Abstract
Endoplasmic reticulum (ER) stress transducers transduce signals from the ER to the cytoplasm and nucleus when unfolded proteins accumulate in the ER. BBF2 human homolog on chromosome 7 (BBF2H7) and old astrocyte specifically induced substance (OASIS), ER-resident transmembrane proteins, have recently been identified as novel ER stress transducers that have roles in chondrogenesis and osteogenesis, respectively. However, the molecular mechanisms that regulate the activation of BBF2H7 and OASIS under ER stress conditions remain unresolved. Here, we showed that BBF2H7 and OASIS are notably unstable proteins that are easily degraded via the ubiquitin-proteasome pathway under normal conditions. ER stress conditions enhanced the stability of BBF2H7 and OASIS, and promoted transcription of their target genes. HMG-CoA reductase degradation 1 (HRD1), an ER-resident E3 ubiquitin ligase, ubiquitinated BBF2H7 and OASIS under normal conditions, whereas ER stress conditions dissociated the interaction between HRD1 and BBF2H7 or OASIS. The stabilization of OASIS in Hrd1−/− cells enhanced the expression of collagen fibers during osteoblast differentiation, whereas a knockdown of OASIS in Hrd1−/− cells suppressed the production of collagen fibers. These findings suggest that ER stress stabilizes OASIS family members and this is a novel molecular mechanism for the activation of ER stress transducers.
Similar content being viewed by others
Main
Eukaryotic cells have adapted to deal with unfolded proteins that accumulate in the endoplasmic reticulum (ER) via diverse signals from the ER lumen to the cytoplasm and nucleus. This system is termed as the unfolded protein response (UPR).1 The three major transducers of the UPR are inositol requiring 1 (IRE1),2 PKR-like endoplasmic reticulum kinase (PERK)3 and activating transcription factor 6 (ATF6),4, 5 which all sense the presence of unfolded proteins in the ER lumen and transduce signals to the cytoplasm and nucleus.
Recently, novel types of ER stress transducers, ER-resident transcription factors that share a region of high sequence similarity with ATF6, have been identified.6, 7 These transcription factors have a transmembrane domain that allows them to associate with the ER, and possess both a transcription-activation domain and a basic leucine zipper (bZIP) domain. These new types of ER stress transducers are called the old astrocyte specifically induced substance (OASIS) family proteins and include Luman/LZIP/cyclic AMP-response element-binding protein 3 (CREB3),8 OASIS/CREB3L1,9 BBF2 human homolog on chromosome 7 (BBF2H7)/CREB3L2,10 cyclic AMP-response element-binding protein H (CREBH)/CREB3L3,11 and androgen-induced bZIP (AIbZIP)/CREB4/Tisp40/CREB3L4.12, 13 OASIS family members (except Luman) reveal unique cell or tissue-specific expression patterns. OASIS, BBF2H7, CREBH and AIbZIP are preferentially expressed in osteoblasts and astrocytes,9, 14 chondrocytes,15 liver cells,11 and testis and prostate,12, 16 respectively, indicating that each member functions by regulating the UPR in specific organs and tissues.
ATF6 and OASIS family members are activated through regulated intramembrane proteolysis (RIP).17, 18 Upon activation, these transcription factors are transported from the ER to the Golgi apparatus and sequentially cleaved by site-1 protease (S1P) and site-2 protease (S2P) to release the N-terminal fragment, which translocates into the nucleus to activate the transcription of target genes.18 Comparison of the sequences between OASIS family members and ATF6 shows that the lumenal domains of each OASIS family member have little homology with that of ATF6. The lumenal domain in ATF6 has important sites for binding to ER chaperone BiP (immunoglobulin heavy chain-binding protein) and Golgi localization signaling that are important for sensing the unfolded proteins and the translocation from the ER to the Golgi, respectively.5 On the other hand, all deletion mutants for the lumenal domain of OASIS reveal that proteolytic processing and translocation to the Golgi remain intact, indicating that OASIS does not have significant sequences for Golgi localization in its lumenal domain.19 Therefore, the sensing of unfolded proteins and the translocation system from the ER to the Golgi could be quite different between OASIS family members and ATF6. The detailed molecular mechanisms that regulate the activation of OASIS family members remain unresolved.
Oasis–/– mice exhibit severe osteopenia caused by a decrease in the levels of type I collagen, which is a major component of the bone matrix.14 In osteoblasts, activated OASIS directly binds to the Col1a1 promoter region and induces its transcription for osteogenesis. ER stress, which is necessary for the activation of OASIS, occurs during differentiation from mesenchymal stem cells to mature osteoblasts. Bbf2h7–/– mice reveal severe chondrodysplasia caused by the failure of chondrocyte differentiation and production of extracellular matrix proteins.15 In chondrocytes, activated BBF2H7 directly binds to the Sec23a promoter region and facilitates its transcription. Sec23a has crucial roles in coat protein complex II (COPII) vesicle formation and anterograde transport from the ER to the Golgi. During chondrocyte differentiation, mild ER stress occurs and activates BBF2H7. The phenotypes of Oasis–/– and Bbf2h7–/– mice indicate the possibility that a novel molecular mechanism exists for activation of these ER stress transducers in response to ER stress.
In this study, we investigated the mechanisms of activation of BBF2H7 and OASIS in response to ER stress. We propose a novel molecular mechanism, in which activation of these ER stress transducers prevents their degradation in response to ER stress.
Results
Full-length OASIS and BBF2H7 are easily degraded under normal conditions
The overall structures of OASIS and BBF2H7 are similar to that of ATF6α, containing a transmembrane domain and a bZIP motif in the cytoplasmic portion (Figure 1a). To determine whether OASIS or BBF2H7 directly interact with BiP under normal conditions, as does ATF6α, we performed immunoprecipitation with an anti-KDEL antibody that recognizes BiP in MC3T3-E1 or HeLa cells (Figure 1b). ATF6α co-immunoprecipitated with BiP, as previously reported,5 whereas neither OASIS nor BBF2H7 co-immunoprecipitated with BiP (Figure 1b, lanes 2 and 4), indicating that the activation mechanisms in response to ER stress are quite different between ATF6α and OASIS or BBF2H7.
Full-length BBF2H7 and OASIS are easily degraded under normal conditions and are elevated under ER stress conditions. (a) Predicted peptide features of mouse BBF2H7, OASIS and ATF6α. (b) Endogenous BiP interacts with endogenous ATF6α but not with OASIS or BBF2H7. Co-immunoprecipitation (IP) of BiP and ATF6α, OASIS or BBF2H7 in MC3T3-E1 (left) or HeLa (right) cells. The cell lysates were immunoprecipitated with the anti-KDEL antibody, which recognizes BiP, and then immunoblotted (IB) with anti-ATF6α, anti-OASIS (left) or anti-BBF2H7 (right). (c and d) Expression of exogenous OASIS (c) and BBF2H7 (d). MEF tet-off-FLAG-OASIS or -BBF2H7 cells were cultured in medium containing 1 μg/ml DOX (lane 1). Flag-tagged full-length (F) or activated N-terminal (N) OASIS or BBF2H7 expression was induced by culturing the cells in DOX-free medium for 12 h in the absence or presence of the ER stressor thapsigargin (Tg; 1 μM), and/or the proteasomal inhibitors MG132 (3 μM) or lactacystin (lactacys; 5 μM). Xbp1 and β-actin mRNA levels were measured by RT-PCR. The spliced form of Xbp1 mRNA (Xbp1-s) appeared upon treatment with the ER stressor but not with the proteasomal inhibitor. Xbp1-u represents the unspliced form of Xbp1. Equal amounts of total protein or RNA were analyzed in each lane. (e) Expression of endogenous BBF2H7 and ATF6α in HeLa cells. Cells were incubated with 3 μM MG132, 3 μM lactacystin, 1 mM DTT (an ER stressor), or DTT with MG132 for 4 h, and lysates were immunoblotted with anti-ATF6α or anti-BBF2H7. Xbp1 and β-actin mRNA levels were measured by RT-PCR. (f) Expression of endogenous BBF2H7 and OASIS in C6 glioma cells. Cells were incubated with 3 μM MG132 or 1 mM DTT for the indicated times. (Upper) The lysates were western blotted with anti-BBF2H7 or anti-OASIS. (Lower) The levels of Xbp1, Bbf2h7, Oasis and β-actin mRNAs were measured by RT-PCR. (g) C6 glioma cells were incubated in the presence or absence of 3 μM MG132 for 8 h, then co-stained with anti-BBF2H7 or anti-OASIS and anti-KDEL. MG132 treatment increased the immunoreactivity of BBF2H7 and OASIS both in the ER and in the nucleus
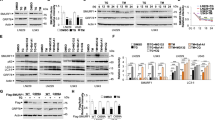
To investigate the regulation of the activation of OASIS and BBF2H7 through RIP in response to ER stress, we established stable mouse embryonic fibroblast (MEF) tet-off cell lines in which the expression of FLAG-tagged full-length OASIS or BBF2H7 was negatively controlled by the presence of doxycycline (DOX) in the culture medium (Figures 1c and d). To investigate RIP in detail, we selected cells that showed low expression levels of OASIS and BBF2H7 when induced by the absence of DOX, and the expression level of OASIS or BBF2H7 did not cause ER stress. Western blotting of these cell lines with an anti-FLAG antibody showed that FLAG-tagged full-length OASIS (OASIS(F)) and BBF2H7 (BBF2H7(F)), and small amounts of the active, cleaved forms of OASIS (OASIS(N)) and BBF2H7 (BBF2H7 (N)), were present in the absence of DOX (Figure 1c, lane 2; Figure 1d, lane 2). The absence of DOX did not cause ER stress because the spliced form of X-box binding protein 1 (XBP1) (Xbp1-s) was not detected. Interestingly, treatment with the ER stressor thapsigargin enhanced the expression of not only the active-form, but also the full-length OASIS and BBF2H7 (Figure 1c, lane 3; Figure 1d, lane 5). We hypothesized that increased expression of both the full length and active form of OASIS and BBF2H7 under ER stress conditions was caused by an increase in the stabilities of OASIS and BBF2H7. Therefore, to determine whether the full length and active form of OASIS and BBF2H7 are degraded, we performed western blotting on MEF tet-off cell lines exposed to the proteasomal inhibitors MG132 or lactacystin (Figure 1c, lane 4; Figure 1d, lanes 3 and 4). Not only the active form of OASIS and BBF2H7, but also full-length OASIS and BBF2H7, were drastically increased; although treatment with the proteasomal inhibitors did not cause ER stress. These results suggest that both the full length and active form of OASIS and BBF2H7 are unstable proteins that are easily degraded under normal conditions.
We next investigated the stability of endogenous BBF2H7 and ATF6α in HeLa cells (Figure 1e). Although the amount of full-length ATF6α (ATF6α(F)) was unchanged in the presence of MG132 and lactacystin (Figure 1e, lanes 2 and 3), full-length ATF6α was reduced in the presence of the ER stressor dithiothreitol (DTT) (Figure 1e, lane 4). This result corresponds with the increase in ATF6α(N) that is activated through RIP. In contrast, full-length BBF2H7 was significantly increased in the presence of either a proteasomal inhibitor or an ER stressor (Figure 1e, lanes 2–4). Furthermore, by co-treatment with a proteasomal inhibitor and ER stressor, both exogenous and endogenous full-length BBF2H7 and OASIS were increased synergistically (Figure 1c, lane 5; Figure 1d, lane 6; Figure 1e, lane 5); although the full-length ATF6α expression decreased (Figure 1e, lane 5). These results suggest that full-length BBF2H7 and OASIS are unstable proteins compared with ATF6α, and that these molecules are upregulated under ER stress conditions. To confirm that endogenous full-length BBF2H7 and OASIS are increased in the presence of either a proteasomal inhibitor or an ER stressor in other types of cells, we examined C6 glioma cells (Figure 1f). As expected, the levels of full-length BBF2H7 and OASIS were dramatically increased by treatment with either MG132 or DTT in a time-dependent manner; however, Bbf2h7 and Oasis mRNAs were not increased.
Next, we performed immunofluorescence staining in the absence or presence of a proteasomal inhibitor to examine the stability and subcellular localization of BBF2H7 and OASIS (Figure 1g). Immunofluorescence with the anti-BBF2H7 and anti-OASIS antibodies showed weak fine reticular staining surrounding the nucleus. The staining pattern was very similar to that obtained with the antibody against KDEL, an ER marker. When cells were treated with MG132, intense BBF2H7 and OASIS signals were observed in the nucleus and cytosol. These results confirmed that treatment with proteasomal inhibitors increased the amount of OASIS and BBF2H7 in both the full-length inactive form at the ER membrane and the N-terminal active form in the nucleus.
ER stress inhibits the ubiquitination and proteasome-mediated degradation of BBF2H7 and OASIS
Next, we compared the degradation rate of full-length BBF2H7 with that of ATF6α in HeLa cells during inhibition of translation using cycloheximide (CHX), to exclude the possibility that the increased full-length BBF2H7 and OASIS in the presence of proteasomal inhibitors was caused by an increase in protein induction (Figure 2a). BBF2H7 was rapidly degraded and its expression level was reduced to 20% in 2 h. In contrast, ATF6α was more stable than BBF2H7 and its expression level remained at 60% over the 2-h period. To determine whether ER stress affected the stability of BBF2H7 and OASIS, we next investigated the degradation rate of full-length BBF2H7 and OASIS in the presence or absence of an ER stressor during inhibition of translation using CHX (Figure 2b). Without ER stress, BBF2H7 and OASIS were rapidly degraded in C6 glioma cells, and their expression levels were reduced to 20 and 10% in 2 h, respectively. Treatment with any of the three distinct ER stressors (thapsigargin, DTT or tunicamycin) significantly suppressed the degradation rate of both BBF2H7 and OASIS. The expression levels of BBF2H7 and OASIS remained at 35–65% and 40–90% over 2 h, respectively. This suggests that, although BBF2H7 and OASIS are unstable and are easily degraded by the proteasome under normal conditions, they are stabilized by ER stress.
ER stress enhances the stability of BBF2H7 and OASIS, which are easily degraded via the ubiquitin/proteasome pathway under normal conditions. (a) CHX chase assay. (Left) HeLa cells were treated with 1 μg/ml CHX and then incubated for the indicated periods. Total cell lysates were analyzed by western blotting using anti-ATF6α, anti-BBF2H7 and anti-β-actin antibodies. (F) indicates full length. (Right) Densitometric analysis of ATF6α and BBF2H7 protein levels relative to the amount present at time 0 (mean±S.D.; n=3; *P<0.05; Student's t-test). (b) CHX chase assay with ER stress. (Upper) C6 glioma cells were treated with 1 μM thapsigargin (Tg) for 3 h, 3 μg/ml tunicamycin (Tm) for 5 h or 1 mM DTT for 2 h for BBF2H7. The pretreatment of OASIS was 1 μM Tg for 7 h, 3 μg/ml Tm for 5 h or 1 mM DTT for 4 h. The cells were then treated with 1 μg/ml CHX for the indicated periods with or without the ER stressor Tg, Tm or DTT. Total cell lysates were western blotted using anti-BBF2H7, anti-OASIS and anti-β-actin antibodies. (Lower) Densitometric analysis of protein levels in the upper panel. Levels of BBF2H7 and OASIS at each time point were plotted relative to the amount present at time 0 (mean±S.D.; n=3; *P<0.05; Student's t-test). (c) C6 rat glioma cells were treated with 1 μM thapsigargin or 3 μM MG132 for 4 h. (Left) Equal amounts of proteins were immunoprecipitated (IP) with anti-BBF2H7 (left) or anti-OASIS (right), and then immunoblotted (IB) with anti-ubiquitin and anti-BBF2H7 or anti-OASIS. (d) C6 rat glioma cells were treated with 3 μM MG132 and an ER stressor—1 μM Tg, 3 μg/ml Tm or 1 mM DTT—simultaneously for 4 h. Equal amounts of protein were immunoprecipitated with anti-BBF2H7 (upper) or anti-OASIS (lower), and then immunoblotted with anti-ubiquitin and anti-BBF2H7 or anti-OASIS. (e) HEK293T cells were transiently transfected with FLAG-tagged BBF2H7 (upper) or FLAG-tagged OASIS (lower) and HA-tagged wild-type ubiquitin (HA-Ub-wt) or the dominant-negative mutant form of ubiquitin (HA-Ub-K48R), and then treated with 3 μM MG132 for 4 h. Cell lysates were immunoprecipitated (IP) with an anti-FLAG and then immunoblotted (IB) with an anti-ubiquitin
To ascertain directly that BBF2H7 and OASIS are degraded by the proteasome, and that ER stress inhibits their degradation, we examined the ubiquitination of BBF2H7 and OASIS with or without ER stressor treatment in C6 glioma cells by immunoprecipitation (Figures 2c and d). Although the ubiquitination of BBF2H7 and OASIS was not detected either with or without thapsigargin treatment (Figure 2c, lanes 2, 3, 6 and 7), MG132 treatment led to the ubiquitination of BBF2H7 and OASIS (Figure 2c, lanes 4 and 8; Figure 2d, lanes 1 and 5). Moreover, the ubiquitination of BBF2H7 and OASIS disappeared upon co-treatment with MG132 and various ER stressors (Figure 2d, lanes 2–4 and 6–8), indicating that BBF2H7 and OASIS were rapidly ubiquitinated and degraded under normal conditions; however, ER stress inhibited the ubiquitination and degradation of BBF2H7 and OASIS. The reason that no ubiquitination of BBF2H7 and OASIS was detected under normal conditions without MG132 treatment is not known (Figure 2c, lanes 2 and 6); however, there is a possibility that the ubiquitinated proteins are degraded so rapidly that it may be difficult to detect, as previously shown.20
To confirm directly whether BBF2H7 and OASIS were ubiquitinated under normal conditions, FLAG-tagged BBF2H7 or OASIS and human influenza hemagglutinin (HA)-tagged wild-type ubiquitin (HA-Ub-wt) or dominant-negative mutant form of ubiquitin (HA-Ub-K48R) were transiently co-transfected into HEK293T cells, and immunoprecipitated with an anti-FLAG antibody and subsequently immunoblotted with an anti-ubiquitin antibody (Figure 2e). Compared with HA-Ub-wt, the HA-Ub-K48R mutant decreased OASIS or BBF2H7 ubiquitination, confirming that these proteins are easily degraded under normal conditions via the ubiquitin-proteasome pathway.
BBF2H7 and OASIS are ubiquitinated by HRD1, an ER-associated E3 ubiquitin ligase
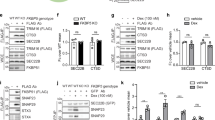
To clarify which E3 ubiquitin ligase ubiquitinates BBF2H7 and OASIS under normal conditions,21 we silenced several ER-associated E3 ubiquitin ligase genes and examined the expression of BBF2H7 and OASIS in ATDC5 and MC3T3-E1 cells. Among the E3 ligases examined, the expression of full-length BBF2H7 and OASIS was dramatically increased only in the HMG-CoA reductase degradation 1 (HRD1)-knockdown cells (Figure 3a). A lack of HRD1 in ATDC5 cells also decreased BBF2H7 ubiquitination (Figure 3b).
HRD1, an ER-associated E3 ubiquitin ligase, ubiquitinates BBF2H7 and OASIS under normal conditions. (a) ATDC5 (left) and MC3T3-E1 (right) cells were treated with the indicated siRNAs and immunoblotted to examine the levels of full-length BBF2H7 (left) or OASIS (right) and β-actin. Note that only HRD1-knockdown cells demonstrated enhanced expression of BBF2H7 and OASIS. (b) HRD1 depletion prevents ubiquitination of BBF2H7. ATDC5 cells were transiently transfected with control or HRD1 siRNA. Cell lysates were immunoprecipitated (IP) with anti-BBF2H7, and immunoblotted (IB) with anti-ubiquitin or anti-BBF2H7. As input, the cell lysates were directly immunoblotted with anti-BBF2H7, anti-HRD1 or anti-β-actin. (c) Schematic representation of the HRD1 and OASIS constructs. wt, wild-type; ΔM, deleted for the transmembrane domain; MR, transmembrane and RING domains only. Numbers in parentheses indicate the corresponding amino-acid residues. (d) Interaction of OASIS with HRD1 and its mutants. OASIS and HRD1 mutants were co-immunoprecipitated in HEK293T cells transiently transfected with FLAG-OASIS and either an empty vector (Mock), or myc-tagged wild-type or mutant HRD1. Cell lysates were immunoprecipitated with anti-FLAG and immunoblotted with anti-myc. The interaction was not detected with MR-HRD1 (lane 9). (e) Interaction of HRD1 with OASIS and its mutants. HRD1 and OASIS mutants were co-immunoprecipitated in HEK293T cells transiently transfected with wt-HRD1-myc and either an empty vector (Mock) or FLAG-tagged wild-type (Full) or deletion mutants of OASIS. The cell lysates were immunoprecipitated with anti-FLAG and immunoblotted with anti-myc. The interaction was not detected with OASIS (1–374). (f) Interaction of endogenous HRD1 with BBF2H7 or OASIS. Co-immunoprecipitation of HRD1 and BBF2H7 (left) or OASIS (right) was performed in C6 glioma cells with or without treatment with 1 μM thapsigargin (Tg) for 10 h. Cell lysates were immunoprecipitated with anti-BBF2H7 or anti-OASIS and immunoblotted with anti-HRD1 and anti-BBF2H7 or anti-OASIS
To determine whether HRD1 interacts with OASIS, myc-tagged HRD1 and FLAG-tagged OASIS were transiently co-transfected into HEK293T cells and immunoprecipitated with an anti-FLAG antibody. The overexpressed HRD1 protein co-immunoprecipitated with the overexpressed OASIS (Figure 3d, lane 7). To investigate which HRD1 region interacts with OASIS, a variety of HRD1 mutants tagged with a myc tag were transfected into HEK293T cells and immunoprecipitated with FLAG-tagged OASIS (Figures 3c and d). ΔM-HRD1 (ΔMembrane, Δ42–204 aa; lacking four of the five membrane-spanning domains) and C329S-HRD1 (a RING finger mutant) co-immunoprecipitated with OASIS (Figure 3d, lanes 8 and 10), whereas MR-HRD1 (Membrane-RING, aa 1–340 only; defective in the proline-rich region) was not co-immunoprecipitated (Figure 3d, lane 9). This indicates that HRD1 interacts with OASIS through the proline-rich region.
To determine which OASIS region interacts with HRD1, FLAG-tagged wild type and deletion mutants of OASIS were transfected into HEK293T cells and immunoprecipitated with myc-tagged HRD1 (Figures 3c and e). Full-length wild-type OASIS, Δ98, Δ97–197, Δ198–291, Δ292–374 and 1–430S1 OASIS co-immunoprecipitated with HRD1, whereas OASIS (1–374), lacking the transmembrane and lumenal domains, did not co-immunoprecipitate with HRD1 (Figure 3e, lane 3). This indicates that OASIS interacts with HRD1 through the transmembrane region. Taken together, the transmembrane region of OASIS interacts with the proline-rich region of HRD1.
We further demonstrated the physiological interaction between endogenous HRD1 and BBF2H7 or OASIS in C6 glioma cells (Figure 3f). The interaction was detected under normal conditions (Figure 3f, lanes 4 and 9), but not in cells treated with an ER stressor (Figure 3f, lanes 5 and 10). These results suggest that BBF2H7 and OASIS are ubiquitinated by HRD1 under normal conditions, but that ER stress conditions dissociate this interaction, enhancing the stability of these proteins.
Recently, it was reported that ATF6α is also degraded by HRD1.22 Under normal conditions, ATF6α is ubiquitinated by HRD1 with an interaction with Wolfram syndrome 1 (WFS1).22 Under ER stress conditions, ATF6α dissociates from HRD1 and WFS1, allowing translocation to the Golgi and cleavage by RIP. To determine whether BBF2H7 or OASIS also interact with WFS1, FLAG-tagged BBF2H7, OASIS or ATF6α and HA-tagged WFS1 were transiently co-transfected into HEK293T cells and immunoprecipitated with an anti-FLAG antibody. ATF6α co-immunoprecipitated with WFS1 under normal conditions (Supplementary Figure S1, lane 3), but not in cells treated with an ER stressor (Supplementary Figure S1, lane 4), as previously reported,22 whereas neither BBF2H7 nor OASIS co-immunoprecipitated with WFS1 (Supplementary Figure S1, lanes 7, 8, 11 and 12), confirming that the activation mechanisms in response to ER stress are quite different between ATF6α and OASIS or BBF2H7.
Stabilization of BBF2H7 and OASIS by ER stress enhances transcriptional activation of their target genes
To investigate whether increased BBF2H7 and OASIS under ER stress conditions enhanced activation of their target genes, we used a reporter assay in ATDC5 or MC3T3-E1 cells overexpressing full-length BBF2H7 or OASIS, respectively (Figure 4a). The reporter constructs carried the Sec23a or Col1a1 promoter upstream of the firefly luciferase gene, respectively.14, 15 In ATDC5 cells, treatment with either a proteasome inhibitor or an ER stressor promoted the expression of luciferase to ∼2.5- to 3-fold higher than that in untreated cells (Figure 4a, left panel). In MC3T3-E1 cells, treatment with either a proteasome inhibitor or an ER stressor promoted 1.7- to 2.2-fold higher expression than in untreated cells (Figure 4a, right panel). These results indicate that increasing BBF2H7 and OASIS by inhibiting their degradation enhances the activation of their target genes.
Stabilization of BBF2H7 and OASIS under ER stress conditions enhances transcription of their target genes for chondrogenesis and osteogenesis. (a) Reporter assay with overexpression of BBF2H7 and OASIS. (Left) ATDC5 cells were transiently transfected with a luciferase reporter driven by a 1.0-kb Sec23a promoter (pGL3-Sec23a-luc), a Renilla vector and an empty vector (control) or a vector expressing BBF2H7. (Right) MC3T3-E1 cells were transiently transfected with a luciferase reporter driven by a 2.3-kb Col1a1 promoter (pGL3-Col1a1-luc), a Renilla vector and an empty vector (control) or a vector expressing OASIS. The relative luciferase activities were determined with or without (NT) treatment with 5 μM lactacystin (Lact), 1 μM thapsigargin (Tg), 3 μg/ml tunicamycin (Tm) or 1 mM DTT and/or 5 μM MG132 for 12 h. (b) Reporter assay with the knockdown of BBF2H7 and OASIS. (Left) ATDC5 cells were transiently transfected with BBF2H7 siRNA, and then transfected with a luciferase reporter driven by the Sec23a promoter (pGL3-Sec23a-luc) and a Renilla vector. (Right) MC3T3-E1 cells were transiently transfected with OASIS siRNA, and then transfected with a luciferase reporter driven by the Col1a1 promoter (pGL3-Col1a1-luc) and a Renilla vector. The relative luciferase activities in the cells were determined with or without treatment with 5 μM MG132 or 1 μM Tg for 12 h (mean±S.D.; n=3; *P<0.05; Student's t-test)
Next, we silenced BBF2H7 or OASIS using siRNA and determined the expression of reporter genes after treatment with MG132 or thapsigargin, to investigate whether decreased endogenous BBF2H7 or OASIS reduced the expression of their target genes. As expected, knockdown of BBF2H7 or OASIS significantly suppressed the reporter activities (Figure 4b). These results suggest that the stabilization of BBF2H7 and OASIS by ER stress enhances transcriptional activation of their target genes.
Stabilization of OASIS in HRD1-deficient cells accelerates osteogenesis
To investigate whether stabilized OASIS functions physiologically, we examined osteogenesis in Hrd1−/− MEFs, which are known to differentiate into osteoblasts following treatment with bone morphogenetic protein 2 (BMP2).23 Van Gieson staining showed that collagen fibers were significantly increased in Hrd1−/− MEFs compared with wild-type MEFs 10 days after BMP2-induced differentiation into osteoblasts (Figure 5a). Moreover, the enhanced expression of collagen fibers was significantly decreased in Hrd1−/− MEFs by knocking-down OASIS levels using siRNA. Electrophoretic analysis of type I collagen extracted from cells also showed that Hrd1−/− MEFs, which contain increased levels of OASIS, expressed significantly higher levels of type 1 collagen (Col1α1 and Col1α2) compared with wild-type MEFs (Figure 5b, lanes 2 and 4). Type I collagen extracted from OASIS-siRNA-treated Hrd1−/− MEFs were reduced to a level similar to that observed in wild-type MEFs (Figure 5b, lanes 2 and 6). To clarify the role of OASIS in differentiating osteoblasts to the ER stress-related genes, the expression levels of BiP between wild-type, Hrd1−/− and OASIS-siRNA-treated Hrd1−/− MEFs were compared. The expression level of BiP did not differ between these MEFs (Figure 5b, lanes 2, 4 and 6). These results indicate that enhanced stabilization of OASIS under ER stress conditions has a physiological role in promoting osteogenesis through the production of type I collagen.
Increased OASIS in HRD1-deficient cells enhanced the expression of collagen fibers. (a) Wild-type, Hrd1−/− and OASIS-siRNA-treated Hrd1−/− MEFs were cultured with ascorbic acid (50 μg/ml) and β-glycerophosphate (5 mM), with or without BMP2 (100 ng/ml) for 10 days. (Left) Cultures were stained with van Gieson for the detection of collagen fibers. Hrd1−/− cultures showed an increase in collagen fibers in the presence of BMP2, whereas OASIS-siRNA-treated Hrd1−/− MEFs cultures showed a decrease in collagen fibers. (Right) Quantitative analysis of van Gieson staining of wild-type, Hrd1−/− and OASIS-siRNA-treated Hrd1−/− MEFs with or without BMP2. Cultures were stained with van Gieson, and the intensities were measured. Levels of staining were plotted in the presence of BMP2 relative to the levels in the absence of BMP2 (mean±S.D.; n=3; *P<0.05; Student's t-test). (b) Wild-type, Hrd1−/− and OASIS-siRNA-treated Hrd1−/− MEFs were cultured with ascorbic acid (50 μg/ml) and β-glycerophosphate (5 mM), with or without BMP2 (100 ng/ml) for 10 days. The cell lysates were analyzed by western blotting using the anti-OASIS, anti-KDEL or anti-β-actin antibody and by electrophoretic analysis of type I collagen (Col1α1 and Col1α2) with Coomassie brilliant blue (CBB) staining
Discussion
In this study, we have demonstrated a novel molecular mechanism for the activation of the ER stress transducers BBF2H7 and OASIS. First, BBF2H7 and OASIS were easily degraded under normal conditions via the ubiquitin-proteasome pathway, but this was inhibited by ER stress, which enhanced their stability (Figures 1 and 2). Second, BBF2H7 and OASIS were ubiquitinated by an interaction with the ER-associated E3 ubiquitin ligase HRD1; this was inhibited under ER stress conditions (Figure 3). Third, stabilization of BBF2H7 or OASIS enhanced transcription of their target genes (Figure 4). Finally, stabilization of OASIS in Hrd1−/− MEFs enhanced the production of collagen fibers during osteogenesis, whereas OASIS-siRNA-treated Hrd1−/− MEFs reduced production of these fibers (Figure 5).
HRD1 is an ER-resident E3 ubiquitin ligase that ubiquitinates misfolded proteins accumulated in the ER.24, 25, 26, 27 These ubiquitinated proteins are degraded in the proteasome. Although it is not clear whether BBF2H7 and OASIS are misfolded under normal conditions, their degradation may be important in regulating protein expression levels. Degradation of BBF2H7 and OASIS may be necessary to optimize protein expression levels in a physiological situation. An additional significance of this degradation is that the switch from degradation to stabilization could be a novel mechanism for the activation of these ER stress transducers. The transport of BBF2H7 and OASIS to the Golgi under ER stress conditions, permitted by the dissociation with HRD1, protects these proteins from degradation and could trigger their activation. The transcription factor NF-E2-related factor 2 (Nrf2), which has a central role in the inducible expression of cytoprotective genes in response to oxidative and electrophilic stress, is also easily degraded via the ubiquitin-proteasome pathway under normal conditions.28 In the presence of reactive oxygen species or electrophiles, Nrf2 degradation ceases, and stabilized Nrf2 accumulates in nuclei and activates its target genes for cytoprotection. BBF2H7 and OASIS have a similar activation mechanism as ER stress transducers. Although this rapid degradation appears to be inefficient for a living cell under normal conditions, this system may be useful to response to stress.
In the present study, we were not able to define the mechanisms of dissociation between HRD1 and BBF2H7 or OASIS under ER stress conditions. There are at least two possibilities that could explain this. First, ER stress increases the amounts of unfolded proteins, HRD1 degrades these unfolded proteins, and then the degradation of BBF2H7 and OASIS is relatively decreased. In this first model, under ER stress, unfolded proteins and BBF2H7 or OASIS are competitors as substrates for HRD1. Second, ER stress modulates novel unknown proteins, which interact with BBF2H7 or OASIS and lead to degradation under normal conditions. When ER stress occurs, the unknown proteins dissociate from BBF2H7 or OASIS, stabilizing them. In this second model, the unknown proteins can be referred to as anchor or escort proteins, as are known for sterol regulatory element-binding proteins (SREBPs). The anchor and escort proteins for SREBPs have been identified as Insig and SREBP-cleavage activating protein (SCAP), respectively.29, 30 Under normal conditions, the SREBP/SCAP complex is retained in the ER through binding of Insig to SCAP. Upon a cholesterol level decrease, a conformational change in SCAP causes its dissociation from Insig, thereby allowing translocation of the SREBP/SCAP complex to the Golgi.31, 32 Identification of the anchor and escort proteins for BBF2H7 and OASIS will facilitate a more precise understanding of how BBF2H7 and OASIS sense ER stress.
Recent reports showed that ATF6α is also degraded by HRD1.22 Although the activation model of ATF6α could be similar to that of BBF2H7 and OASIS, we have revealed three critical differences between ATF6α and BBF2H7 or OASIS. First, ATF6α is a more stable protein than BBF2H7 (Figure 2a). Second, ATF6α directly binds to BiP under normal conditions, whereas BBF2H7 and OASIS do not interact with BiP (Figure 1b). Third, ATF6α binds to WFS1 under normal conditions, whereas BBF2H7 and OASIS do not interact with WFS1 (Supplementary Figure S1). Therefore, although the degradation of ATF6α by HRD1 has a physiological role to prevent its hyperactivation,22 it does not have a normal activating role. Our findings suggest that although ATF6α, BBF2H7 and OASIS act as ER stress transducers, they use different molecular mechanisms: ATF6α is activated by dissociation from BiP, whereas BBF2H7 and OASIS are activated by inhibiting their degradation. The existence of such a difference in activation machinery between ATF6α and BBF2H7 or OASIS is unknown. Further studies are needed to clarify the advantage of the activation machinery by inhibiting degradation in response to ER stress.
Materials and Methods
Plasmids
The expression vectors for pcDNA-FLAG-OASIS and pcDNA-FLAG-BBF2H7 have been described previously.9, 10 The expression vectors for mouse OASIS truncated mutants with the FLAG tag at the N-terminus were generated by PCR. OASIS (1–430, S1) was deleted from the S1P site to the C-terminus. The expression vectors for HA-tagged wild-type ubiquitin (HA-Ub-wt) and the dominant-negative mutant form of ubiquitin (HA-Ub-K48R) have been described previously33 and were kindly provided by Dr. Lee-Yuan Liu-Chen (Temple University School of Medicine). The expression vectors for wild-type human HRD1 and its truncation mutants tagged with myc and polyhistidine (6 × His) epitopes at the C-terminus have been described previously.34 The expression vector for HA-tagged WFS1 has been described previously.22 A fragment of the mouse full-length OASIS, or the full-length BBF2H7, with a FLAG tag at the N-terminus was inserted into the pTRE2hyg vector (Clontech, Palo Alto, CA, USA) to create pTRE2hyg-FLAG-OASIS and pTRE2hyg-FLAG-BBF2H7 for tet-off cell lines. Reporter plasmids pGL3-Sec23a-luc and pGL3-Col1a1-luc have been described previously.14, 15
Cell culture and reagents
C6 (rat glioma), HEK293T and HeLa cells were grown in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal calf serum (FCS). MC3T3-E1 (mouse osteoblast) cells were maintained in α-modified Eagle’s medium (MP Biomedicals, Irvine, CA, USA) containing 10% FCS. ATDC5 (mouse chondrocyte) cells were maintained in DMEM/F12 (a mixture of DMEM and Ham's F12 medium) containing 10% FCS. We used thapsigargin, an inhibitor of an ER Ca2+-ATPase, tunicamycin, an inhibitor of protein glycosylation, and DTT, an inhibitor of disulphide bond formation, as ER stress inducers (all from Sigma, St. Louis, MO, USA); lactacystin (Sigma) and MG132 (Calbiochem, La Jolla, CA, USA) as proteasome inhibitors and CHX (Sigma) as a translation inhibitor. MEF tet-off cells (Clontech) were grown in DMEM containing 10% FCS, 100 μg/ml G418, 100 μg/ml hygromycin B and 1 μg/ml DOX (all from Clontech). MEF tet-off cells were transfected by electroporation (900 V, 30 ms, 1 pulse) with pTRE2hyg-FLAG-OASIS or pTRE2hyg-FLAG-BBF2H7. A stable transformant was isolated by adding 800 μg/ml hygromycin B to the culture medium. Expression of FLAG-OASIS or FLAG-BBF2H7 was confirmed by immunoblotting before and after removal of DOX from the culture medium. A stable transformant, in which OASIS or BBF2H7 was detected only after the removal of DOX, and whose growth rate was similar regardless of the presence or absence of DOX, was selected and used for MEF tet-off-FLAG-OASIS or MEF tet-off-FLAG-BBF2H7 cells. MEF(Hrd1+/+) and MEF(Hrd1−/−) cells, which have been described previously35 and were kindly provided by Dr. Toshihiro Nakajima (Tokyo Medical University), were grown in DMEM containing 10% FCS. For osteoblast differentiation, MEFs were grown in recombinant human BMP2 (100 ng/ml; Sigma), ascorbic acid (50 μg/ml) and β-glycerophosphate (5 mM) for 10 days. MEF(Hrd1−/−) cells were transfected with OASIS siRNA twice using the Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA, USA). Van Gieson staining was performed according to a standard protocol. The extraction procedure for collagen was performed as described previously.36 The density of each band was quantified using Image J (National Institutes of Health, Bethesda, MD).
Antibodies, western blotting, immunoprecipitation and immunofluorescence
Rabbit polyclonal and mouse monoclonal anti-OASIS antibodies were generated as described previously.9, 14 A mouse monoclonal antibody secreting hybridoma line was established by fusing splenic cells immunized with mouse recombinant BBF2H7 (amino acids 1–292) and the SP2/0-Ag14 mouse myeloma cell line. The mouse monoclonal antibody against BBF2H7 was purified from hybridoma culture supernatant using a MAbTrap kit (GE Healthcare, Buckinghamshire, UK). Rabbit antisera were raised against recombinant BBF2H7 (amino acids 1–292) fused to glutathione S-transferase and affinity-purified using a HiTrap NHS-activated HP column (Amersham Biosciences, Piscataway, NJ, USA). For western blotting and immunofluorescence, the following antibodies were used: anti-β-actin (Sigma; 1 : 3000), anti-ATF6α (BioAcademia, Osaka, Japan; 1 : 1000), anti-ubiquitin (Cell Signaling, Danvers, MA, USA; 1 : 1000), anti-HRD1 (Santa Cruz, Santa Cruz, CA, USA; 1 : 500), anti-HRD1 C-terminal (Abgent, San Diego, CA, USA; 1 : 1000), anti-myc PL14 (MBL, Woburn, MA, USA; 1 : 1000), anti-HA (Millipore, Billerica, MA, USA; 1 : 1000), anti-FLAG M2 (Sigma; 1 : 1000) and anti-KDEL (MBL; 1 : 100). We also used an anti-HRD1 antibody, which have been described previously35 and was kindly provided by Dr. Toshihiro Nakajima (Tokyo Medical University). For western blotting, cells were lysed in hot SDS, as described previously,37 before electrophoresis using 7.5–12.5% SDS-polyacrylamide gels. The protein concentration of each sample was quantified by the Lowry assay (DC protein assay; Bio-Rad, Hercules, CA, USA), and protein-equivalent samples were subjected to western blotting.
For immunoprecipitation, cells were incubated in a lysis buffer (50 mM Tris-HCl, pH 7.5, 140 mM NaCl, 5 mM EDTA and 1% NP-40 with complete protease inhibitors). Cell lysates were incubated overnight with the antibody for immunoprecipitation. The immune complexes were incubated with Protein G Magnetic Beads (Millipore) for 2 h, and rinsed three times with wash buffer (50 mM Tris-HCl, pH 7.5, 140 mM NaCl, 5 mM EDTA and 1% NP-40) and boiled with Laemmli SDS-polyacrylamide gel electrophoresis sample buffer for 5 min. Total cell extracts and immunoprecipitates were subjected to western blotting.
For immunofluorescence, C6 glioma cells were grown on CELLocate Coverslips (Eppendorf, Hamburg, Germany) in 60-mm dishes. Cells were fixed with cold methanol for 30 min and permeabilized in 0.5% NP-40 for 10 min. Rabbit anti-BBF2H7 or anti-OASIS polyclonal antibody was used overnight at 4°C. For co-staining of the ER, we used the mouse anti-KDEL antibody. Primary antibodies were visualized with fluorescein-conjugated goat anti-rabbit IgG and Alexa-conjugated goat anti-mouse IgG antibodies for 2 h at room temperature. Stained cells were viewed using a confocal microscope (FV1000D; Olympus, Tokyo, Japan).
RT-PCR
Total RNA was extracted from cells using a QIAGEN RNA kit (QIAGEN, Hilden, Germany). RT-PCR assays were performed according to our published procedures.38 The oligonucleotides used for RT-PCR were as follows: BBF2H7-5′, 5′-GGAGCAGAGCGTCCTGCAGTG-3′ and BBF2H7-3′, 5′-CCACCTCCTCGTCATTGAAGCTGTC-3′; OASIS-5′, 5′-CCTTGTGCCTGTCAAGATGGAG-3′ and OASIS-3′, 5′-GCAGCAGCCATGGCAGAGGAG-3′; human-XBP1-5′, 5′-CAGCGCTTGGGGATGGATGC-3′ and human-XBP1-3′, 5′-CCATGGGGAGATGTTCTGGA-3′; mouse-XBP1-5′, 5′-ACACGCTTGGGAATGGACAC-3′; rat-XBP1-5′, 5′-ACACGCTTGGGGATGGATGC-3′ and mouse/rat-XBP1-3′, 5′-CCATGGGAAGATGTTCTGGG-3′; β-actin-5′, 5′-TCCTCCCTGGAGAAGAGCTAC-3′ and β-actin-3′, 5′-TCCTGCTTGCTGATCCACAT-3′.
RNA interference
ATDC5 or MC3T3-E1 cells were transfected by siRNA electroporation (1650 V, 20 ms, 1 pulse) for gp70, HRD1, Fbxw7, Fbs2, CHIP, parkin, OASIS or BBF2H7. After the pulse, cells were transferred into prewarmed medium. The transfected cells were incubated at 37°C for 48 h and then harvested for western blotting. The sequences or reagents for siRNAs were as follows: HRD1 (Ambion, Austin, TX, USA, Cat. #: 4390771, ID: s92311, CAUCGUCUCUCUCAUGUUUttAAACAUGAGAGAGACGAUGcg); Fbxw7 (Ambion, Cat. #: 4390771, ID: s78406, GGACACCAGUCAUUAACGAttUCGUUAAUGACUGGUGUCCtg); gp78 (Santa Cruz, sc-44579); CHIP (Santa Cruz, sc-44731); parkin (Santa Cruz, sc-42159); and Fbs2 (Santa Cruz, sc-145133). The sequences for the OASIS and BBF2H7 siRNAs were described previously.9, 10
Luciferase assay
ATDC5 or MC3TCE1 cells plated onto 24-well plates were transfected with 0.1 μg of a reporter plasmid carrying the firefly luciferase gene and 0.01 μg of the reference plasmid pRL-SV40 carrying the Renilla luciferase gene under the control of the SV40 enhancer and promoter (Promega, Madison, WI, USA) with or without 0.1 μg of the BBF2H7 or OASIS expression plasmid. After 24 h, cells were treated with 5 μM MG132, 5 μM lactacystin, 1 μM thapsigargin, 3 μg/ml tunicamycin and/or 1 mM DTT for 12 h. The cells were lysed in 200 μl of Passive Lysis buffer (Promega). Firefly luciferase and Renilla luciferase activities were measured in 10 μl of cell lysate using the Dual-Luciferase Reporter Assay System (Promega) and a luminometer (Berthold Technologies, Bad Wildbad, Germany). Relative activity was defined as the ratio of firefly luciferase activity to Renilla luciferase activity. For the luciferase assay with BBF2H7 or OASIS siRNA, the siRNA-transfected cells were incubated at 37°C for 48 h. The reporter plasmid and reference plasmid were then transfected for 24 h and the cells were stimulated by ER stressors or proteasomal inhibitors.
Statistical analysis
Statistical comparisons were made using Student’s t-test. P<0.05 was considered statistically significant and is displayed as one asterisk.
Abbreviations
- AIbZIP:
-
androgen-induced bZIP
- ATF6:
-
activating transcription factor 6
- BBF2H7:
-
BBF2 human homologue on chromosome 7
- BiP:
-
immunoglobulin heavy chain-binding protein
- BMP2:
-
bone morphogenetic protein 2
- bZIP:
-
basic leucine zipper
- Col1:
-
type I collagen
- COPII:
-
coat protein complex II
- CREB:
-
cyclic AMP-response element-binding protein
- CREBH:
-
cyclic AMP-response element-binding protein H
- DOX:
-
doxycycline
- DTT:
-
dithiothreitol
- ER:
-
endoplasmic reticulum
- HA:
-
human influenza hemagglutinin
- HRD1:
-
HMG-CoA reductase degradation 1
- IRE1:
-
inositol requiring 1
- MEF:
-
mouse embryonic fibroblast
- Nrf2:
-
NF-E2-related factor 2
- OASIS:
-
old astrocyte specifically induced substance
- PERK:
-
PKR-like endoplasmic reticulum kinase
- RIP:
-
regulated intramembrane proteolysis
- S1P:
-
site-1 protease
- S2P:
-
site-2 protease
- SCAP:
-
SREBP-cleavage activating protein
- SREBPs:
-
sterol regulatory element-binding proteins
- UPR:
-
unfolded protein response
- WFS1:
-
Wolfram syndrome 1
- XBP1:
-
X-box binding protein 1
References
Ron D, Walter P . Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol 2007; 8: 519–529.
Calfon M, Zeng H, Urano F, Till JH, Hubbard SR, Harding HP et al. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature 2002; 415: 92–96.
Harding HP, Zhang Y, Ron D . Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature 1999; 397: 271–274.
Yoshida H, Okada T, Haze K, Yanagi H, Yura T, Negishi M et al. ATF6 activated by proteolysis binds in the presence of NF-Y (CBF) directly to the cis-acting element responsible for the mammalian unfolded protein response. Mol Cell Biol 2000; 20: 6755–6767.
Shen J, Chen X, Hendershot L, Prywes RER . stress regulation of ATF6 localization by dissociation of BiP/GRP78 binding and unmasking of Golgi localization signals. Dev Cell 2002; 3: 99–111.
Kondo S, Saito A, Asada R, Kanemoto S, Imaizumi K . Physiological unfolded protein response regulated by CREB/ATF family members, transmembrane bZIP transcription factors. IUBMB Life 2011; 63: 233–239.
Asada R, Kanemoto S, Kondo S, Saito A, Imaizumi K . The signaling from endoplasmic reticulum-resident bZIP transcription factors involved in diverse cellullar physiology. J Biochem 2011; 149: 507–518.
DenBoer LM, Hardy-Smith PW, Hogan MR, Cockram GP, Audas TE, Lu R . Luman is capable of binding and activating transcription from the unfolded protein response element. Biochem Biophys Res Commun 2005; 331: 113–119.
Kondo S, Murakami T, Tatsumi K, Ogata M, Kanemoto S, Otori K et al. OASIS, a CREB/ATF-family member, modulates UPR signaling in astrocytes. Nat Cell Biol 2005; 7: 186–194.
Kondo S, Saito A, Hino S-I, Murakami T, Ogata M, Kanemoto S et al. BBF2H7, a novel transmembrane bZIP transcription factor, is a new type of endoplasmic reticulum stress transducer. Mol Cell Biol 2007; 27: 1716–1729.
Zhang K, Shen X, Wu J, Sakaki K, Saunders T, Rutkowski DT et al. Endoplasmic reticulum stress activates cleavage of CREBH to induce a systemic inflammatory response. Cell 2006; 124: 587–599.
Nagamori I, Yabuta N, Fujii T, Tanaka H, Yomogida K, Nishimune Y et al. Tisp40, a spermatid specific bZip transcription factor, functions by binding to the unfolded protein response element via the Rip pathway. Genes Cells 2005; 10: 575–594.
Stirling J, O’Hare P . CREB4, a transmembrane bZip transcription factor and potential new substrate for regulation and cleavage by S1P. Mol Biol Cell 2006; 17: 413–426.
Murakami T, Saito A, Hino S-I, Kondo S, Kanemoto S, Chihara K et al. Signalling mediated by the endoplasmic reticulum stress transducer OASIS is involved in bone formation. Nat Cell Biol 2009; 11: 1205–1211.
Saito A, Hino S-I, Murakami T, Kanemoto S, Kondo S, Saitoh M et al. Regulation of endoplasmic reticulum stress response by a BBF2H7-mediated Sec23a pathway is essential for chondrogenesis. Nat Cell Biol 2009; 11: 1197–1204.
Cao G, Ni X, Jiang M, Ma Y, Cheng H, Guo L et al. Molecular cloning and characterization of a novel human cAMP response element-binding (CREB) gene (CREB4). J Hum Genet 2002; 47: 373–376.
Brown MS, Ye J, Rawson RB, Goldstein JL . Regulated intramembrane proteolysis: a control mechanism conserved from bacteria to humans. Cell 2000; 100: 391–398.
Bailey D, O’Hare P . Transmembrane bZIP transcription factors in ER stress signaling and the unfolded protein response. Antioxid Redox Signal 2007; 9: 2305–2321.
Murakami T, Kondo S, Ogata M, Kanemoto S, Saito A, Wanaka A et al. Cleavage of the membrane-bound transcription factor OASIS in response to endoplasmic reticulum stress. J Neurochem 2006; 96: 1090–1100.
Chen C, Sun X, Ran Q, Wilkinson KD, Murphy TJ, Simons JW et al. Ubiquitin-proteasome degradation of KLF5 transcription factor in cancer and untransformed epithelial cells. Oncogene 2005; 24: 3319–3327.
Pickart CM . Back to the future with ubiquitin. Cell 116: 181–190.
Fonseca SG, Ishigaki S, Oslowski CM, Lu S, Lipson KL, Ghosh R et al. Wolfram syndrome 1 gene negatively regulates ER stress signaling in rodent and human cells. J Clin Invest 2010; 120: 744–755.
Tohmonda T, Miyauchi Y, Ghosh R, Yoda M, Uchikawa S, Takito J et al. The IRE1α-XBP1 pathway is essential for osteoblast differentiation through promoting transcription of Osterix. EMBO Rep 2011; 12: 451–457.
Amano T, Yamasaki S, Yagishita N, Tsuchimochi K, Shin H, Kawahara K et al. Synoviolin/Hrd1, an E3 ubiquitin ligase, as a novel pathogenic factor for arthropathy. Genes Dev 2003; 17: 2436–2449.
Christianson JC, Shaler TA, Tyler RE, Kopito RR . OS-9 and GRP94 deliver mutant alpha1-antitrypsin to the Hrd1-SEL1L ubiquitin ligase complex for ERAD. Nat Cell Biol 2008; 10: 272–282.
Omura T, Kaneko M, Okuma Y, Orba Y, Nagashima K, Takahashi R et al. A ubiquitin ligase HRD1 promotes the degradation of Pael receptor, a substrate of Parkin. J Neurochem 2006; 99: 1456–1469.
Kaneko M, Koike H, Saito R, Kitamura Y, Okuma Y, Nomura Y . Loss of HRD1-mediated protein degradation causes amyloid precursor protein accumulation and amyloid-beta generation. J Neurosci 2010; 30: 3924–3932.
Taguchi K, Motohashi H, Yamamoto M . Molecular mechanisms of the Keap1–Nrf2 pathway in stress response and cancer evolution. Genes Cells 2011; 16: 123–140.
DeBose-Boyd RA, Brown MS, Li WP, Nohturfft A, Goldstein JL, Espenshade PJ . Transport-dependent proteolysis of SREBP: relocation of site-1 protease from Golgi to ER obviates the need for SREBP transport to Golgi. Cell 1999; 99: 703–712.
Yang T, Espenshade PJ, Wright ME, Yabe D, Gong Y, Aebersold R et al. Crucial step in cholesterol homeostasis: sterols promote binding of SCAP to INSIG-1, a membrane protein that facilitates retention of SREBPs in ER. Cell 2002; 110: 489–500.
Brown AJ, Sun L, Feramisco JD, Brown MS, Goldstein JL . Cholesterol addition to ER membranes alters conformation of SCAP, the SREBP escort protein that regulates cholesterol metabolism. Mol Cell 2002; 10: 237–245.
Radhakrishnan A, Sun LP, Kwon HJ, Brown MS, Goldstein JL . Direct binding of cholesterol to the purified membrane region of SCAP: mechanism for a sterol-sensing domain. Mol Cell 2004; 15: 259–268.
Li JG, Haines DS, Liu-Chen LY . Agonist-promoted Lys63-linked polyubiquitination of the human kappa-opioid receptor is involved in receptor down-regulation. Mol Pharmacol 2008; 73: 1319–1330.
Omura T, Kaneko M, Onoguchi M, Koizumi S, Itami M, Ueyama M et al. Novel functions of ubiquitin ligase HRD1 with transmembrane and proline-rich domains. J Pharmacol Sci 2008; 106: 512–519.
Yamasaki S, Yagishita N, Sasaki T, Nakazawa M, Kato Y, Yamadera T et al. Cytoplasmic destruction of p53 by the endoplasmic reticulum-resident ubiquitin ligase 'Synoviolin'. EMBO J 2007; 26: 113–122.
Domenicucci C, Goldberg HA, Hofmann T, Isenman D, Wasi S, Sodek J . Characterization of porcine osteonectin extracted from foetal calvariae. Biochem J 1988; 253: 139–151.
Erickson AH, Blobel G . Early events in the biosynthesis of the lysosomal enzyme Cathepsin D. J Biol Chem 1979; 254: 11771–11774.
Imaizumi K, Tsuda M, Imai Y, Wanaka A, Takagi T, Tohyama M . Molecular cloning of a novel polypeptide, DP5, induced during programmed neuronal death. J Biol Chem 1997; 272: 18842–18848.
Acknowledgements
We thank Dr. Toshihiro Nakajima (Tokyo Medical University) for kindly providing MEF(Hrd1+/+) and MEF(Hrd1−/−) cells and anti-HRD1 antibody, Dr. Lee-Yuan Liu-Chen (Temple University School of Medicine) for kindly providing the ubiquitin expression plasmids and S Nakagawa for technical support. This work was partly supported by grants from the Japan Society for the Promotion of Science KAKENHI (#22020030, #22800049), the Takeda Science Foundation and the Pharmacological Research Foundation Tokyo.
S Kondo, SH and KI designed the experiments. S Kondo, SH, AS, S Kanemoto, NK, RA, SI, HI, MO and HM performed the experiments. S Kondo and KI wrote the manuscript. MK, YN and FU provided substantial input into the writing of the manuscript.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by M Piacentini
Supplementary Information accompanies the paper on Cell Death and Differentiation website
Supplementary information
Rights and permissions
About this article
Cite this article
Kondo, S., Hino, SI., Saito, A. et al. Activation of OASIS family, ER stress transducers, is dependent on its stabilization. Cell Death Differ 19, 1939–1949 (2012). https://doi.org/10.1038/cdd.2012.77
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cdd.2012.77
Keywords
This article is cited by
-
Modulation of osteogenic differentiation by Escherichia coli-derived recombinant bone morphogenetic protein-2
AMB Express (2022)
-
TRMT6/61A-dependent base methylation of tRNA-derived fragments regulates gene-silencing activity and the unfolded protein response in bladder cancer
Nature Communications (2022)
-
Expression analysis and functional characterization of thioredoxin domain-containing protein 11
Molecular Biology Reports (2022)
-
OASIS/CREB3L1 is a factor that responds to nuclear envelope stress
Cell Death Discovery (2021)
-
Pituitary cell translation and secretory capacities are enhanced cell autonomously by the transcription factor Creb3l2
Nature Communications (2019)