Abstract
Unrepaired DNA double-strand breaks (DSBs) cause genetic instability that leads to malignant transformation or cell death. Cells respond to DSBs with the ordered recruitment of signaling and repair proteins to the sites of DNA lesions. Coordinated protein SUMOylation and ubiquitylation have crucial roles in regulating the dynamic assembly of protein complexes at these sites. However, how SUMOylation influences protein ubiquitylation at DSBs is poorly understood. We show herein that Rnf4, an E3 ubiquitin ligase that targets SUMO-modified proteins, accumulates in DSB repair foci and is required for both homologous recombination (HR) and non-homologous end joining repair. To establish a link between Rnf4 and the DNA damage response (DDR) in vivo, we generated an Rnf4 allelic series in mice. We show that Rnf4-deficiency causes persistent ionizing radiation-induced DNA damage and signaling, and that Rnf4-deficient cells and mice exhibit increased sensitivity to genotoxic stress. Mechanistically, we show that Rnf4 targets SUMOylated MDC1 and SUMOylated BRCA1, and is required for the loading of Rad51, an enzyme required for HR repair, onto sites of DNA damage. Similarly to inactivating mutations in other key regulators of HR repair, Rnf4 deficiency leads to age-dependent impairment in spermatogenesis. These findings identify Rnf4 as a critical component of the DDR in vivo and support the possibility that Rnf4 controls protein localization at DNA damage sites by integrating SUMOylation and ubiquitylation events.
Similar content being viewed by others
Main
DNA double-strand breaks (DSBs) represent the most destructive type of chromosomal lesion that pose a major threat to genome stability, if not correctly sensed and repaired. Cells counteract DNA damage via a sophisticated DNA damage signaling response, which impacts on diverse cellular processes, such as DNA repair itself, cell cycle progression, DNA replication, and transcription.1, 2 Most cellular DSBs are repaired by homologous recombination (HR) or NEHJ,3 and defects in either pathway exhibit increased sensitivity to ionizing radiation (IR).4
Following the DSBs induction, multiple DNA damage signaling and repair factors become concentrated at, or excluded from, specific areas of the nucleus. In particular, a protective and DNA repair-stimulating microenvironment is formed around the DSB sites. These so-called IR-induced foci (IRIF) concentrate multiple DNA repair factors in the vicinity of the strand breaks, promote checkpoint signaling, and constitute a barrier toward further DNA decay.5 Protein recruitment to such structures occurs in a highly dynamic and hierarchical manner, and is controlled by post-translational modifications within DSB-flanking chromatin.6 Phosphorylation of multiple DSB-signaling components by the ATM/ATR/DNA-PK kinases have a central role in promoting this response; one of such key events being ATM-mediated phosphorylation of H2AX on ser139, an epigenetic mark commonly referred to as γ-H2AX.7 Phosphorylation of γ-H2AX provides a direct binding platform for the MDC1 scaffold protein.8
Nonproteolytic ubiquitylation also has an important role in orchestrating protein interactions with DSB sites.9 ATM-mediated phosphorylation of MDC1 provides a docking site for the ubiquitin ligase RNF8.10, 11 RNF8 promotes the polyubiquitylation of core histones H2A and H2AX to trigger a second, ubiquitin-dependent round of protein recruitment to DSB-flanking chromatin. In addition to RNF8, other E3 ligases such as HERC2 and RNF168 are also recruited to these sites, to assist RNF8 in promoting the formation of nonproteolytic, K63-linked polyubiquitin chains on histones and possibly on other targets at sites of DNA damage. These modifications generate permissive conditions for recruitment of downstream DNA repair factors such as 53BP1 and BRCA1-containing protein complexes.12 Although several other E3 ligases are recruited to IRIF,12 including RAD18, BRCA1, BMI1, the identity of their substrates within these structures, remains largely unknown.
The small ubiquitin-like modifier (SUMO) has also been implicated in genome stability and repair responses.13 The SUMO E3 ligases PIAS1 and PIAS4 are required for the recruitment of BRCA1 and 53BP1 to IRIF, and both SUMO1 and SUMO2/3 accumulate in the DSB-associated chromatin areas.14, 15 Interestingly, these studies uncovered a more direct function of SUMO in facilitating DSB-induced histone ubiquitylation upstream of 53BP1 and BRCA1 recruitment. However, the molecular mechanisms that govern the relationship between SUMOylation and ubiquitylation remain unknown.
RNF4 is currently the only known mammalian member of the SUMO-targeted ubiquitin ligase family.16 Given that these proteins selectively ubiquitylate SUMOylated proteins they are well suited to have an important role in the cross talk between SUMO and ubiquitin pathways.17 RNF4 possesses four amino-terminal SUMO-interacting motifs (SIMs) and a C-terminal RING-finger domain that together facilitate the specific ubiquitylation of poly-SUMOylated substrates.18 The yeast and Drosophila orthologues of RNF4 have been implicated in the maintenance of genome stability and DNA repair.19, 20 Because of the early embryonic lethality of Rnf4-null mice, it has not been possible to study the role of Rnf4 in the DNA damage response (DDR) in mammals. Here, we describe a mouse Rnf4 allelic series, which allowed us to establish the first genetic link between Rnf4 and the DDR in mammals.
Results
Generation of an allelic series of Rnf4 in mice
Complete genetic ablation of Rnf4 function is deleterious for embryonic development.21 In order to study the physiological function of mammalian Rnf4 and in particular its role in DDR in vivo, we therefore generated an allelic series in the Rnf4 gene. Our approach is on the basis of use of a gene-trap vector design that contains loxP sites flanking the splice acceptor (SA) of the gene-trapping element (Figure 1a). Removal of the SA results in splicing around the vector to restore normal wild-type transcript at a reduced level. This strategy has recently been used successfully to generate Cop1 hypomorphic mice.22
Generation of an allelic series of Rnf4. (a) The structure of the pGT01xf gene trap vector along with the structures of the wild-type, trapped (assimilated to a null allele) and hypomorphic Rnf4 alleles. The hypomorphic allele is generated by Cre-mediated conversion of the trapped allele. Exons are indicated by black boxes. SA, splicing acceptor site; En2 intr1, intron 1 of Engrailed2; β-geo, lacz/neomycin reporter/selection cassette. (b) A representative PCR analysis of E9.5 embryos with the indicated genotypes (upper panel). The three primers used are described in the Materials and Methods section. Discrimination of the wild-type, null and hypomorphic alleles by PCR using two different primer sets (lower panel). (c) RT-qPCR showing relative mRNA expression using total RNA isolated from E9.5 embryos of the indicated genotypes. The data were normalized to the level of expression in control wild-type (+/+) embryos, which is set to 1. The data represent the mean±S.D. of 2 independent experiments. (d) wild-type (+/+) and homozygous trapped Rnf4−/− (−/−) embryos at E10.5 (upper panel). wild-type (+/+) and embryos harboring two hypomorphic allele (h/h) at E12.5 (lower panel). (e) Control (+/+) mice and mice homozygous for the hypomorphic mutation (h/h) at P120
Two different ES (Embryonic Stem cell) clones (RRR624 and HMA301) were used to produce independent Rnf4 mutant mouse lines. As both lines produced identical phenotypes, we only describe one of these lines here. In the RRR624 clone, the pGT01xf vector was inserted in intron 1 (Figure 1a). Accordingly, the trapped allele produces a hybrid transcript containing a part of the Rnf4 5′UTR fused to the β-galactosidase protein. This allele is therefore expected not to produce any functional Rnf4 protein and as such to correspond to a null mutation. The Rnf4-trapped allele is thereafter referred to as Rnf4−.
Rnf4+/− mice are viable and fertile. However, none of the 123 offspring from heterozygous intercrosses were Rnf4− homozygous (supplementary Table S1). Although expected, Mendelian ratios were observed in embryos isolated at E9.5, the proportion of homozygous embryos declined at later stages such that no live homozygous embryos were identified among 42 embryos at E12.5 (Supplementary Table S1 and Figure 1b). At E10.5, all Rnf4−/− embryos were developmentally delayed (Figure 1d).
We crossed Rnf4+/− mice with a Cre-deleter strain in an attempt to generate a hypomorphic allele. We reasoned that low levels of Rnf4 activity may rescue the recessive lethality associated with the null allele. We chose the Sox2-Cre transgenic line as it promotes high-efficiency recombination in all epiblast-derived tissues.23 A PCR-based genotyping strategy confirmed efficient Cre-mediated excision of the SA site; the non-recombined allele could not be detected by PCR in E9.5 Cre-positive homozygous embryos for the trap allele (Figure 1b). Viable mice heterozygous for the converted, putative hypomorphic allele (Rnf4hypo/+) were obtained and intercrossed. Importantly, viable Rnf4hypo/hypo mice were found, but at a reduced Mendelian distribution at P4 (Supplementary Table S2). Both male and female Rnf4hypo/hypo viable mice weighted ∼20% less than wild-type mice both at P20 and P120 (Figure 1e).
Timed harvests revealed that, although growth retarded, several Rnf4hypo/hypoembryos are viable at E12.5 (Supplementary Table S2 and Figure 1c). In order to confirm hypomorphic expression of RNF4 from the Cre-recombined allele, we harvested E9.5 embryos of various genotypes and performed reverse transcription quantitative PCR (RT-qPCR) analyses (Figure 1c). As predicted, the presence of one Cre-recombined allele (thereafter referred to as Rnf4hypo) leads to the expression of low levels of wild-type Rnf4. Rnf4 expression was reduced by 95% in Rnf4hypo/hypo embryos. In summary, our genetic approach allows the generation of viable Rnf4-mutant mice (Rnf4hypo/hypo) mice with a 95% decrease in Rnf4 expression.
Rnf4-deficiency leads to increased sensitivity to DNA damage
In contrast to Rnf4−/−, Rnf4hypo/hypo mouse embryonic fibroblast (MEF) cultures could be established. When serially transferred on a 3-day schedule (3T3), Rnf4hypo/hypo MEFs proliferated at slower rates than wild-type controls starting from passage 3 onwards (Figure 2a) and eventually underwent apoptosis. Clonogenic assays further confirmed a role for Rnf4 in the control of cellular growth (Figure 2b). RT-qPCR analysis confirmed that Rnf4 expression levels are reduced to ∼5% of wild type in Rnf4hypo/hypo MEFs (Figure 2c).
Rnf4-deficiency leads to increased sensitivity to DNA damage. (a) Proliferation of Rnf4 +/+ and Rnf4 h/h MEF cultures on a 3T3 schedule. The relative cumulative cell numbers is shown on a linear scale on the y axis. (b) Colony formation assay using Rnf4 +/+ and Rnf4 h/h MEFs. 5.000 cells of each genotypes were plated on 10cm dishes. (c) RT-qPCR showing relative mRNA expression using total RNA isolated from MEFs with the indicated genotypes. The data were normalized to the level of expression in control wild-type (Rnf4 +/+) MEFs, which is set to 1. The data represent the mean±S.D. of 3 independent experiments. (d) Accumulation at the G2 phase of Rnf4 +/+ and Rnf4 h/h MEFs, which have been infected with lentivirus directing the expression of the Puro selection marker alone (Empty) or the Puro marker and flag-tagged RNF4 (Flag-RNF4), following IR exposure (5 Gy). Cell cycle profiles were examined by PI staining and representative data are shown on the left panel. Right panel shows the relative population of cells in G2 phase of cell cycle before and after IR treatment from three independent experiments (±S.D.). *p<0.05. (e) Western blot analysis confirms expression of Flag-tagged RNF4 in cells infected with lentiviruses directing Flag-RNF4 expression. γ-Tubilin (γ-Tub) serves here as a loading control. (f) FragEL assay of thymus sections (scale bar, 100 μm) performed 24 h after whole-body irradiation (5 Gy). The number of FragEL positive cells per field was measured on ten different consecutive slides. *P<0.05. (g) MEFs with the indicated genotypes were left untreated or were irradiated with 5 Gy of gamma irradiation, apoptosis was measured after 24 H using Annexin-V/PI staining, followed by FACS analysis. Data from one representative experiment is shown on the left panel. The mean percentages of apoptotic cells from three independent experiments±S.D. are shown on the right panel. *P<0.05. (h) Active Caspase-3 staining of skin sections (scale bar, 50 μm) performed on untreated or 24 h after whole-body exposure to 5 Gy. The number of active caspase-3 positive cells per field was measured on ten different consecutive slides. **P<0.01
It is well documented that increased ROS-induced DNA damage limits the in vitro growth of MEFs in atmospheric oxygen concentrations. Our results therefore indicate that Rnf4-deficiency may sensitize MEFs to ROS-induced DNA damage. To directly determine whether Rnf4-deficiency confers increased sensitivity to DNA damage, we subjected Rnf4hypo/hypo MEFs and mice to 5 Gy of γ-radiation. DNA-damage signaling activates several cell cycle checkpoints and deficiency in genes involved in DDR such as H2AX,24 Mdc1,25 Rnf8,11, 26 and 53bp127 leads to a defect in the G2/M checkpoint. Cell cycle analyses of Rnf4-deficient and wild-type MEFs 18 h post-IR indicated that a higher proportion of the Rnf4-deficient cells is found at the G2/M phase (Figure 2d). To further demonstrate that this phenotype is a direct consequence of Rnf4-deficiency, we re-established Rnf4 expression levels in Rnf4hypo/hypo MEFs using a lentiviral-based approach. The proportion of Rnf4hypo/hypo MEFs expressing flag-tagged exogenous RNF4 found at the G2/M phase was comparable to that found in Rnf4+/+ cells (Figures 2d and e). Together these observations indicate that Rnf4 is required for enforcing the radiation-induced G2/M DNA damage checkpoint in MEFs.
Whole-body γ-radiation induces apoptosis in a number of tissues, especially in radiosensitive organs such as the thymus and small intestine. We measured apoptosis in the thymus (Figure 2f) and small intestine (data not shown) of Rnf4hypo/hypo mice after whole-body radiation. The absolute number of thymocytes was not significantly reduced and the distribution of the CD4−, CD8−, CD4+, CD8+, and CD4+ and CD8+ thymocyte populations was not affected in Rnf4-deficient mice (data not shown). However, a significant increase in the number of apoptotic cells was evident in the thymus (and small intestine) of γ-irradiated Rnf4hypo/hypo mice compared with wild-type controls (Figure 2f). Decreased Rnf4 expression also led to increased sensitivity to IR-induced apoptosis in non-radiosensitive cells/tissues; a significant fraction of Rnf4-deficient MEFs and keratinocytes underwent apoptosis in response to IR exposure (Figures 2g and h). Together these studies demonstrate that Rnf4 deficiency leads to increased radiosensitivity.
Rnf4-deficiency leads to increased IR-induced DNA damage and sustained DNA damage signaling
We observed an increase in phosphorylated H2AX (γ-H2AX) levels in Rnf4-deficient MEFs upon passaging (Figure 3a). A significant increase in the number and size of γ-H2AX foci was also observed in these cells compared with wild-type controls (Figure 3b). Moreover, whereas γ-H2AX foci were rapidly induced following IR and subsequently resolved progressively over a period of 24-h recovery in wild-type control cells, Rnf4-deficiency significantly delayed recovery (Figure 3b). This defect is a direct consequence of Rnf4-deficiency, as it is not observed in Rnf4hypo/hypo MEFs reconstituted with flag-tagged exogenous RNF4 (Supplementary Figure S1).
RNF4 deficiency leads to increased IR-induced DNA damage and sustained DNA Damage Signaling. (a) Western blot analysis of γ-H2AX levels in Rnf4 +/+ and Rnf4 h/h MEFs. Vinculin (Vinc.) serves as loading control. (b) Left panel shows representative immunofluorescence images of γ-H2AX staining in untreated or IR-treated (5 Gy) Rnf4 +/+ and Rnf4 h/h MEFs. Right panel shows graphical representation of the number of γH2AX foci x total area/cell in untreated or IR-treated (5 Gy) Rnf4 +/+ and Rnf4 h/h MEFs. More than 200 nuclei were quantified for each genotype and time points. *P<0.05. (c) Pulsed-field gel showing the analysis of untreated or IR-treated (5 Gy) Rnf4 +/+ and Rnf4 h/h MEFs. DSBs were visualized by DNA fragments that migrate into the gel (left panel). Quantification of DSBs was done using image J software, relative intensity of fragmented DNA is shown on y-axis and the error bars represent the standard deviations of two independent experiments. *P<0.05; **P<0.01. (right panel). (d) MEFs with the indicated genotypes and treatment were examined 6 hour post-IR by immunoblotting using the indicated antibodies. (e) mRNA expression of established p53 target genes was detected using RT-qPCR, RNA isolated after 24 H from MEFs with the indicated genotypes and treatment. The data were normalized to the level of expression in untreated control, which is set to 1. The data represent the mean±S.D. of 2 independent experiments. (f) Immunohistochemistry for p-ATM and p53 on small intestine sections from Rnf4 +/+ and Rnf4 hypo/hypo mice. Mice were either untreated (NT) or exposed to 5 Gy of IR. Staining was performed 24 H after treatment
In order to obtain a direct evidence for the presence of DNA damage, we analyzed untreated and IR-induced cells for DSBs by PFGE (Figure 3c). In the absence of IR, Rnf4-deficient cells exhibited ∼1.2 times more DSBs compared with wild-type littermate controls. The modest—but reproducible—increase in DSBs observed in the absence of exogenously induced DNA damage is consistent with the basal increase in γ-H2AX levels. Importantly, 24 h after IR exposure, there were approximately twice as many DSBs in Rnf4-deficient cells compared with wild-type cells. These analyses indicate that Rnf4 deficiency impairs IR-induced DSBs repair.
The ATM-Chk2-p53 signaling pathway has essential roles in response to DSBs.28 IR-induced Ser-18 phosphorylation of p53 was elevated in Rnf4-deficient cells compared with controls, and this led to an increase in its transcriptional activity as demonstrated by a significant increase in the mRNA expression levels of some of its well-established target genes (Figures 3d and e). An increase in DDR signaling in Rnf4-compromised cells was confirmed in vivo by IHC (Figure 3f). A readily detectable increase in the levels of γ-H2AX, phosphorylated ATM, Ser-18 phosphorylation of p53, and pan-p53 was observed in the small intestine of Rnf4-deficient mice compared with wild-type control littermates, following whole-body irradiation (Figure 3f and data not shown). Collectively, these data indicate that Rnf4 has a fundamental role in DSB repair and signaling.
RNF4 is recruited to IR-induced DNA damage foci
The SUMO-E3 ligases PIAS1 and PIAS4 are recruited to DNA damage sites, promote the SUMOylation of several key DNA repair factors, including 53BP1 and BRCA1, and are required for effective ubiquitin-adduct formation mediated by RNF8 and RNF168 at those sites.14 RNF4, currently the only known mammalian SUMO-dependent E3 ligase, might also promote ubiquitylation of a number of DNA-damage-induced SUMOylated proteins at sites of DNA damage. To test this, we assessed the subcellular localization of exogenous V5-tagged RNF4 upon transfection in the commonly used murine fibroblastic cell line NIH-3T3 (Figure 4a). V5-RNF4 localized diffusely to the nucleoplasm in the untreated cells but it concentrated in foci co-localizing with the DSB marker γ-H2AX upon exposure to IR.
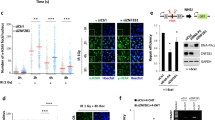
RNF4 is recruited to sites of DNA damage in a SIM-dependent manner. (a) Representative micrographs of mock and V5-RNF4 transfected NIH3T3 cells stained with anit-V5 antibody, anti γ-H2AX antibody and DAPI. Cells were untreated or exposed to 5 Gy of IR and fixed after 2 hr. (b) Endogenous RNF4 accumulates at sites of laser-induced DNA damage that are marked by γH2AX. U2-OS cells were micro-irradiated with a multi-photon laser. After 1,4 and 6 min(s) cells were stained with Dapi and immunostained for γH2AX and RNF4. Scale bars, 10 μm. (c) GFP-RNF4 wt/ΔSIM recruitment to DNA damage induced by multi-photon irradiation. RNF4 wild-type, but not RNF4 lacking functional SIMs, accumulates in DNA damage-containing laser tracks. U2-OS cells expressing GFP-RNF4 wt or GFP-RNF4 ΔSIM were subjected to laser micro-irradiation and the accumulation of RNF4 was monitored at the indicated time points after irradiation. The average of two independent experiments −/+ s.e.m. is presented and representative images for the indicated time points are shown. Scale bar, 10 μm. (d) Western blot analysis showing similar levels of expression of GFP-RNF4 and ΔSIMs-mutant in the U2-OS transfected cells
To determine the subcellular localization of endogenous RNF4, we raised specific antibodies against human RNF4. We used multi-photon laser micro-irradiation to induce local DNA damage in living U2-OS cells,29 and showed rapid accumulation of endogenous RNF4 in laser tracks that were marked by γ-H2AX (Figure 4b).
To determine the role of SUMOylation in RNF4-targeting, we mutated the four SUMO interaction motifs (SIMs) in RNF4. Wild-type RNF4 fused to GFP rapidly accumulated in laser tracks generated by either multi-photon or UV-A laser irradiation (Figure 4c and Supplementary Figure S2). In contrast, recruitment of RNF4 SIM mutant (ΔSIM) to laser tracks was significantly reduced (Figure 4c), indicating that RNF4 is recruited to sites of DNA damage in a SUMOylation-dependent manner. Low-level accumulation of the ΔSIM mutant could be explained by RING–RING interactions between mutant and endogenous RNF4. Notably, the kinetics of RNF4 recruitment did not fully resemble those reported earlier,30, 31 which may be because of the nature of the lesions induced by the different laser methodologies that were employed.
SUMOylated MDC1 and SUMOylated BRCA1 are regulated by RNF4
We used SILAC technology to study SUMOylation dynamics in response to IR and search for relevant RNF4 SUMO targets. Consistent with previous reports,14, 32 we identified 53BP1 and MDC1 as IR-induced SUMO-2 target proteins. To determine whether these SUMO conjugates are regulated by RNF4, knockdown experiments were performed in cells stably expressing low levels of His-SUMO-2. Cells were treated with IR (10 Gy) and the proteasome inhibitor MG132 (10 μ M) combined or were left untreated. SUMOylated proteins were purified under denaturing conditions and analyzed by immunoblotting (Figure 5). We confirmed the SUMOylation of 53BP1 and MDC1 in response to IR. Furthermore, a significant increase in IR-induced MDC1 SUMOylation was observed upon RNF4 knockdown, indicating that SUMOylated MDC1 is regulated by RNF4 (Figure 5a). In contrast, knockdown of RNF4 resulted in a decrease of 53BP1 SUMOylation (Figure 5b).
MDC1 and BRCA1 are SUMO2 target proteins that are regulated by RNF4. (a–e) U2-OS cells stably expressing His-SUMO2 were either infected with a Mission library lentivirus to knockdown RNF4 or with a negative control lentivirus. Three days after infection cells were either mock-treated or treated with MG-132 (10 μ M) to inhibit the proteasome. One hour after the start of MG-132 treatment, cells were either mock-treated or irradiated with IR (10 Gy). The damaged and undamaged cells were subsequently incubated for 4 hours and lysed. His-SUMO2 conjugates were purified and eluted samples were loaded on 3-8% Tris-acetate gels and analyzed by immunoblotting using antibodies directed against MDC1 (a), 53BP1 (b), BRCA1 (c) and SUMO2/3 (d) to detect the SUMOylated forms of these proteins in response to DNA damage and proteasome inhibition. (e) Total cell lysates were loaded on a 4-12% Bis-Tris gel and analyzed by immunoblotting, using anti-RNF4 antibody to assess the efficiency of RNF4 knockdown
BRCA1, a critical HR component, has been identified as a DDR component that is regulated by SUMOylation14, 15 in response to proteasome inhibition.33 A significant increase in BRCA1 SUMOylation was observed upon knockdown of RNF4, indicating that SUMOylated BRCA1 is also regulated by RNF4 (Figure 5c). Under these conditions, no increase in BRCA1 SUMOylation upon irradiation was observed. Similar to the RNF4 target protein PML,18, 33 SUMOylated BRCA1 was targeted for degradation by the proteasome, explaining why BRCA1 was missing in our SILAC screen. The observed pattern of SUMO conjugation is compatible with SUMO polymers being attached to BRCA1. Combined, these results identify BRCA1 and MDC1 as RNF4-regulated SUMO-2 target proteins.
RNF4 deficiency affects both HR and NHEJ
To directly establish the implication of RNF4 in DSB repair, we used various GFP-reporter-based assays. For HR, we used a human cancer cell line (HEK293) with an integrated copy of the DR-GFP reporter,34 and assessed repair efficiency following RNF4 knockdown. With this reporter, a DSB is introduced into the chromosome by expressing the I-SceI endonuclease, and, if HDR occurs, GFP is expressed, which is quantifiable by flow cytometry. Strikingly, the depletion of RNF4 significantly decreased HR-mediated DNA repair (Figure 6a). Western blotting analysis confirmed the silencing of RNF4 in shRNF4-1- and sh-RNF4-2-expressing cells. Non-homologous end-joining (NHEJ) was measured in human cancer cells (H1299) using a recently developed assay.35 These cells stably express the IRES-TK-EGFP reporter cassette, which contains two cleavage sites for the I-SceI endonuclease.35 Transfection of the I-SceI expression plasmid into these cells specifically cleaves the two I-SceI sites in the substrate, and produces DSBs with incompatible ends. NHEJ of the two broken DNA strands results in the deletion of the herpes simplex virus-thymidine kinase open reading frame and leads to the production of a transcript that enables translation of EGFP. The efficiency of NHEJ was assessed by monitoring EGFP production in cells expressing sh-RNF4-2 or a control sh-RNA (sh-Luc). A significant decrease in NHEJ was observed in RNF4 KD cells compared with control cells, indicating that RNF4 is also required for efficient NHEJ-mediated repair (Figure 6b). The ability to repair a linearized GFP reporter construct by NHEJ was also measured in Rnf4-deficient MEFs. This assay confirms that the efficiency of NHEJ-mediated repair is decreased in Rnf4-depleted cells (Figure 6c). Together, these data indicate that RNF4 contributes to both HR and NHEJ-mediated DNA repair.
RNF4 is required for HR-repair and Rad51 recruitment to DSBs. (a) Graphical representation of the efficiency of Sce–I induced HR in 293 DR-GFP expressing the indicated shRNAs (upper panel). Immunoblot showing RNF4 levels in 293 DR-GFP cells tranfected with the indicated shRNAs; Vinculin serves as loading control (lower panel). (b) Graphical representation of the efficiency of NHEJ DSBs repair, as measured by the proportion of EGFP-positive cells expressing the indicated shRNAs. (c) Graphical representation of the efficiency of NHEJ DSBs repair in Rnf4 +/+ and Rnf4 h/h MEFs. (d) Representative micrographs of MEFs stained with anti-Rad51 antibody and DAPI. Rnf4 +/+ and Rnf4 h/h were untreated (NT) or exposed to 5 Gy of IR and fixed at the indicated time post IR. Bars, 10 μm. (e) Representative micrographs of MEFs stained with anti-Rad51 and anti γ-H2 AX antibodies. Rnf4 +/+ and Rnf4 h/h were untreated (NT) or exposed to 5 Gy of IR and fixed 6h post IR
Rnf4 is required for the recruitment of Rad51 to IR-induced DNA damage sites
Rad51 is recruited to DSBs and is one of the key enzymes in HR repair.36 We therefore examined whether Rad51 co-localizes normally at γ-H2AX-marked damage in Rnf4 deficient cells. Strikingly, whereas as expected, there was a substantial increase in Rad51 foci in Rnf4 wild-type cells, with a peak between 3 and 6 h post-IR; Rad51 staining is much more diffuse and only marginally overlap with γ-H2AX foci in Rnf4-deficient cells (Figures 6d and e). In contrast, the recruitment of 53bp1 to IR-induced γH2AX-foci was only marginally affected (data not shown). These observations demonstrate a critical role for Rnf4 in the recruitment for and/or retention in repair foci induced by IR.
Impaired spermatogenesis in Rnf4-hypomorphic mice
Programmed DSBs initiated by the meiosis-specific protein Spo11 are essential for meiotic recombination.37 The repair of these breaks is mediated by HR. Defects in this process lead to impaired spermatogenesis and infertility. We examined the effect of Rnf4 deficiency on meiosis and fertility. Testicular sizes were comparable between 6-week-old Rnf4hypo/hypo and wild-type littermates, and histological examination did not reveal any obvious abnormalities at that stage. However, at 17 weeks of age, Rnf4hypo/hypo males had smaller testicles than wild-type littermates (Figure 7a). Histological analysis revealed signs of testicular degeneration and atrophy with prominent interstitial compartment with large aggregates of Leydig cells separating degenerated seminiferous tubules characterized by severe vacuolation of Sertoli cells and depletion of germ cells (Figure 7b). The concentration of mature spermatozoa was also drastically decreased in the epididymis of 17-week-old Rnf4hypo/hypo males compared with controls (Figure 7c). These data demonstrate that Rnf4 is required for spermatogenesis in an age-dependent manner.
Impaired spermatogenesis in RNF4-hypomorphic mice. (a) Comparison of testes size in 17-week-old Rnf4 +/+ (left) and Rnf4 h/h (right) male mice. H&E, 12.5x. (b) Comparative microscopic examination of testes in 17-week-old Rnf4 +/+ and Rnf4 h/h (central and right) male mice. Normal testicular histology (left); prominent interstitial compartment with large aggregates of Leydig cells (*) separating seminiferous tubules (central); groups of degenerated seminiferous tubules (arrows) characterized by severe vacuolation of sertoli cells and depletion of germ cells. There was no evidence of tubular degeneration seen in sections from RNF4 +/+ mice. H&E, 100x. (c) Comparative microscopic examination of epididymides in 17-week-old Rnf4 +/+ and Rnf4 h/h males. Normal luminal content with high number of mature spermatozoa was observed in control mice; reduced concentration of spermatozoa associated with degeneration/necrosis of multinucleated spermatids was seen in Rnf4-deficient mice. H&E, 100x
Discussion
To assess the physiological function of Rnf4 in DDR, we have generated a mouse model for Rnf4 deficiency. We show that similar to the effect of null or hypomorphic mutations of genes involved in DSB signaling, such as H2Ax,24 Mdc1,25 Rnf8,38 Rnf168,39 Rnf4 hypomorphic mice are growth retarded and radiosensitive. In addition, we observed increased cell death and a reduced number of spermatocytes in the seminiferous tubules of these mice. This meiotic defect is also reminiscent of loss of DDR proteins such as H2Ax,24 Mdc1,25 Rnf8,38 and Rnf168,39 and can be explained by the observation that Spo-11-mediated generation of DSBs and their repair by HR are required for proper meiosis.37 Together, these data establish an important genetic link between Rnf4 and the DDR in mammalian cells. Our findings are consistent with and complementary to two recent biochemical studies that have implicated RNF4 in DSB repair.30, 31
Consistent with a critical role in DSB repair, RNF4 is recruited to IRIF. It has indeed been shown before that SUMO1, SUMO2/3 and the SUMO E3 ligases PIAS1 and PIAS4 are recruited to IRIF and required for ubiquitin-adduct formation at sites of DNA damage and subsequent DSB repair. This raises the possibility that RNF4 is recruited to IRIF by binding to PIAS1/4-substrate(s) and that, in turn, RNF4 promotes the ubiquitylation of these and/or other substrates which have a critical role in DSB repair. In this context, it is interesting that RNF4 recruitment to laser tracks depends on the integrity of its SIM domains; an observation which is consistent with previous findings.30, 31 In support of this model, Galanty et al.31 showed that RNF4 recruitment to DSBs is impaired in PIAS1 and PIAS4 KD cells.
Another unresolved question is what is/are the target(s) of RNF4-dependent ubiquitylation and whether RNF4-mediated ubiquitylation at DNA lesions promotes proteolytic degradation. One mechanism through which RNF4 could promote damage repair is by inducing the degradation and subsequent removal of its substrates, allowing proper assembly of downstream factors at the sites of damage. In this context, we identified BRCA1 as an RNF4-regulated SUMO-2 conjugate. Interestingly, Yin et al.30 recently showed that depletion of BRCA1 abrogates RNF4 the accumulation at sites of DNA damage. One possible scenario is therefore that SUMOylation of BRCA1 creates an important docking site for RNF4 which, in turn, could promote the degradation and clearance of BRCA1 from sites of DNA damage.
Notably, although the regulatory function of RNF4 appears to be ubiquitin-mediated degradation of poly-SUMOylated proteins such as PML,18 RNF4-mediated ubiquitylation does not always lead to degradation.40 Our SILAC screen uncovered MDC1 and 53BP1 as IR-induced SUMO-2 target proteins. These proteins could also be involved in the early phase of RNF4 recruitment to sites of DNA damage. However, it is unlikely that they are subsequently ubiquitylated and degraded by the proteasome, as proteasome inhibition combined with IR reduced the SUMOylation of 53BP1 and had little effect on the SUMOylation of MDC1.
Among other potential RNF4 substrates are RPA1, the 70-kDa subunit of the RPA complex, and BLM, both of which are SUMOylated in response to IR and replication stress.14, 41, 42 Our findings that Rnf4-deficiency leads to impaired recruitment of Rad51 to IRIF, decreased HR-mediated repair and hypogonadism in mice strongly support a role for Rnf4 in HR-mediated DNA repair. A growing body of evidence supports HR as the dominant pathway for repair of replication-associated one-ended DSBs, which form at collapsed replication forks.43 The role of Rnf4 in replication-stress-induced DNA damage is currently being investigated in our laboratory.
Although RNF4 is yet to be linked to human pathologies, the pleiotropic defects associated with RNF4 inactivation including growth retardation, radiosensitivity, and impaired spermatogenesis highlight important physiological functions of RNF4.
Materials and Methods
Generation of Rnf4-mutant alleles and genotyping
An ES clone (RRR624) with a single gene-trap insertion of pGT01lxf in the Rnf4 locus was obtained from the International Gene Trap Consortium. The pGT01lxf vector contains an SA site flanked by loxP sites and a β-geo cassette for positive selection. This clone was used to generate mice with the unmodified trapped allele (Rnf4–) that were subsequently bred to Sox2-Cre transgenic mice to remove the SA and generate a Rnf4-hypomorphic allele (Rnf4hypo). Genotyping of the Rnf4-trapped allele (Rnf4–) was done by PCR using a 3-primer-based method. Two primers are in intron 1 (one forward: 5′-GGCTTCCTACCACCCTAACC-3′ and one reverse: 5′-CTGGCCTCAAACTCAGA TCC-3′), and one primer is in the trapping vector (5′-TCTAGGACAAGAGGGCGAGA-3′). Genotyping of the Rnf4-hypomorphic allele was done by PCR using the following primers: 5′-GCCTGGGGTACCCTATTGGA-3′ and 5′-GGCCTCAGGAAGATCGCA CT-3′. Cre-mediated excision of the SA site results in a shift from a 722 bp for the trapped allele to a 277-bp PCR product for the hypomorphic allele.
Plasmids
Human RNF4 cDNA was amplified by PCR using mammalian genome collection plasmid 42534 and specific extended primers compatible with the Gateway system. The amplified fragment was inserted into pDON207 and GFP- and V5 expression constructs were generated using standard Gateway technology (Invitrogen, Grand Island, NY, USA). A plasmid encoding the RNF4 SIM mutant was generated, replacing leucines, isoleucines, and valines in the four SIM domains of RNF4 (I36, L38, V39, I46, V47, L49, V57, V58, V59, L61, V67, V68, I69, V70) for alanines.
Cell culture
MEFs were generated from day 12.5 embryos using the standard procedures. MEFs were cultured in DMEM (Gibco Invitrogen Corporation, Grand Island, NY, USA) supplemented with 10% FCS. 10% fetal bovine serum, 2 mM GlutaMAX (Invitrogen), and 100 U/ml penicillin and 100 mg/ml streptomycin. 293 DR cells were cultured in same conditions but they were grown on poly-lysine-coated plates.
Apoptosis (FACS analysis)
Early passage MEFs were left untreated or treated with IR (5 Gy). 24 h later, cells were harvested and resuspended in 500 μl of 1X binding buffer. 5 μl of Annexin V-FITC (BD Biosciences, San Jose, CA, USA) and 5 μl of propidium iodide was added. Cells were incubated at room temperature for 5 min in the dark. Using appropriate settings cells were analyzed by FACS using BD FACSCanto.
Cell Cycle Analysis
Early-passage MEFs were left untreated or treated with IR (5 Gy). Twenty-four hours later, cells were stained with PI as per the manufacturer’s instruction using Cyclotest Plus DNA Regent Kit (Becton–Dickinson, San Jose, CA, USA).
Real-time RTq-PCR assays
Total RNA was prepared from cell pellets using the RNeasy Mini Kit (QIAGEN, Valencia, CA, USA), according to the manufacturer’s protocol. Total RNA (1 μg) was reverse transcribed in a final volume of 20 μl using a SuperScript kit (Invitrogen). These assays were performed, following the manufacturer’s specifications (PE Applied Biosystems, Foster city, CA, USA). Primer pairs and TaqMan probes were designed by Applied Biosystems (assays on demand). Each sample was analyzed in triplicate. To measure the Rnf4 mRNA expression, we used the Syber green detection assay (Applied Biosystems, Foster city, CA, USA), according to the manufacturer’s instruction. The sequence of the forward and reverse primers used is 5′-CTGGAGTCGGTACGTCCTTG-3′ and 5′-GGAGCCAACTAGATGTGCAG-3′, respectively.
Western blotting analysis
Ten microgram of whole-cell extract were fractionated by SDS-polyacrylamide gel electrophoresis and transferred to nitrocellulose. The primary antibodies used were as follows: anti–phosphorylated H2AX (Cell Signaling Technology, Danvers, MA, USA); anti-p53 (1C12, Cell Signaling Technology), anti-phospho-Ser18-P53 (Cell Signaling Technology), anti-BRCA1 (SC-646, Santa-Cruz Biotechnology, Santa Cruz, CA, USA), anti-MDC1 (A300-052A, Bethyl Laboratories, Montgomery, TX, USA), anti 53BP1 (A300-272A, Bethyl Laboratories), anti SUMO-2/3 (8A2, Abcam, Cambridge, UK), anti-GFP (Roche, Mannheim, Germany) and anti-vinculin (clone hVIN-1, Sigma-Aldrich, St Louis, MO, USA). Anti-RNF4 antibodies were raised in rabbit using bacterially produced RNF4. Secondary antibodies used include peroxidase-conjugated goat anti-rabbit IgG and anti-mouse IgG (Pierce, Rockford, IL, USA). Proteins were detected using ChemiDoc-It 500 (UVP, Upland, CA, USA). VisionWorksLS software (UVP) was used to calculate signal levels.
Immunofluorescence microscopy
Early passage MEFs were grown on glass slides were left untreated or treated with IR (5 Gy), and fixed with 4% PFA for 10 min at room temperature. Fixed MEFs were blocked with antibody dilution buffer (5% BSA and 0.05% Triton X-100 in PBS) and incubated with rabbit anti-Rad51 antibody (Abcam), rabbit anti-phospho-H2AX (Ser139) antibody (Millipore) and/or rabbit anti-phospho-H2AX (Cell Signaling Technology) for 1 hour at RT. Labeling was detected using Alexa Fluor 488-labeled goat-anti-rabbit immunoglobulin or Alexa Fluor 555-labeled goat-anti-mouse immunoglobulin secondary antibodies (Molecular Probes). Cells were mounted with mount solution vectashield (vector laboratories) containing DAPI. The slides were observed under Nikon A1R Eclips Ti microscope equipped with digital camera. Images were acquired under × 400 magnification. NIH3T3 cells were either mock-transfected or transfected with wild-type RNF4 V-5 tagged expression vectors. Forty-eight hours post-transfection, cells were irradiated (5 Gy), fixed 2 h later and stained with anti-V5 antibody (Invitrogen), as described earlier.
Multi-photon laser micro-irradiation
Laser micro-irradiation was carried out on a Leica SP5 confocal microscope equipped with an environmental chamber set to 37 °C and 5% CO2. U2-OS cells were grown on glass coverslips. DNA damage-containing tracks (1.5 x n μm) were generated with a Mira modelocked Ti:Sapphire laser (λ=800 nm, pulse-length=200 fs, repetition rate=76 MHz, output power=80 mW ). Cells were irradiated and fixed at the indicated time points and immunostained with γ-H2AX and RNF4 antibodies. GFP-RNF4 wt/ΔSIM expression vectors were transiently transfected in U2-OS cells and live cell imaging was performed after DNA damage induction to monitor the recruitment of RNF4.
UV-A laser micro-irradiation
U2-OS cells expressing GFP-tagged versions of RNF4 were grown on 18 mm coverslips and treated with 10 μ M 5′-bromo-2-deoxyuridine (BrdU) for 24 h. For micro-irradiation, the cells were placed in a Chamlide TC-A live-cell imaging chamber that was mounted on the stage of a Leica DM IRBE widefield microscope stand (Leica, Wetzlar, Germany) integrated with a pulsed nitrogen laser (Micropoint Ablation Laser System; Photonic Instruments, Inc., Belfast, Ireland). The pulsed nitrogen laser (16 Hz, 364 nm) was directly coupled to the epifluorescence path of the microscope and focused through a Leica 40x HCX PL APO 1.25-0.75 oil-immersion objective. Living cells were irradiated with the laser output at 80 in CO2-independent Leibovitz’s L-15 medium supplemented with 10% FCS at 37 °C. Images were taken at the indicated time points after micro-irradiation using Andor IQ software (Andor, Belfast, Northern Ireland) and an Andor DL658M camera (Andor).
Pulse Field Gel Electophoresis
Cells were untreated or treated with 5 Gy and 24 h later, PFGE was performed and relative intensity of DNA breakage was determined, as described earlier.44
Histology and Fragel Assay
Rnf4 +/+ and Rnf4 h/h littermate mice were left untreated or treated with IR (5 Gy). Mice were sacrificed after 24 h, and small intestine and thymus were fixed in 4%PFA, sectioned (5–7 μm), processed and embedded in paraffin using the standard immunohistochemistry protocol. Formalin-fixed and paraffin-embedded tissues were stained with either appropriate antibodies. In order to detect the apoptotic cells, the Klenow FragEL DNA Fragmentation Kit (Calbiochem, Darmstadt, Germany) was used, according to the manufacturer’s instructions. Skin sections were prepared, as described above and stained for active Caspase-3.
RNF4 knockdown
One million cells were seeded in a 15-cm dish and the next day, cells were either infected with shRNA TRCN0000017054 directed against RNF4 or control non-targeting shRNA SHC002 viruses at MOI 2 (Sigma-Aldrich). After changing the media on the third day, cells were incubated for another 3 days to obtain proper knockdown of RNF4. Cells were harvested and SUMO purifications were performed.
NHEJ and HR assay
Efficiency of NHEJ repair was measured in human cells using an assay that was recently described.35 RNF4 was KD, as described above in the engineered H1299 cells and then transfected with I-SceI-expression vector (pCBA Sce), using Lipofectamine 2000 reagent (Invitrogen). NHEJ was also measured in Rnf4 +/+ and Rnf4 h/h MEFs, following transfection with HindIII-linearized pBabe-GFP plasmid together with pBabe-HcRed as a control of transfection efficiency. NHEJ was measured by the rate of re-circulation 48 h after transfection using FACSCanto (Becton–Dickinson). HR efficiency was measured in 293HEK with a stable integration of the transgenic reporter DR-GFP, as previously described.34 RNF4 was KD in these cells, as described above, and subsequently transfected with I-SceI-expression vector (pCBA Sce), using Turbofect (Invitrogen). Forty-eight hours after the last transfection, GFP-positive cells were assayed by FACSCanto (Becton-Dickinson).
Purification of the SUMO conjugates
His-SUMO conjugates were purified essentially, as described previously.33 U2-OS cells were washed and collected in ice-cold PBS. Small aliquots of cells were lysed in 1% SDS, 1% NP-40, 100 mM Tris/Hcl, pH 7.5, 150 mM Nacl to determine the protein concentration. Guanidinium lysis buffer (6 M guanidinium-HCl, 0.1 M Na2HPO4/NaH2PO4, 0.01 M Tris/HCl, pH 8.0 and competing imidazole) was added to the cell pellet to lyse the cells, after which, the cells were sonicated to reduce the viscosity. His-SUMO conjugates were enriched on nickel-nitrilotriacetic acid-agarose beads (Qiagen) after which the beads were washed using wash buffers A to D. Wash buffer A: 6 M guanidinium-HCl, 0.1M Na2HPO4/NaH2PO4, 0.01M Tris/HCl, pH 8.0, 10 mM β-mercaptoethanol, 0.3% Triton X-100. Wash buffer B: 8 M urea, 0.1 M Na2HPO4/NaH2PO4, 0.01 M Tris/HCl, pH 8.0, 10 mM β-mercaptoethanol, 0.3% Triton X-100. Wash buffer C: 8M urea, 0.1 M Na2HPO4/NaH2PO4, 0.01 M Tris/HCl, pH 6.3, 10 mM β-mercaptoethanol, 0.3% Triton X-100. Wash buffer D: 8 M urea, 0.1 M Na2HPO4/NaH2PO4, 0.01 M Tris/HCl, pH 6.3, 10 mM β-mercaptoethanol, 0.1% Triton X-100. Samples were eluted in 7 M urea, 0.1 M NaH2PO4/Na2HPO4, 0.01 M Tris/HCl, pH 7.0, 500 mM imidazole.
Abbreviations
- DSBs:
-
double-strand breaks
- HR:
-
homologous recombination
- NHEJ:
-
non-homologous end joining
- IR:
-
ionizing radiation
- UV:
-
ultraviolet
- IRIF:
-
ionizing radiation-induced foci
- DDR:
-
DNA damage response
- STUbl:
-
SUMO-targeted ubiquitin ligase
- SA:
-
splice acceptor
- PCR:
-
polymerase chain reaction
- MEFs:
-
mouse embryonic fibroblasts
- RT-qPCR:
-
reverse transcription quantitative PCR
- ROS:
-
reactive oxygen species
- PFGE:
-
pulsed-field gel electrophoresis
- IHC:
-
immunohistochemistry
- RING:
-
really interesting new gene
- H&E:
-
hematoxylin and eosin
- (E)GFP:
-
(enhanced) green fluorescent protein
- Hypo:
-
hypomorphic
- SIM:
-
SUMO interaction motif
References
Jackson SP, Bartek J . The DNA-damage response in human biology and disease. Nature 2009; 461: 1071–1078.
Ciccia A, Elledge SJ . The DNA damage response: making it safe to play with knives. Mol Cell 2010; 40: 179–204.
Wyman C, Kanaar R . DNA double-strand break repair: all's well that ends well. Annu Rev Genet 2006; 40: 363–383.
Rothkamm K, Kruger I, Thompson LH, Lobrich M . Pathways of DNA double-strand break repair during the mammalian cell cycle. Mol Cell Biol 2003; 23: 5706–5715.
Misteli T, Soutoglou E . The emerging role of nuclear architecture in DNA repair and genome maintenance. Nat Rev Mol Cell Biol 2009; 10: 243–254.
Polo SE, Jackson SP . Dynamics of DNA damage response proteins at DNA breaks: a focus on protein modifications. Genes Dev 2011; 25: 409–433.
Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM . DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem 1998; 273: 5858–5868.
Stucki M, Clapperton JA, Mohammad D, Yaffe MB, Smerdon SJ, Jackson SP et al. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell 2005; 123: 1213–1226.
Panier S, Durocher D . Regulatory ubiquitylation in response to DNA double-strand breaks. DNA Repair (Amst) 2009; 8: 436–443.
Mailand N, Bekker-Jensen S, Faustrup H, Melander F, Bartek J, Lukas C et al. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell 2007; 131: 887–900.
Huen MS, Grant R, Manke I, Minn K, Yu X, Yaffe MB et al. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell 2007; 131: 901–914.
Bekker-Jensen S, Mailand N . The ubiquitin- and SUMO-dependent signaling response to DNA double-strand breaks. FEBS Lett 2011; 585: 2914–2919.
Bergink S, Jentsch S . Principles of ubiquitin and SUMO modifications in DNA repair. Nature 2009; 458: 461–467.
Galanty Y, Belotserkovskaya R, Coates J, Polo S, Miller KM, Jackson SP . Mammalian SUMO E3-ligases PIAS1 and PIAS4 promote responses to DNA double-strand breaks. Nature 2009; 462: 935–939.
Morris JR, Boutell C, Keppler M, Densham R, Weekes D, Alamshah A et al. The SUMO modification pathway is involved in the BRCA1 response to genotoxic stress. Nature 2009; 462: 886–890.
Sun H, Leverson JD, Hunter T . Conserved function of RNF4 family proteins in eukaryotes: targeting a ubiquitin ligase to SUMOylated proteins. EMBO J 2007; 26: 4102–4112.
Prudden J, Pebernard S, Raffa G, Slavin DA, Perry JJ, Tainer JA et al. SUMO-targeted ubiquitin ligases in genome stability. EMBO J 2007; 26: 4089–4101.
Tatham MH, Geoffroy MC, Shen L, Plechanovova A, Hattersley N, Jaffray EG et al. RNF4 is a poly-SUMO-specific E3 ubiquitin ligase required for arsenic-induced PML degradation. Nat Cell Biol 2008; 10: 538–546.
Kosoy A, Calonge TM, Outwin EA, O’Connell MJ . Fission yeast Rnf4 homologs are required for DNA repair. J Biol Chem 2007; 282: 20388–20394.
Barry KC, Abed M, Kenyagin D, Werwie TR, Boico O, Orian A et al. The Drosophila STUbL protein Degringolade limits HES functions during embryogenesis. Development 2011; 138: 1759–1769.
Hu XV, Rodrigues TM, Tao H, Baker RK, Miraglia L, Orth AP et al. Identification of RING finger protein 4 (RNF4) as a modulator of DNA demethylation through a functional genomics screen. Proc Natl Acad Sci USA 2011; 107: 15087–15092.
Migliorini D, Bogaerts S, Defever D, Vyas R, Denecker G, Radaelli E et al. Cop1 constitutively regulates c-Jun protein stability and functions as a tumor suppressor in mice. J Clin Invest 2011; 121: 1329–1343.
Hayashi S, Lewis P, Pevny L, McMahon AP . Efficient gene modulation in mouse epiblast using a Sox2Cre transgenic mouse strain. Mech Dev 2002; 119 (Suppl 1): S97–S101.
Celeste A, Petersen S, Romanienko PJ, Fernandez-Capetillo O, Chen HT, Sedelnikova OA et al. Genomic instability in mice lacking histone H2AX. Science 2002; 296: 922–927.
Lou Z, Minter-Dykhouse K, Franco S, Gostissa M, Rivera MA, Celeste A et al. MDC1 maintains genomic stability by participating in the amplification of ATM-dependent DNA damage signals. Mol Cell 2006; 21: 187–200.
Kolas NK, Chapman JR, Nakada S, Ylanko J, Chahwan R, Sweeney FD et al. Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science 2007; 318: 1637–1640.
Ward IM, Minn K, van Deursen J, Chen J . p53 Binding protein 53BP1 is required for DNA damage responses and tumor suppression in mice. Mol Cell Biol 2003; 23: 2556–2563.
Bartek J, Bartkova J, Lukas J . DNA damage signalling guards against activated oncogenes and tumour progression. Oncogene 2007; 26: 7773–7779.
Lukas C, Falck J, Bartkova J, Bartek J, Lukas J . Distinct spatiotemporal dynamics of mammalian checkpoint regulators induced by DNA damage. Nat Cell Biol 2003; 5: 255–260.
Yin Y, Seifert A, Chua JS, Maure JF, Golebiowski F, Hay RT et al. SUMO-targeted ubiquitin E3 ligase RNF4 is required for the response of human cells to DNA damage. Genes Dev 2012; 26: 1196–1208.
Galanty Y, Belotserkovskaya R, Coates J, Jackson SP . RNF4, a SUMO-targeted ubiquitin E3 ligase, promotes DNA double-strand break repair. Genes Dev 2012; 26: 1179–1195.
Luo K, Zhang H, Wang L, Yuan J, Lou Z . Sumoylation of MDC1 is important for proper DNA damage response. EMBO J 2012; 31: 3008–3019.
Schimmel J, Larsen KM, Matic I, van Hagen M, Cox J, Mann M et al. The ubiquitin-proteasome system is a key component of the SUMO-2/3 cycle. Mol Cell Proteomics 2008; 7: 2107–2122.
Pierce AJ, Hu P, Han M, Ellis N, Jasin M . Ku DNA end-binding protein modulates homologous repair of double-strand breaks in mammalian cells. Genes Dev 2001; 15: 3237–3242.
Ogiwara H, Ui A, Otsuka A, Satoh H, Yokomi I, Nakajima S et al. Histone acetylation by CBP and p300 at double-strand break sites facilitates SWI/SNF chromatin remodeling and the recruitment of non-homologous end joining factors. Oncogene 2011; 30: 2135–2146.
San Filippo J, Sung P, Klein H . Mechanism of eukaryotic homologous recombination. Annu Rev Biochem 2008; 77: 229–257.
Longhese MP, Bonetti D, Guerini I, Manfrini N, Clerici M . DNA double-strand breaks in meiosis: checking their formation, processing and repair. DNA Repair (Amst) 2009; 8: 1127–1138.
Li L, Halaby MJ, Hakem A, Cardoso R, El Ghamrasni S, Harding S et al. Rnf8 deficiency impairs class switch recombination, spermatogenesis, and genomic integrity and predisposes for cancer. J Exp Med 2010; 207: 983–997.
Bohgaki T, Bohgaki M, Cardoso R, Panier S, Zeegers D, Li L et al. Genomic instability, defective spermatogenesis, immunodeficiency, and cancer in a mouse model of the RIDDLE syndrome. PLoS Genet 2011; 7: e1001381.
Fryrear KA, Guo X, Kerscher O, Semmes OJ . The Sumo-targeted ubiquitin ligase RNF4 regulates the localization and function of the HTLV-1 oncoprotein Tax. Blood 2011; 119: 1173–1181.
Eladad S, Ye TZ, Hu P, Leversha M, Beresten S, Matunis MJ et al. Intra-nuclear trafficking of the BLM helicase to DNA damage-induced foci is regulated by SUMO modification. Hum Mol Genet 2005; 14: 1351–1365.
Dou H, Huang C, Singh M, Carpenter PB, Yeh ET . Regulation of DNA repair through deSUMOylation and SUMOylation of replication protein A complex. Mol Cell 2010; 39: 333–345.
Arnaudeau C, Lundin C, Helleday T . DNA double-strand breaks associated with replication forks are predominantly repaired by homologous recombination involving an exchange mechanism in mammalian cells. J Mol Biol 2001; 307: 1235–1245.
Ouyang KJ, Woo LL, Zhu J, Huo D, Matunis MJ, Ellis NA . SUMO modification regulates BLM and RAD51 interaction at damaged replication forks. PLoS Biol 2009; 7: e1000252.
Acknowledgements
We thank Odessa Van Goethem for excellent technical assistance and Martijn Luijsterburg for help with the UV-A laser micro-irradiation experiments. This work was supported by the Belgian federation against cancer (BFK), the Association for International Cancer Research (AICR), the Flemish Organisation for Scientific Research (FWO ZKC2592-00_WO1), and by the Netherlands Organisation for Scientific Research (NWO).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by G Melino
Supplementary Information accompanies the paper on Cell Death and Differentiation website
Rights and permissions
About this article
Cite this article
Vyas, R., Kumar, R., Clermont, F. et al. RNF4 is required for DNA double-strand break repair in vivo. Cell Death Differ 20, 490–502 (2013). https://doi.org/10.1038/cdd.2012.145
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cdd.2012.145
Keywords
This article is cited by
-
SENP5 promotes homologous recombination-mediated DNA damage repair in colorectal cancer cells through H2AZ deSUMOylation
Journal of Experimental & Clinical Cancer Research (2023)
-
RNF4 silencing induces cell growth arrest and DNA damage by promoting nuclear targeting of p62 in hepatocellular carcinoma
Oncogene (2022)
-
Crosstalk between SUMOylation and ubiquitylation controls DNA end resection by maintaining MRE11 homeostasis on chromatin
Nature Communications (2022)
-
RNF4~RGMb~BMP6 axis required for osteogenic differentiation and cancer cell survival
Cell Death & Disease (2022)
-
DNA damage-induced degradation of Sp1 promotes cellular senescence
GeroScience (2022)