noxa induction in response to ABT-737.
pRb and E2F-1 bind to the Noxa promoter upon ABT-737 treatment
The regulation of E2F-1 transcriptional activity by pRb is, in great part, due to a physical interaction between these two proteins.27, 28 It is well established that the binding of the hypophosphorylated pRb to E2F-1 inhibits the ability of the latter to induce the expression of genes involved in cell-cycle progression.38 By contrast, recent data showed that hyperphosphorylated pRb can maintain an interaction with E2F-139 and that pRb may also function, in certain instances, as a binding partner for E2F-1 to positively upregulate pro-apoptotic genes, such as p73.40, 41 To further understand how, upon ABT-737 treatment, E2F-1 and pRb contribute to induce Noxa expression and cell death, we studied the interactions between E2F-1 and the forms of pRb, in addition to their individual and combined presence on the noxa promoter.
By co-immunoprecipitation assays using antiE2F-1 antibodies and lysates from ABT-737 or mock-treated cell, we observed that E2F-1 interacted with full-length pRb in U251 cells treated with ABT-737 or not (Figure 6, top panel), even in its phosphorylated form (Figure 6, bottom panel). The p68Rb form, but not the p48Rb form, co-immunoprecipitated with anti E2F-1 antibodies upon ABT-737 treatment (Figure 6, bottom panel). This is consistent with the fact that p68Rb still harbors the domain of interaction with E2F-1, whereas p48Rb does not (Figure 5b).38 The use of pRb antibodies (that recognized both full-length and p68Rb forms) in reverse immunoprecipitation experiments confirmed that E2F-1 co-immunopreciptated with pRb in cells treated with ABT-737 or not (Figure 6, top panel).
E2F-1 interacts with full-length and truncated p68Rb upon ABT-737 treatment. U251 cells were treated by ABT-737 at 2 μM for 48 h and cells lysates were immunoprecipated (IP) with an anti-E2F-1 (antibody (Ab) C-20) or an anti-pRb antibody (Ab 4H1). The presence of E2F-1, full-length pRb or p68Rb (Ab XZ55), p48Rb (Ab G3-245) truncated forms and phosphorylated form of pRb (Ab pS807-pS811) were analyzed by western blot
The data above suggest that pRb may cooperate with E2F-1 to induce Noxa transcription upon ABT-737 treatment. Thus, we studied the ability of these two proteins to occupy noxa and p73 promoter regions. Chromatin immunoprecipitation (ChIP) experiments showed that both E2F-1 and pRb were present on noxa and p73 promoter regions and that this occupancy was increased by ABT-737 treatment (Figure 7a). Also, in correlation with the expression patterns of these genes following ABT-737 treatment, RNA polymerase II was observed bound to these promoters. The recruitment of pRb on these promoters appeared specific as it was not recruited on the promoter of plk1 (a E2F-1 target gene involved in cell-cycle progression), whereas E2F-1 was present as expected (Figure 7a).
pRb and E2F-1 are recruited together on noxa and p73 promoters upon ABT-737 treatment. (a–c) U251 cells were treated or not by ABT-737 at 2 μM for 48 h and/or the pan caspase inhibitor QVD-OPh at 10 μM. Soluble chromatin was immunoprecipitated (a and c), or used for serial chromatin immunoprecipitation assays (Re-ChIP) (b), with indicated antibodies directed against pRb (antibody (Ab) 4H1), E2F-1 (Ab C-20) and RNA polymerase II (Pol II). DNA was amplified using primers that cover the E2F-1-binding sites of the indicated promoters and amplicons visualized on agarose gel. (d) U251 cells were treated or not by ABT-737 at 2 μM for 48 h. Chromatin and soluble nuclear fractions were extracted and western blot analysis of pRb (Ab XZ55) was performed. (e) BT549 cells were infected for 48 h, or not (Untr), with retroviruses expressing either GFP alone (vector: pMIG), p48Rb or p68Rb truncated forms and treated with ABT-737 at 2 μM (top panel) or with the pan caspase inhibitor QVD-OPh at 10 μM (bottom panel). Noxa and/or pRb (Abs XZ55 and G3-245) expression were analyzed by western blot
To further investigate whether E2F-1 and pRb occupy promoter regions in ABT-737-treated cells as binding partners, serial chromatin immunoprecipitation assays were performed (Figure 7b). E2F-1 and pRb were indeed recruited together on noxa and p73 promoters upon ABT-737. Moreover, the pRb complexes present on these promoters were associated with RNA polymerase II stabilization in response to ABT-737. These results indicate that pRb-E2F1-RNA polymerase II complexes bind specifically to noxa and p73 promoter regions, supporting the notion that these complexes function as transcriptional inducers of these genes in response to ABT-737.
The antibodies used in the ChIP assays cannot discriminate full-length pRb from its p68Rb-truncated form. We thus performed numerous assays to investigate whether caspase-cleaved p68Rb contributes to E2F-1 transcriptional activation of pro-apoptotic genes upon ABT-737 treatment. First, we performed ChIP assays in cells pretreated with a pan caspase inhibitor before ABT-737 (Figure 7c) to evaluate a role for caspases in pRb recruitment to critical promoters. We observed that the recruitment of pRb, and that of RNA polymerase II, on p73 and noxa promoters in ABT-737-treated cells were strongly reduced by caspase inhibition. Second, we analyzed the presence of pRb forms in the chromatin and in the soluble fractions of nuclear extracts of U251 cells treated with ABT-737 or not. The full-length pRb form was detected in both fractions of untreated cells while p68Rb strongly accumulated in the chromatin fraction of treated cells (Figure 7d). Third, we directly tested the ability of p68Rb to induce Noxa expression independently from caspases. Breast cancer BT549 pRb-negative cell line was infected with retroviruses allowing expression p48Rb or p68Rb truncated forms alongside with a GFP marker (Figure 7e). The ectopic expression of p68Rb was sufficient by itself to trigger a dramatic accumulation of Noxa in BT549 cells treated with a pan-caspase inhibitor. Induction of Noxa by ectopic p68Rb was also patent in ABT-737-treated BT549 cells. By contrast, no Noxa induction was observed in uninfected cells or in cells expressing ectopic p48Rb (Figure 7e).
As the recruitment of pRb on the noxa and p73 promoter together with RNA polymerase II is caspase-dependent, as the p68Rb form is chromatin bound and as its ectopic expression leads to Noxa induction, our data strongly argue that caspase cleavage of pRb into p68Rb contributes to Noxa transcription in ABT-737-treated cells.
Nutlin-3a sensitizes p53-mutated breast cancer cells to ABT-737 by upregulating Noxa in a pRb- and E2F-1-dependent manner
As treatment with the Mdm2 inhibitor Nutlin-3a was shown to enhance the pRb/E2F-1 pathway,33, 34 we reasoned that it may increase cell sensitivity to ABT-737, provided pRb is present. We found that combination of Nutlin-3a to ABT-737, but not treatment with anyone alone, induced dramatic cell death in the breast cancer cell lines MDA-MB-231 and MDA-MB-468, cells that carry p53 mutations (R280K and R273H, respectively) and constitutively express low levels of pRb (Figures 8a and b). A strong induction of Noxa protein expression, associated with a cleavage of pRb, was detected in both the cell lines treated with ABT-737 and Nutlin-3a (Figure 8b). Moreover, silencing of either Bax, Noxa, pRb or E2F-1 lead to a decrease of MDA-MB-231 cell death in response to the combination treatment ABT-737/Nutlin-3a (Figure 8c). Silencing of E2F-1 and pRb impeded Noxa induction in ABT-737/Nutlin-3a-treated cells, suggesting that they regulate Noxa expression under these conditions (Figure 8c). By contrast, neither cell death nor induction of Noxa were observed in ABT-737/Nutlin-3a-treated BT549 cells, which lack pRb expression, consistent with the notion that cell death induced by ABT-737/Nutlin-3a involves pRb (Figures 8a and d).
Nutlin-3a sensitizes MDA-MB-231 and MDA-MB-468 cells to ABT-737 by upregulating Noxa. (a and b) MDA-MB-231, MDA-MB-468 and BT549 cells were treated by ABT-737 at 2 μM and/or Nutlin-3a at 10 μM for 48 h. (a) Cell death was assessed by trypan bleu staining and (b) Noxa and pRb (antibody (Ab) XZ55) expression levels were analyzed by western blot. (c) MDA-MB-231 cells were transfected with the indicated siRNA. Forty-eight hours later, cells were treated by ABT-737 at 2 μM and Nutlin-3a at 10 μM for 48 h. Western blot analysis of Bax, Noxa, E2F-1 and pRb was performed and cell death was assessed as in a. (d) BT549 cells were treated by ABT-737 at 2 μM and/or Nutlin-3a at 10 μM for 48 h and Noxa expression was analyzed by western blot. Data presented in a and c are mean±S.E.M. of three independent experiments
Thus, cell death induced by ABT-737 combined with Nutlin-3a may recruit pRb and E2F-1 and relies on their ability to increase Noxa expression.
Discussion
We report here that inhibition of Bcl-2 and Bcl-xL by ABT-737 enhances a caspase-dependent increase in Noxa induction mediated by E2F-1 and, less expectedly, by its regulator protein pRb. Transcriptional upregulation of Noxa was previously described in response to treatment with BH3-mimetics.24, 42 As Noxa potently binds to Mcl-1,6 its ability to promote ABT-737 induction of cell death generally relies on its capacity to inhibit Mcl-1 that is not targeted by ABT-737.13, 14, 15, 16, 17 Silencing of Mcl-1 increased cell death rates in ABT-737-treated U251 cells (without affecting Noxa expression in untreated or ABT-737-treated cells, Supplementary Figures S3A and S3B). This is consistent with the notion that, in these cells, Mcl-1 represents a major restrain to ABT-737 efficiency that Noxa induction contributes to overcome. The novel perspective that emerges from our study is that caspases impact on mitochondrial permeability by triggering Noxa-mediated Mcl-1 inhibition in response to inhibition of Bcl-2 and Bcl-xL and that this occurs via transcription-dependent mechanisms that involve E2F-1 and pRb.
The transcription factor E2F-1 is a key actor of this caspase-dependent induction of Noxa, as evidenced by its role in cell death induced by ABT-737, by its influence on Noxa expression and its ability to occupy the noxa promoter. E2F-1 was already described to promote apoptosis in response to either DNA damage or oncogenic stress, in part, through regulating the transcription of genes encoding for BH3-only proteins.25, 27, 28, 30 In U251 cells, ABT-737 had no effect on Puma expression, which appeared dispensable for efficient induction of apoptosis (Supplementary Figures S3A and S3C). By contrast, the E2F-1 target gene p73 was enhanced upon ABT-737 treatment (Supplementary Figure S1A) and its silencing affected cell death and also impeded the expression of Noxa (Supplementary Figure S1B). Thus, p73 may by itself contribute to cell death in response to ABT-737. This suggests that E2F-1 (and pRb as discussed below) may regulate Noxa transcription (and cell death) not only directly but also indirectly via its transcriptional target p73.
A recent work showed that pRb participates (even in its phosphorylated form) to form a transcriptionally active complex with E2F-1 to regulate expression of pro-apoptotic genes.40, 41 Our data are mostly consistent with these data, as they show that, upon ABT-737 treatment, pRb interacts with E2F-1, is recruited to both noxa and p73 promoters and contributes to Noxa expression and to cell death induction, contrary to current understanding of the regulation of E2F-1 by pRb. Strikingly, our data establish that caspase cleavage of pRb may favor these molecular events. One feature of pRb is that it can provide a link between caspase activation and E2F-1 activity as it is cleaved by caspases during chemotherapy-induced cell death, at two distinct sites.37 Upon ABT-737 treatment also, pRb is cleaved at these two sites and gives rise to the p68Rb and p48Rb forms. Cleavage of pRb was proposed to promote apoptosis via the release of E2F-1 from cleaved pRbs.37, 43 Nevertheless, the p68Rb-truncated form keeps the domain of interaction with E2F-1 and, in agreement, we found that p68Rb interacts with E2F-1 upon ABT-737 treatment. As caspase activity is required to upregulate Noxa and as a siRNA-targeting pRb expression impedes the induction of Noxa, we propose that p68Rb has an active role in the E2F-1 upregulation of Noxa (Supplementary Figure S4). We suggest that it does so by engaging complexes in which E2F-1 is transcriptionally active and that encompass RNA polymerase II. Supporting this, we found that the p68Rb truncated form is present in the chromatin fraction of ABT-737-treated cells, that caspase inhibition affects the recruitment of pRb and RNA polymerase II on noxa and p73 promoters and that ectopic expression of p68Rb in the BT549 pRb-negative cells promotes a strong induction of Noxa independently from caspases.
In conclusion, our studies put forth the notion that, upon inhibition of Bcl-2/Bcl-xL, caspases can trigger Mcl-1 inhibition through induction of Noxa. They do so by generating a cleaved form of p68Rb that exerts a previously unforeseen positive regulation of the noxa promoter together with E2F-1, with which it interacts. Enhancement of this transcriptional pathway may constitute an efficient way to overcome cancer cell resistance to ABT-737, independently of p53, as exemplified by the fact that the combination of ABT-737 with Nutlin-3a induces Noxa and promotes cell death in breast cancer cells carrying mutant p53, provided pRb is expressed.
Materials and Methods
Cell lines and cell culture
The U251, MDA-MB-231, MDA-MB-468 and BT 549 cell lines obtained from ATCC, were grown in DMEM and RPMI medium, respectively. HCT116 p53−/− cells, grown in McCoy’s 5A, were kindly provided by Dr B. Vogelstein (The John Hopkins Kimmel Cancer Center, Baltimore, MD, USA). When specified, ABT-737 was used at 2 μM, Nutlin-3a (Sigma, St Louis, MO, USA) at 10 μM, zVAD-FMK (Promega, Madison, WI, USA) at 50 μM and QVD-OPh (R&D System, Minneapolis, MN, USA) at 10 μM.
Plasmids, siRNAs and transfection
The E2F-1 cDNA from pSG5LHAE2F1 vector (Addgene, Cambridge, MA, USA, ref. 10736)44 was cloned into pcDNA3Flag from pcDNA3Flagp53 (Addgene ref. 10838)45 and the resulting plasmid, named pE2F1Flag, was sequenced. The E132 E2F-1, which carries a point mutation within the DNA-binding domain, was expressed from a plasmid obtained from Addgene (ref. 24224).46 Following siRNAs were used: siBax (HSC.RNAI.N138761.10.1), siBcl-2 (HSC.RNAI.N000633.6.1), siBcl-xL (BCL2L1 ON-TARGETplus L-003458-00), siE2F1 (HSC.RNAI.N005225.10.3), siNoxa (AC2Z4U4), siPuma (ON-TARGETplus smart pool t-004380-00-0005), sip73 (HSC.RNAI.N0055427.10.8), sipRb (sc-29468) and SiScramble (siScr) (SC37007). Plasmids and siRNAs were transfected according to manufacturer’s instructions (Invitrogen, Carlsbad, CA, USA) using Lipofectamine 2000 and lipofectamine RNAi Max, respectively.
Retroviral production
The bicistronic retroviral pMIG vector containing an internal ribosome entry site upstream of the enhanced GFP gene,47 was used to introduce cDNAs corresponding to p48Rb or p68Rb using BglII/HpaI and BamHI/PmeI sites, respectively. Retroviruses were produced in Phoenix cells maintained in DMEM with 10% (vol/vol) FBS. Briefly, 10 Phoenix cells were transfected with 45 μg of pMIG (mock, encoding p48Rb or p68Rb) and 5 μg of pVSVG (encoding the vesicular stomatitis virus G protein) complexes with PEI.6 Forty-eight hours after transfection, viral supernatants were collected. MOI was determined by flow cytometry and BT549 cells were infected at MOI of 1.
Cellular assays
Cell viability was determined by a trypan blue staining procedure. Caspase activities were measured using the Caspase Glow assay system according to the manufacturer’s protocol (Promega) and were normalized to the activity measured under control condition. Immunoprecipitations were performed at 4 °C on cell extracts (500 μg) from cells lysed in lysis buffer (HEPES 10 mM; NaCl 150 mM; 1% CHAPS; pH7.4; protease inhibitors; 0.5% Nonidet P-40). Cell extracts were precleared with 20μl of protein G-agarose (Sigma-Aldrich, St Louis, MO, USA) for 1 h, and cleared extracts were immunoprecipitated with 4 μg of the indicated antibodies overnight followed by the addition of 50 μl of protein G-agarose for 1 h. Immunoprecipitated proteins were washed in lysis buffer and once with 10 mM Tris, pH 8, 100 mM EDTA and then eluted with heated sample buffer. To extract proteins from the chromatin and soluble nuclear fraction, cells were collected with a rubber scraper. The pellet was resuspended in solution A (10 mM Hepes pH7.9, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM DTT, 0.5 mM PMSF) and submitted to three freezing/thawing cycles. The mixture was centrifuged at 2000 r.p.m. for 5 minutes. The pellet was washed in solution A and then resuspended in solution C (20 mM Hepes pH 7.9, 25% glycerol, 1.5 mM MgCl2, 420 mM NaCl, 0.2 mM EDTA, 0.5 mM DTT, 0.5 mM PMSF). The mixture was incubated for 30 min in ice and then centrifuged at 12 000 r.p.m. for 15 min. The supernatant represents the soluble nuclear fraction. The pellet, corresponding to the chromatin fraction, was resuspended in lysis buffer (SDS: 1%; EDTA: 10 mM; Tris-Hcl ph 8,1: 50 mM; protease inhibitors Na3VO4 1 mM, NaF 100 × ) and sonicated. The mixture was centrifuged at 13 000 r.p.m. and the supernatant that corresponds to the chromatin fraction was collected.
RNA extraction and quantitative real-time PCR
Total RNAs were extracted with RNAeasy mini kit (Qiagen, Valencia, CA, USA). The quality of the RNAs was assessed by analysis on an Agilent 2100 bioanalyser (Agilent Biotechnologies, Santa Clara, CA, USA). RNAs were retro-transcribed using Superscript transcriptase (Superscript II, Invitrogen). Following forward and reverse primers were used: p73 (5′-CTTCAACGAAGGACAGTCTG-3′ and 5′-AAGTTGTACAGGATGGTGGT-3′), Noxa (5′-GCTGGAAGTCGAGTGTGCTA-3′ and 5′-CCTGAGCAGAAGAGTTTGGA-3′). Real-time PCR was carried out as previously described.48
Immunoblot analysis and antibodies
After two washings with cold PBS, cells were lysed in ice-cold lysis buffer (SDS: 1%; EDTA: 10 mM; Tris-Hcl ph 8,1: 50 mM; protease inhibitors Na3VO4 1 mM, NaF 100 × ) and extracts were sonicated. Protein extracts were separated by SDS-PAGE, transferred onto a PVDF membrane (Millipore, Billerica, MA, USA) and revealed with a chemiluminescence kit (Millipore). Following antibodies were used: actin (MAB1501R, Millipore), β-tubulin (T0198, Sigma), Bax (A3533, Dako, Glostrup, Denmark), Bcl-2 (1017-1, Epitomics, Burlingame, CA, USA), Bcl-xL (1018-1, Epitomics), E2F-1 (3742, Cell Signaling, Danvers, MA, USA), Mcl-1 (sc-819, Santa Cruz Biotechnologies, Santa Cruz, CA, USA), Noxa (ALX-804-408, Enzo Life Science, New York, NY, USA), pRb XZ55 (554144, BD Pharmingen, San Diego, CA, USA), pRb G3-245 (554136, BD Pharmingen), phospho-pRb (S807-811) (558389, BD Pharmingen), p73 (5B429, Imgenex, San Diego, CA, USA), Puma (4743, Sigma), and Flag (F1804, Sigma). The following antibodies against E2F-1 C-20 (sc-193, Santa cruz Biotechnologies), pRb 4H1 (9309, Cell Signaling) and polymerase II (sc-899, Santa Cruz Biotechnologies) were used for immunoprecipitations and ChIP assays.
ChIP and serial ChIP assay
Single (ChIP) and serial ChIP experiments were performed essentially as previously described.49 The following primers were used: NOXA IS: 5′-CGTCTAGTTTCCCTACGTC-3′; 5′-AGATGCCAACTACACACG-3′, p73-1100: 5′-TGAGCCATGAAGATGTGC-3′; 5′-GCTGCTTATGGTCTGATGCTTATG-3′,39 p73 control 5′-CAATTGTCCCCCTCTTCTGA-3′; 5′-GTGGCAGAAGGGTGCTTAAA-3′ and PLK1 IS 5′-GTTTTCCCCGGCTGGGTCCG-3′; 5′-AAGCTGCGCTGCAGACCTCG-3′.
Data analysis
Data were from at least three independent experiments. Statistical analysis of data was performed using one-tailed Student’s t-test on GraphPad Prism (La Jolla, CA, USA). Error bars represent S.E.Ms. The following symbols are used: *, ** and *** that correspond to a P value inferior to 0.05, 0.01 and 0.001, respectively.
Abbreviations
- BH:
-
Bcl-2 homology
- ChIP:
-
chromatin immunoprecipitation
References
Adams JM, Cory S . The Bcl-2 apoptotic switch in cancer development and therapy. Oncogene 2007; 26: 1324–1337.
Lalier L, Cartron P-F, Juin P, Nedelkina S, Manon S, Bechinger B et al. Bax activation and mitochondrial insertion during apoptosis. Apoptosis 2007; 12: 887–896.
Chipuk JE, Moldoveanu T, Llambi F, Parsons MJ, Green DR . The BCL-2 family reunion. Mol Cell 2010; 37: 299–310.
Kelly PN, Strasser A . The role of Bcl-2 and its pro-survival relatives in tumourigenesis and cancer therapy. Cell Death Differ 2011; 18: 1414–1424.
Chonghaile TN, Letai A . Mimicking the BH3 domain to kill cancer cells. Oncogene 2009; 27: S149–S157.
Chen L, Willis SN, Wei A, Smith BJ, Fletcher JI, Hinds MG et al. Differential targeting of prosurvival Bcl-2 proteins by their BH3-Only ligands allows complementary apoptotic function. Mol Cell 2005; 17: 393–403.
Shamas-Din A, Brahmbhatt H, Leber B, Andrews DW . BH3-only proteins: orchestrators of apoptosis. Biochim Biophys Acta 2011; 1813: 508–520.
Aranovich A, Liu Q, Collins T, Geng F, Dixit S, Leber B et al. Differences in the mechanisms of proapoptotic BH3 proteins binding to Bcl-XL and Bcl-2 quantified in live MCF-7 cells. Mol Cell 2012; 45: 754–763.
Oltersdorf T, Elmore SW, Shoemaker AR, Armstrong RC, Augeri DJ, Belli BA et al. An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature 2005; 435: 677–681.
Del Gaizo Moore V, Brown JR, Certo M, Love TM, Novina CD, Letai A . Chronic lymphocytic leukemia requires BCL2 to sequester prodeath BIM, explaining sensitivity to BCL2 antagonist ABT-737. J Clin Invest 2007; 117: 112–121.
Gallenne T, Gautier F, Oliver L, Hervouet E, Noël B, Hickman JA et al. Bax activation by the BH3-only protein Puma promotes cell dependence on antiapoptotic Bcl-2 family members. J Cell Biol 2009; 185: 279–290.
Gautier F, Guillemin Y, Cartron PF, Gallenne T, Cauquil N, Le Diguarher T et al. Bax activation by engagement with, then release from, the BH3 binding site of Bcl-xL. Mol Cell Biol 2011; 31: 832–844.
van Delft MF, Wei AH, Mason KD, Vandenberg CJ, Chen L, Czabotar PE et al. The BH3 mimetic ABT-737 targets selective Bcl-2 proteins and efficiently induces apoptosis via Bak/Bax if Mcl-1 is neutralized. Cancer Cell 2006; 10: 389–399.
Tagscherer KE, Fassl A, Campos B, Farhadi M, Kraemer A, Böck BC et al. Apoptosis-based treatment of glioblastomas with ABT-737, a novel small molecule inhibitor of Bcl-2 family proteins. Oncogene 2008; 27: 6646–6656.
Okumura K, Huang S, Sinicrope FA . Induction of Noxa sensitizes human colorectal cancer cells expressing Mcl-1 to the small-molecule Bcl-2/Bcl-xL inhibitor, ABT-737. Clin Cancer Res 2008; 14: 8132–8142.
Zall H, Weber A, Besch R, Zantl N, Häcker G . Chemotherapeutic drugs sensitize human renal cell carcinoma cells to ABT-737 by a mechanism involving the Noxa-dependent inactivation of Mcl-1 or A1. Mol Cancer 2010; 9: 164.
Zhang L, Lopez H, George NM, Liu X, Pang X, Luo X . Selective involvement of BH3-only proteins and differential targets of Noxa in diverse apoptotic pathways. Cell Death Differ 2011; 18: 864–873.
Quinn BA, Dash R, Azab B, Sarkar S, Das SK, Kumar S et al. Targeting Mcl-1 for the therapy of cancer. Expert Opin Investig Drugs 2011; 10: 1397–1411.
Lakhani SA, Masud A, Kuida K, Porter GA, Booth CJ, Mehal WZ et al. Caspases 3 and 7: key mediators of mitochondrial events of apoptosis. Science 2006; 311: 847–851.
Tait SWG, Parsons MJ, Llambi F, Bouchier-Hayes L, Connell S, Muñoz-Pinedo C et al. Resistance to caspase-independent cell death requires persistence of intact mitochondria. Dev Cell 2010; 18: 802–813.
Llambi F, Moldoveanu T, Tait SWG, Bouchier-Hayes L, Temirov J, McCormick LL et al. A unified model of mammalian BCL-2 protein family interactions at the mitochondria. Mol Cell 2011; 44: 517–531.
Buron N, Porceddu M, Brabant M, Desgué D, Racoeur C, Lassalle M et al. Use of human cancer cell lines mitochondria to explore the mechanisms of BH3 peptides and ABT-737-induced mitochondrial membrane permeabilization. PLoS One 2010; 5: e9924.
Song JH, Kandasamy K, Kraft AS . ABT-737 induces expression of the death receptor 5 and sensitizes human cancer cells to TRAIL-induced apoptosis. J Biol Chem 2008; 283: 25003–25013.
Song JH, Kandasamy K, Zemskova M, Lin Y-W, Kraft AS . The BH3 mimetic ABT-737 induces cancer cell senescence. Cancer Res 2011; 71: 506–515.
Stevens C, La Thangue NB . The emerging role of E2F-1 in the DNA damage response and checkpoint control. DNA Repair (Amst) 2004; 3: 1071–1079.
Polager S, Ginsberg D . p53 and E2f: partners in life and death. Nat Rev Cancer 2009; 9: 738–748.
Polager S, Ginsberg D . E2F—at the crossroads of life and death. Trends Cell Biol 2008; 18: 528–535.
Iaquinta PJ, Lees JA . Life and death decisions by the E2F transcription factors. Curr Opin Cell Biol 2007; 19: 649–657.
Stiewe T, Pützer BM . Role of the p53-homologue p73 in E2F1-induced apoptosis. Nat Genet 2000; 26: 464–469.
Hershko T, Ginsberg D . Up-regulation of Bcl-2 homology 3 (BH3)-only proteins by E2F1 mediates apoptosis. J Biol Chem 2004; 279: 8627–8634.
Zhao Y, Tan J, Zhuang L, Jiang X, Liu ET, Yu Q . Inhibitors of histone deacetylases target the Rb-E2F1 pathway for apoptosis induction through activation of proapoptotic protein Bim. Proc Natl Acad Sci USA 2005; 102: 16090–16095.
Hao H, Dong Y, Bowling MT, Gomez-Gutierrez JG, Zhou HS, McMasters KM . E2F-1 induces melanoma cell apoptosis via PUMA up-regulation and Bax translocation. BMC Cancer 2007; 7: 24.
Ambrosini G, Sambol EB, Carvajal D, Vassilev LT, Singer S, Schwartz GK . Mouse double minute antagonist Nutlin-3a enhances chemotherapy-induced apoptosis in cancer cells with mutant p53 by activating E2F1. Oncogene 2007; 26: 3473–3481.
Peirce SK, Findley HW . The MDM2 antagonist nutlin-3 sensitizes p53-null neuroblastoma cells to doxorubicin via E2F1 and TAp73. Int J Oncol 2009; 34: 1395–1402.
An B, Dou QP . Cleavage of retinoblastoma protein during apoptosis: an interleukin 1 beta-converting enzyme-like protease as candidate. Cancer Res 1996; 56: 438–442.
Boutillier AL, Trinh E, Loeffler JP . Caspase-dependent cleavage of the retinoblastoma protein is an early step in neuronal apoptosis. Oncogene 2000; 19: 2171–2178.
Fattman CL, Delach SM, Dou QP, Johnson DE . Sequential two-step cleavage of the retinoblastoma protein by caspase-3/-7 during etoposide-induced apoptosis. Oncogene 2001; 20: 2918–2926.
Dick FA . Structure-function analysis of the retinoblastoma tumor suppressor protein—is the whole a sum of its parts? Cell Div 2007; 2: 26.
Cecchini MJ, Dick FA . The biochemical basis of CDK phosphorylation-independent regulation of E2F1 by the retinoblastoma protein. Biochem J 2011; 434: 297–308.
Julian LM, Palander O, Seifried LA, Foster JEG, Dick FA . Characterization of an E2F1-specific binding domain in pRB and its implications for apoptotic regulation. Oncogene 2008; 27: 1572–1579.
Ianari A, Natale T, Calo E, Ferretti E, Alesse E, Screpanti I et al. Proapoptotic function of the retinoblastoma tumor suppressor protein. Cancer Cell 2009; 15: 184–194.
Albershardt TC, Salerni BL, Soderquist RS, Bates DJP, Pletnev AA, Kisselev AF et al. Multiple BH3 mimetics antagonize anti-apoptotic MCL1 by inducing the endoplasmic reticulum stress response and up-regulating BH3-only protein NOXA. J Biol Chem 2011; 28: 24882–24895.
Katsuda K, Kataoka M, Uno F, Murakami T, Kondo T, Roth JA et al. Activation of caspase-3 and cleavage of Rb are associated with p16-mediated apoptosis in human non-small cell lung cancer cells. Oncogene 2002; 21: 2108–2113.
Sellers WR, Novitch BG, Miyake S, Heith A, Otterson GA, Kaye FJ et al. Stable binding to E2F is not required for the retinoblastoma protein to activate transcription, promote differentiation, and suppress tumor cell growth. Genes Dev 1998; 12: 95–106.
Gjoerup O, Zaveri D, Roberts TM . Induction of p53-independent apoptosis by simian virus 40 small t antigen. J Virol 2001; 75: 9142–9155.
Helin K, Wu CL, Fattaey AR, Lees JA, Dynlacht BD, Ngwu C et al. Heterodimerization of the transcription factors E2F-1 and DP-1 leads to cooperative trans-activation. Genes Dev 1993; 7: 1850–1861.
Pear WS, Miller JP, Xu L, Pui JC, Soffer B, Quackenbush RC et al. Efficient and rapid induction of a chronic myelogenous leukemia-like myeloproliferative disease in mice receiving P210 bcr/abl-transduced bone marrow. Blood 1998; 92: 3780–3792.
Braun F, Bertin-Ciftci J, Gallouet A-S, Millour J, Juin P . Serum-nutrient starvation induces cell death mediated by Bax and Puma that is counteracted by p21 and unmasked by Bcl-xL inhibition. PLoS One 2011; 6: e23577.
Barré B, Perkins ND . The Skp2 promoter integrates signaling through the NF-kappaB, p53, and Akt/GSK3beta pathways to regulate autophagy and apoptosis. Mol Cell 2010; 38: 524–538.
Acknowledgements
We thank O. Micheau and B. Mignotte for their kind gift of the pMIG plasmid and of pRb cDNA, respectively. We thank members of the lab ‘Cell Survival and Tumor Escape in Breast Cancer’ for their technical advice and for fruitful comments. JB-C and JLP are supported by grants from the Ministère de la Recherche et de l’Enseignement Supérieur and from INSERM Region, respectively. PJ is supported by ARC (no. SFI20101201568), Fondation de France (Tumor committee), Région Pays de Loire and Canceropole Grand Ouest (2010-12798).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by RA Knight
Supplementary Information accompanies the paper on Cell Death and Differentiation website
Rights and permissions
About this article
Cite this article
Bertin-Ciftci, J., Barré, B., Le Pen, J. et al. pRb/E2F-1-mediated caspase-dependent induction of Noxa amplifies the apoptotic effects of the Bcl-2/Bcl-xL inhibitor ABT-737. Cell Death Differ 20, 755–764 (2013). https://doi.org/10.1038/cdd.2013.6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cdd.2013.6
Keywords
This article is cited by
-
Constitutive p53 heightens mitochondrial apoptotic priming and favors cell death induction by BH3 mimetic inhibitors of BCL-xL
Cell Death & Disease (2016)
-
The pro-apoptotic activity of Drosophila Rbf1 involves dE2F2-dependent downregulation of diap1 and buffy mRNA
Cell Death & Disease (2014)
-
Decoding and unlocking the BCL-2 dependency of cancer cells
Nature Reviews Cancer (2013)
-
Metastatic SW620 colon cancer cells are primed for death when detached and can be sensitized to anoikis by the BH3-mimetic ABT-737
Cell Death & Disease (2013)