Abstract
The responses to numerous stress signals are important for cellular growth and survival. The p53 tumor-suppressor protein is stabilized under stress conditions and induces transcription of several genes to regulate cell cycle and apoptosis. Regarding p53 protein accumulation, inhibition of proteasomal degradation of p53 protein, which is mainly mediated by Mdm2, has received much attention. Here, we demonstrate that regulation of translation initiation is also crucial for p53 protein accumulation. Furthermore, we report that heterogeneous nuclear ribonucleoprotein (hnRNP) Q binds to the 5′-untranslated region (UTR) of mouse p53 mRNA and regulates translation efficiency of p53 and apoptosis progression. We also suggest that changes in cytosolic hnRNP Q levels contribute to cell cycle-dependent translational differences in p53 mRNA.
Similar content being viewed by others
Main
The p53 tumor-suppressor protein is a key regulator when cells are exposed to numerous stress signals including ionizing radiation or DNA-damaging drugs.1, 2 p53 can cause cells to attenuate cell cycle progression or even to commit apoptosis if cellular damage is severe.3, 4 Because it has been reported that mutations of p53 are observed in ∼50% of human tumor cases, p53 could be a main target for anticancer research.5 p53 induces transcription of several genes, such as p21 and Bax, to modulate cellular responses to external stimuli.6
As the underlying mechanism of p53 activation under cellular stress conditions has been investigated, a number of studies have shown that p53 accumulation is controlled by posttranslational mechanisms. In particular, Mdm2 can function as an E3 ubiquitin ligase and lead to proteasome-mediated degradation of p53 protein in unstressed cells.7 As a result, p53 has a short half-life, and its level is kept low under normal conditions. In response to various stressful stimuli, however, the p53 protein is phosphorylated and its stability is increased enough to accumulate in the nucleus.8 Moreover, it has been discovered that several posttranslational regulations are important for the stability of p53 and its activity as a transcription factor.9, 10 However, only a few papers reported the translational regulation of p53. In zebrafish, p53 protein enhances the translation of its own mRNA via 3′-untranslated region (UTR) in response to UV irradiation.11 Moreover, HuR enhances 3′UTR-mediated p53 translation under stress condition in human colorectal carcinoma cells.12 ATM-dependent phosphorylation of Mdm2 leads to enhancement of p53 mRNA–Mdm2 interaction and positively regulates p53 translation following DNA damage.13 In addition, RPL26 and nucleolin are p53 5′UTR binding proteins and regulate translation of p53.14
Because of eIF2-α phosphorylation, protein synthesis is globally repressed when cells are exposed to a variety of stress conditions.15 However, certain mechanisms allow a group of mRNAs to be actively translated despite overall suppression of translation initiation. For example, translation of cellular inhibitor of apoptosis 2 (cIAP2) mRNA persists under cellular stress via ribosome shunting.16
Internal ribosomal entry site (IRES)-mediated translation initiation could be another mechanism. IRES element was first discovered in picornavirus.17 In the case of uncapped viral mRNAs, they utilize an IRES-mediated translation initiation for their expression in host cells. However, increasing evidence suggests that mammalian mRNAs also utilize IRES-dependent translation in certain situations such as circadian rhythm,18, 19 chemotoxic stress,20 and apoptosis.21 Cellular IRES elements are found in a limited number of mRNAs.22 p53 is one of the cellular genes that harbor IRES.23
Recently, some evidence has suggested that p53 mRNA is regulated posttranscriptionally. It has been reported that 3′-end processing of p53 pre-mRNA by heterogeneous nuclear ribonucleoprotein (hnRNP) H/F contributes to p53-mediated apoptosis.24 Also, conserved cytoplasmic polyadenylation elements in p53 mRNA are known to influence mRNA stability and protein synthesis.25 It has also been found that the interaction between the 5′ coding region of p53 mRNA and hnRNP C1/C2 is strongly enhanced in response to the DNA-damaging drug cisplatin.26 In line with these findings, we suggest an important posttranscriptional mechanism in the regulation of p53 protein synthesis. Under both normal and stress circumstances, hnRNP Q binds to the 5′UTR of mouse p53 mRNA and regulates translation efficiency, without affecting mRNA levels. In addition, we present evidence that hnRNP Q can mediate apoptosis progression. Moreover, we suggest that the change of hnRNP Q localization, dependent on cell cycle phase, could be one possible mechanism explaining differences in cell cycle-dependent translation efficiency of p53 mRNA.
Results
p53 protein induction is mediated by posttranscriptional regulation
To explore the mechanisms of p53 protein accumulation, we induced p53 protein by treating NIH3T3 fibroblasts with sodium nitroprusside (SNP) or etoposide. As a nitric oxide donor, SNP causes oxidative damage. Etoposide is a topoisomerase II inhibitor, leading to DNA damage. Compared with the vehicle control (Supplementary Figures 1A and B), both SNP and etoposide treatments led to accumulation of p53 rapidly (Figures 1a and c). Interestingly, however, p53 mRNA level remained unchanged for several hours (Figures 1b and d). These data suggest that posttranscriptional regulation, including posttranslational regulation, is critical for p53 accumulation under stress conditions.
Posttranscriptional regulation is important for p53 protein accumulation. (a and c) p53 protein accumulation was confirmed. The levels of mouse p53 protein were analyzed by western blotting after 1 mM sodium nitroprusside (SNP; a) or 100 μM etoposide (c) treatment at the indicated time points. GAPDH protein was used as a loading control. (b and d) Endogenous p53 mRNA levels were verified under 1 mM SNP (b) or 100 μM etoposide (d) treatment conditions. The p53 mRNA levels were determined by quantitative real-time RT-PCR and normalized to mRPL32 mRNA levels. All results are presented as the mean±S.D. of three experiments. Distilled and deionized water (DDW) (b) or dimethylsulfoxide (d) was used for vehicle treatment
To investigate whether translation initiation is necessary for induction of p53 protein, we added rapamycin to the SNP- or etoposide-treated cells. Rapamycin is an mTOR inhibitor and keeps down the cellular translation by suppressing phosphorylation of both S6 ribosomal proteins and 4E-BPs. Intriguingly, accumulation of p53 protein was highly reduced, especially in the early phase of translation interruption, with different elevating patterns to SNP or etoposide (Supplementary Figures 2A and B). Although the influence of translational inhibition on the p53 protein level became weaker with time in SNP-treated cells (Supplementary Figure 2B), these results raise the possibility that translation initiation is also crucial in p53 protein accumulation.
hnRNP Q regulates the induction kinetics of p53
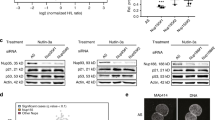
As mentioned earlier, IRES-mediated translation is recognized as more important when overall cap-dependent translation is repressed. Moreover, it has been discovered that p53 mRNA harbors an IRES element in its 5′UTR,27 and polypyrimidine tract-binding protein (PTB) regulates IRES-mediated translation of p53 isoforms.28 Because we previously reported that hnRNP Q regulates IRES-mediated translation of mRev-erb-α and rAANAT (rat arylalkylamine N-acetyltransferase), we also tested whether this protein can function as an IRES trans-acting factor (ITAF) of p53 mRNA. To this end, p53 protein levels were analyzed in either PTB- or hnRNP Q-downregulated cells after SNP treatment. It was already reported that phosphorylation of p53 within or close to the N-terminal MDM2-binding domain (i.e., at Ser-15) increases the stability of p53 by interfering with its ability to bind MDM2.29 In addition, we earlier observed that phophorylation of p53 at Ser-15 was dramatically upregulated 3 h after SNP treatment.30 Taken together, we decided to observe p53 level in an early stage under stress condition to exclude the influence of p53 protein stability issue as much as possible. Interestingly, hnRNP Q downregulation dramatically reduced accumulation of p53 protein as compared with control siRNA (Figure 2a). This result supports the hypothesis that hnRNP Q mediates p53 protein induction via translation initiation regulation.
hnRNP Q regulates the induction kinetics of p53 protein under stress conditions caused by SNP treatment. (a) p53 protein accumulation was suppressed when hnRNP Q was downregulated in the early phase after SNP treatment. PTB reduction was also performed as a positive control. Knockdown of PTB and hnRNP Q was confirmed by western blotting. GAPDH protein was used as a loading control. (b) p53 protein stability was not affected by the level of PTB or hnRNP Q. After the p53 protein level was sufficiently elevated by SNP treatment for 4 h, p53 protein stability was analyzed following treatment with 50 μg/ml cycloheximide (CHX) treatment. Knockdown of PTB and hnRNP Q was confirmed by western blotting. GAPDH protein was used as a loading control. (c and d) hnRNP Q reduction did not alter the endogenous p53 mRNA level in cytosol. (c) RNA polymerase II and 14-3-3 protein were used as nucleus and cytosol marker, respectively. Knockdown of hnRNP Q was confirmed by western blotting. N, nuclear lysate. C, cytosolic lysate. (d) The endogenous cytosolic p53 mRNA levels were determined by quantitative real-time RT-PCR and normalized to mRPL32 mRNA levels at the indicated time points after SNP treatment in control cells or hnRNP Q siRNA-transfected NIH3T3 cells
To exclude the possibility that reduced p53 accumulation in hnRNP Q-downregulated cells could result from a shorter half-life of p53 protein, we analyzed p53 protein degradation rate. The p53 protein level rapidly decreased when cells were treated with the general translation elongation inhibitor cycloheximide (CHX) (Supplementary Figure 1C). Similarly, downregulation of either PTB or hnRNP Q did not change p53 protein stability (Figure 2b). To exclude the possibility that the decrease in the p53 level under hnRNP Q silencing could result from a lowered p53 mRNA level, we analyzed p53 mRNA level. Because mature mRNAs are translated to protein in the cytosol, we analyzed the amount of cytosolic p53 mRNA that was competent to be translated (Figure 2c). As a result, reduction of hnRNP Q did not alter the cytosolic p53 mRNA level (Figure 2d). These results suggest that hnRNP Q mainly regulates p53 protein accumulation in the translation initiation step.
hnRNP Q regulates IRES-mediated translation of p53 mRNA under both normal and stress conditions
We already reported that hnRNP Q regulates translation of rAANAT and mRev-erb-α in a cap-independent manner.18, 19 To verify whether hnRNP Q modulates the IRES-mediated translation of p53 mRNA, a dicistronic pRF vector was used.31 The 5′UTR of mouse p53 mRNA was inserted between the Renilla luciferase (Rluc) and firefly luciferase (Fluc) coding sequences (pRF p53 1–157), and IRES-mediated translation efficiency was measured. In control siRNA-transfected cells, p53 mRNA 5′UTR enhanced the Fluc level approximately eightfold compared with the pRF mock vector. Interestingly, Fluc enhancement was dramatically reduced when hnRNP Q was downregulated (Figures 3a and b). As reported earlier, PTB reduction also decreased Fluc enhancement. This strong correlation between the reporter assay result and the endogenous p53 protein level suggests that hnRNP Q is a crucial translation initiation regulator of p53 mRNA.
hnRNP Q regulates translation of p53 mRNA in both normal and stress conditions. (a) The suppression of IRES activity of p53 when hnRNP Q was reduced is shown. A PTB reduction experiment was also performed as a positive control. The efficiency of IRES-mediated translation of the p53 5′UTR-inserted pRF construct is shown. Because the first cistron Rluc is translated in a cap-dependent manner but the second cistron Fluc is translated by the sequences inserted between the Rluc stop codon and the Fluc start codon, the ratio of Fluc to Rluc means the IRES-mediated translation efficiency. A pRF mock vector was used as a negative control. The IRES activity of the pRF mock vector in control siRNA-transfected cells was set as 1. The activities of firefly and Renilla luciferases are depicted in boxes below the graph. This result is representative of at least three independent experiments. The error bars represent the mean±S.E.M. of duplicate measurements. (b) Knockdown of PTB and hnRNP Q was confirmed by western blotting. GAPDH protein was used as a loading control. (c) Identification of the interaction between the p53 5′UTR and hnRNP Q (68-kDa) protein using UV crosslinking followed by immunoprecipitation. 5′UTR of mouse p53 mRNA was radioactive isotope labeled by in vitro transcription using 32P-UTP. After incubation of 32P-labeled p53 mRNA with NIH3T3 cytosolic extracts, RNA/protein interaction was enhanced by UV and RNases were added. As a result of SDS gel separation, proteins bound to p53 mRNA were detected by autoradiogram. PIS, preimmune serum. (d) Confirmation of the interaction between the p53 5′UTR and hnRNP Q by RNA affinity purification followed by western blotting. (e) p53 protein synthesis was influenced by hnRNP Q knockdown in normal conditions. At 24 h after control or hnRNP Q siRNA transfection, cells were treated with 10 μM MG132. The kinetics of p53 protein was analyzed by western blotting at the indicated time points. Knockdown of hnRNP Q was confirmed by western blotting. GAPDH protein was used as a loading control. (f) At 24 h after transfection with siCon or sihnRNP Q, NIH3T3 cells were treated with 10 μM MG132 for 1 h. Downregulated de novo synthesis of p53 under hnRNP Q silencing was seen in radiogram after immunoprecipitation. (g) The suppression of IRES activity of p53 when hnRNP Q was reduced is confirmed by RNA transfection. Translational efficiencies of p53 5′UTR were measured through luciferase activity. Capped-Rluc was used for transfection efficiency control. The activities of firefly and Renilla luciferases are depicted in boxes on the right side of the graph. The results are expressed as the mean±S.D. of two different experiments
To test the association between hnRNP Q and p53 mRNA, we conducted in vitro binding assay using cytosolic fraction of NIH3T3 fibroblasts. Several proteins clearly bound to the 5′UTR of the mouse p53 mRNA (Figure 3c). To confirm hnRNP Q as a binding factor of p53 mRNA, we performed UV crosslinking followed by immunoprecipitation with an anti-hnRNP Q antibody. As expected, the 68-kDa band was clearly detected in precipitates using anti-hnRNP Q (Figure 3c). In contrast, there was no detectable band in precipitates using preimmune serum. To further confirm the association, we conducted RNA affinity purification followed by western blotting. hnRNP Q was bound to biotinylated p53 mRNA 5′UTR, and the western blot band was diminished by competition with unlabeled p53 mRNA 5′UTR (Figure 3d).
To examine whether hnRNP Q regulates p53 protein level in normal conditions as well, we analyzed p53 protein synthesis rate after hnRNP Q reduction. We treated NIH3T3 cells with the proteasome inhibitor MG132 after siRNA transfection. Compared with control, accumulation of p53 protein was decreased under hnRNP Q silencing (Figure 3e). This observation raised the possibility that hnRNP Q modulates the p53 translation rate, at least in part, regardless of cellular circumstances. To verify the effect of hnRNP Q on synthesis of nascent p53 protein, metabolic labeling followed by immunoprecipitation was performed. As a result, the de novo p53 protein synthesis was reduced under hnRNP Q silencing (Figure 3f). However, overall protein synthesis was rarely affected by hnRNP Q downregulation. To further confirm the translational control of p53 mRNA by hnRNP Q, we conducted a RNA transfection. The RNA transfection method was used to exclude potential problems occurred by DNA transfection, such as possible involvement of cryptic promoter or unexpected splicing. Luciferase activity showed that p53 mRNA 5′UTR-mediated translation of Fluc mRNA was reduced under hnRNP Q knockdown (Figure 3g). Taken together, these data suggest that hnRNP Q controls the translation of p53 mRNA under both normal and stress conditions.
To test whether this kind of regulation also exists in other species, we explored the effect of hnRNP Q on translation of human p53 mRNA. The 5′UTRs of two human p53 mRNA isoforms (NM_000546.5 for p53-1, NM_001126115.1 for p53-2) were inserted between the Rluc and Fluc coding sequences (pRF p53-1 5′UTR and pRF p53-2 5′UTR), and DNA transfection was performed in human neuroblastoma SH-SY5Y cells. Interestingly, Fluc enhancements were reduced ∼30% in both constructs under hnRNP Q knockdown (Supplementary Figure 3A). We also confirmed the association between hnRNP Q and two human p53 mRNA isoforms by RNA affinity purification (Supplementary Figure 3B). Consistent with the result from mouse cell line, p53 protein accumulation was reduced under hnRNP Q silencing in SH-SY5Y (Supplementary Figure 3C).
Downregulation of hnRNP Q leads to delayed cell death by reducing p53 translation
Regulation of cell cycle progression or apoptosis control is a major role of p53 protein. Because the endogenous p53 level was downregulated under hnRNP Q reduction, we analyzed the effect of hnRNP Q on cell cycle changes. However, downregulation of hnRNP Q did not dramatically alter the cell cycle distribution in our condition (Supplementary Figure 4). Next, we tested the influence of hnRNP Q on apoptosis. Because cleaved caspase-3 is a critical executioner of apoptosis, we analyzed the amount of activated caspase-3 under hnRNP Q downregulation. As expected, a reduced p53 protein level was observed in hnRNP Q-downregulated cells (Figure 4a). Correlated with this result, cleaved caspase-3 was also decreased under hnRNP Q silencing, suggesting that hnRNP Q modulates apoptosis kinetics. This result was reproducible with two other hnRNP Q siRNAs (Figure 4b). To confirm the influence of hnRNP Q on apoptosis, we analyzed the apoptotic index by labeling with FITC-labeled Annexin V followed by flow cytometric analysis. As a result, the proportions of apoptotic cells were reduced in hnRNP Q-downregulated cells (Figures 4c and d). To confirm whether hnRNP Q-mediated apoptosis regulation is p53 dependent, we analyzed the apoptotic index in p53/Mdm2 double-knockout mouse embryonic fibroblasts (MEFs). Apoptosis progression was not affected by hnRNP Q silencing in p53-null cells (Figures 4e and f). Collectively, our data suggest that hnRNP Q can regulate cell death kinetics by affecting the p53 protein level.
Reduction of hnRNP Q delayed apoptosis progression through downregulation of the p53 protein level. (a) Caspase-3 activation was suppressed when hnRNP Q was downregulated. At 24 h after control or hnRNP Q siRNA transfection, cells were treated with 50 μM etoposide. Knockdown of hnRNP Q was confirmed by western blotting. The 14-3-3 protein was used as a loading control. (b) Caspase-3 activation was also suppressed when three different kinds of siRNAs against hnRNP Q were used for hnRNP Q downregulation. GAPDH was used as a loading control. (c) Apoptosis was reduced when hnRNP Q was downregulated. At 24 h after control or hnRNP Q siRNA transfection, cells were treated with 100 μM etoposide for 8 h. The apoptotic index was estimated by labeling with FITC-labeled Annexin V. (d) Knockdown of hnRNP Q was confirmed by western blotting. GAPDH protein was used as a loading control. (e) The apoptotic index was estimated by labeling with FITC-labeled Annexin V in p53/Mdm2 double-knockout MEFs. At 24 h after control or hnRNP Q siRNA transfection, cells were treated with 200 μM etoposide for 8 h. (f) Knockdown of hnRNP Q was confirmed by western blotting. GAPDH protein was used as a loading control
Next, we tested whether reduction of hnRNP Q diminishes the mRNA levels of p53 target genes. Overall, mRNA expressions of p21 and Mdm2 were lowered in sihnRNP Q-transfected cells (Supplementary Figures 5A and B). This result supported our hypothesis that hnRNP Q is one of the translational regulators of p53 mRNA.
To identify the cis-acting region, we performed a luciferase assay and in vitro binding assay with deletion constructs (Supplementary Figure 6A). Although there was little change in the IRES activity when the 5′ proximal 86 nucleotides within the 5′UTR were deleted (pRF p53 87–157), additional 23 nucleotides deletion (pRF p53 110–157) reduced the IRES activity to ∼60% of the full-length construct. In line with such findings, we discovered that the 68-kDa protein, which was already confirmed as hnRNP Q (Figures 3c and d), showed reduced binding to the p53 110–157 construct compared with the p53 87–157 construct (Supplementary Figure 6B). To confirm this, the amount of p53 protein generated from the mRNAs containing wild-type or serially deleted 5′UTR was examined (Supplementary Figure 6C). Consistently, exogenous p53 expression was much lowered when nucleotides 87–109 in the 5′UTR of the mouse p53 mRNA were absent. We therefore suggest that the cis-acting element for translation initiation by hnRNP Q resides between nucleotides 87 and 109 in the 5′UTR of the mouse p53 mRNA. Based on previous report that hnRNP Q has a strong affinity for adenosine-rich RNAs,32 we substituted all adenosine in cis-acting region to guanosine and performed luciferase assay and in vitro binding analysis. Interestingly, mutant reporter (pRF p53 1–157 mt) showed reduced IRES activity and weaker association with hnRNP Q as compared with wild-type reporter (Supplementary Figures 6D and E). However, because A–G substitutions did not perfectly inhibit Fluc enhancement and interaction with hnRNP Q, hnRNP Q binding could be affected by other sequences or secondary structure (Supplementary Figure 6F).
p53 protein accumulates in a cell cycle-dependent manner
Previously, it was reported that IRES activity of the human p53 5′UTR (full length) is the highest at the G2/M transition when general cap-dependent translation is downregulated.23 Consistent with this result, IRES activity of mouse p53 5′UTR was more enhanced in G2/M-arrested cells than S-arrested cells (Supplementary Figure 7A). We also found that p53 protein level was higher in G2- or G2/M-arrested cells than in G1- or S-arrested cells. However, the total cellular hnRNP Q level was maintained in all phases, and endogenous p53 mRNA level was also similar in all phases, except that it was upregulated in the S-arrested cells (Supplementary Figures 7B–D). This result was correlated with previous studies that have found that p53 transcription is induced before or during early S phase.33, 34 Because translation is a cytosolic process, we investigated the level of hnRNP Q in the cytoplasm in each cell phase. Interestingly, the cytosolic hnRNP Q levels were also relatively high in the G2- and G2/M-arrested cells, and this phenomenon correlated well with the p53 protein level (Supplementary Figure 7E). We further analyzed p53 protein accumulation kinetics in the S and G2/M arrest condition. As a result, in the G2/M-arrested cells, when the cytoplasm-localized hnRNP Q level was higher, p53 protein accumulated with an increased rate (Supplementary Figures 7F and G). Although we could not totally exclude a side effect of each drug used for cell cycle arrest, these data suggest that cell cycle phase-dependent localization of hnRNP Q in the cytoplasm could control the translation efficiency of p53, at least in part.
Discussion
A number of observations provide evidence that regulation of p53 protein degradation is a crucial step in p53 accumulation. However, we suggest that translational regulation could be also a critical regulation in p53 protein accumulation (Figure 1). We also identified hnRNP Q as a potential key regulator of p53 protein synthesis under both stress and normal conditions (Figures 2 and 3). We further suggest that hnRNP Q could contribute to apoptosis kinetics (Figure 4).
The influence on p53 translation by hnRNP Q seemed not to be constant. In NIH3T3 cells, the IRES activity of p53 reached a peak at 90 min after SNP treatment and gradually decreased (Supplementary Figure 8A). In line with these data, the cytoplasmic hnRNP Q level was also higher at early time points after treatment with SNP or etoposide (Supplementary Figures 8B and C). Contrarily, total level of hnRNP Q remained constant (Supplementary Figures 8D and E). Nevertheless, p53 protein level continuously increased until 6 h after SNP treatment. There are two possible reasons. First, p53 protein degradation could be strongly inhibited over time. We analyzed the p53 translation in early time of stress response, because phophorylation of p53 at Ser-15 was dramatically upregulated 3 h after SNP treatment in SH-SY5Y cells. It will be necessary to investigate the p53 protein stability dynamics in NIH3T3 cells. Second, another factor could mediate translation initiation of p53 mRNA, especially at later time points of a stress condition. Indeed, PTB was gradually relocated to the cytoplasm.
Even though we found that the hnRNP Q-interacting cis-acting region resides between nucleotides 87 and 109 of the 5′UTR of mouse p53 mRNA, we also observed that the IRES activity dramatically increased when the 5′ proximal 51 nucleotides within the 5′UTR were deleted (pRF p53 52–157). It is likely that some translation repressors could bind to the 5′ proximal 51 nucleotides and inhibit protein synthesis.
In this study, we demonstrate that downregulation of hnRNP Q affected apoptosis progression, but cell cycle change was hardly detected. This result could have arisen from the relatively high concentrations of stress-inducing agents. Otherwise, there is another possibility that hnRNP Q may be involved in posttranscriptional regulations of other cell cycle-related genes. In our results, when hnRNP Q was downregulated, p53 accumulation was reduced but expression levels of p21 and Mdm2 seem to be at least 75–80% compared with siRNA control. There could be at least three reasons. First, p53 is not the sole factor that mediates the transcription of p21 and Mdm2. Because we did not monitor the level of other transcription factors under hnRNP Q knockdown, expression levels of p21 and Mdm2 could not be explained only by p53 protein level. Second, p53 protein activity must be considered. Although we confirmed far-less increase of p53 protein level under hnRNP Q downregulation, p53 protein modification was not checked. Increasing evidence has suggested that posttranslational regulations of p53 protein are also important for p53 action. Therefore, it will be necessary to explore the direct or indirect effects of hnRNP Q on p53 protein modification or localization. Last, hnRNP Q could control the transcription or mRNA decay of p21 and Mdm2 in a p53-independent manner.
Although we suggest that hnRNP Q is a translational regulator of p53 mRNA, translational control of p53 seems to be more complex. We observed that ∼50% of IRES activity of mouse p53 remained under hnRNP Q silencing (Figure 3a) and the construct that lacked hnRNP Q binding region (pRF p53 110–157) showed 65–70% of the activity of the full-length construct (Supplementary Figure 6A). Therefore, it is highly possible that other factors are also involved in p53 translation regulation. Indeed, it has been reported that RPL26 and nucleolin are p53 5′UTR binding proteins and regulate translation of p53.14 Although we focused on 5′UTR of p53 mRNA, it has been known that 3′UTR-mediated translational control of p53 is also important.35 It will be necessary to check the interaction between 3′UTR of p53 mRNA and hnRNP Q.
It should also be noted that the influence of hnRNP Q on p53 translation may be dependent on physiological cellular states. SNP is a precursor for nitric oxide and triggers NO-induced36 or NO-independent programmed cell death and increases the percentage of G2/M phase-blocked cells.37 Etoposide triggers phosphorylation of the checkpoint kinase Chk1 and subsequent G2 arrest. In SNP-treated cells, hnRNP Q knockdown strongly reduced the endogenous p53 protein level in the very early phase after SNP treatment (Figure 2a), but this effect was dramatically diminished 4 h after SNP treatment (Figure 2b). In contrast, in etoposide-treated cells, the effect of hnRNP Q on p53 mRNA translation lasted for a much longer time. However, all of the data suggested in common that hnRNP Q regulates the induction kinetics of p53 protein through translation initiation control. It has been reported that reduction in hnRNP Q levels leads to slowed proliferation of human colon cancer cells.38 Although we did not perform a proliferation assay, hnRNP Q may have far more complex roles on diverse genes.
Evidence of posttranscriptional regulation of p53 mRNA has recently been increasing, and we suggest important clues that hnRNP Q is one of the regulators that can regulate p53 translation efficiency and cellular levels and further affect apoptosis progression. Although the mechanisms of hnRNP Q localization change and the existence of cofactors assisting hnRNP Q still need to be determined, we suggest that translational regulation through hnRNP Q could be an important determinant of p53 protein level.
Materials and Methods
Plasmids
To generate pRF p53 1–157, the mouse p53 (accession no. NM_011640.3) 5′UTR was amplified with the forward primer 5′-AAGTCGACTTTCCCCTCCCACGTGCTCA-3′ and the reverse primer: 5′-AACCCGGGCCAGTCTTCGGAGAAGCGTGA-3′ using Pfu polymerase (Solgent, Daejeon, Republic of Korea) and was confirmed by sequencing. The PCR product was digested with SalI and SmaI and inserted into the intercistronic region of a pRF dicistronic vector.19
For in vitro binding followed by UV crosslinking, full-length and deletion fragments of the mouse p53 5′UTR were amplified as described above. The PCR products were digested with EcoRI and XbaI, and then inserted into the pSK’ vector,39 yielding pSK’ p53 5′UTR.
Cell culture
NIH3T3 cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM; Welgene, Daegu, Republic of Korea) supplemented with 10% fetal bovine serum (Hyclone, Logan, Utah, USA) and 1% penicillin–streptomycin (Welgene) in a humidified atmosphere containing 5% CO2 at 37°C. The immortalized fibroblasts from double-knockout p53/Mdm2 mice, a gift from Jaewhan Song (Yonsei University, Seoul, Korea), were grown in DMEM supplemented with 10% fetal bovine serum (Hyclone) and 1% penicillin–streptomycin (Welgene).
Luciferase assay
Firefly and Renilla luciferase activities were determined using the Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA) according to the manufacturer’s instructions. Normalized Fluc activity was determined as the ratio of firefly to Renilla activity.
Transient transfection
For pRF plasmid transfection, NIH3T3 cells were seeded in 24-well plates at a density of 1 × 105 cells per well and incubated for 18 h before transfection. Transfection was carried out using Metafectene (Biontex, Martinsried/Planegg, Germany) according to the manufacturer’s instructions. After incubation for 24 h, cells were harvested for further experiments.
Quantitative real-time RT-PCR
Quantitative real-time RT-PCR was performed as described previously.40 In brief, total RNA was isolated using TRI Reagent (Molecular Research Center, Cincinnati, OH, USA). RNA was reverse transcribed using the ImProm-IITM Reverse Transcription System (Promega) according to the manufacturer’s instructions. For detection and quantification, the StepOnePlus Real-Time PCR System (Applied Biosystems, Carlsbad, CA, USA) was used. The sequences of the forward and reverse primers were as follows: endogenous mouse p53, 5′-GGATGCCCATGCTACAGAGGAGTCT-3′ and 5′-GTCTGAGTCAGGCCCCACTTTCTTG-3′; ribosomal protein L32 (mRPL32), 5′-AACCCAGAGGCATTGACAAC-3′ and 5′-CACCTCCAGCTCCTTGACAT-3′; p21, 5′-TTGCACTCTGGTGTCTGAGC-3′ and 5′-TCTGCGCTTGGAGTGATAGA-3′; MDM2, 5′-TGTGTGAGCTGAGGGAGATG-3′ and 5′-ATCCTGATCCAGGCAATCAC-3′.
In vitro binding assay and immunoprecipitation
In vitro binding assay was performed as described previously.19 In brief, XbaI-linearized pSK’ p53 5′UTR constructs were transcribed using T7 RNA polymerase (Promega) in the presence of [α-32P]UTP. A total of 40 μg of cytosolic extracts were incubated with labeled RNAs at 30°C. After 30 min of incubation, the mixtures were UV irradiated on ice for 15 min with a CL-1000 UV-crosslinker (UVP, Upland, CA, USA). The samples were detected with autoradiography after SDS-PAGE. To confirm the identity of the UV crosslinked protein, 3 μg of a polyclonal antibody against hnRNP Q (Sigma-Aldrich, St. Louis, MO, USA) or preimmune serum were added to RNase-digested reaction mixtures. After 16 h, Protein G agarose beads (Amersham Bioscience, Piscataway, NJ, USA) were added. After a further incubation for 3 h, precipitates were detected with autoradiography after SDS-PAGE.
Flow cytometry
At 24 h after transfection of NIH3T3 cells with indicated siRNAs, etoposide was additionally treated into cells to analyze the apoptotic index. Cells were fixed in a final concentration of 2% paraformaldehyde (PFA). After phosphate-buffered saline (PBS) wash and centrifugation, cells were stained with anti-Annexin V antibody conjugated with allophycocyanin (APC). Annexin V-APC antibody was purchased from BD pharmingen (San Diego, CA, USA), and applied according to the manufacturer’s instruction. Samples containing 5000 cells were analyzed on a FACSCalibur system (BD Biosciences, San Jose, CA, USA).
Cells were prepared as confluency of 70% to synchronize cell cycle. Cells were arrested in G1 phase by treating 500 μM of mimosine (Sigma, St. Louis, MO, USA), in S phase by treating 1 mM of hydroxyurea (Sigma), in G2 phase by treating 50 μM of irinotecan, and in G2/M arrest by treating 50 ng/ml of nocodazole (Sigma) for 16 h. Samples containing 5000 cells were analyzed on a FACSCalibur system (BD Biosciences).
Protein preparation and immunoblot analysis
For immunoblotting, cells were disrupted with complete protein solubilizing buffer containing 1% SDS and 2 M urea in PBS, followed by sonication. Fractionation of NIH3T3 cells into cytosolic and nuclear extracts was performed as previously described.19 Immunoblot analyses were performed with polyclonal anti-polypyrimidine tract binding protein (PTB), monoclonal anti-14-3-3ξ (Santa Cruz, Santa Cruz, CA, USA), polyclonal antibody against hnRNP Q (Sigma-Aldrich), monoclonal antibody against p53 (Cell Signaling, Danvers, MA, USA), polyclonal phosphor-S6 ribosomal protein (Ser 235/236) antibody (Cell Signaling), and polyclonal anti-GAPDH. HRP-conjugated mouse (Thermo scientific, Waltham, MA, USA), rabbit (Promega), or rat (Bethyl Laboratories, Montgomery, TX, USA) secondary antibodies were detected with SUPEX ECL reagent (Neuronex, Daegu, Republic of Korea) and a LAS-4000 system (FUJI FILM, Tokyo, Japan), according to the instructions of the manufacturer.
Metabolic labeling
To determine the de novo protein synthesis, metabolic labeling experiments were carried out as described previously.18 In brief, NIH3T3 cells were seeded in 24-well plates at a density of 1.0 × 105 cells per well a day before labeling. Cells were washed twice with PBS and incubated in methionine- and cysteine-free DMEM (Invitrogen, Carlsbad, CA, USA). After 60 min, 35S-labeled methionine (35S-Met) and Cysteine (35S-Cys) (250 μCi/ml; NEN Life Science products, Boston, MA, USA) were supplemented with either DMSO or 20 nM rapamycin. After 60 min, cells were then harvested for further experiment. p53 (1C12) mouse monoclonal antibody (Cell Signaling, no. 2524) was used for immunoprecipitation.
RNA interference
The sequences of synthesized siRNAs were as follows: siCon, 5′-UUCUCCGAACGUGUCACGUTT-3′ (Samchully Pharm.); siPTB, 5′-ACACCUGUGCCUAGCAAUATT-3′ (Samchully Pharm., Seoul, Republic of Korea); sihnRNP Q, 5′-AGACAGUGAUCUCUCUCAUTT-3′ (Dharmacon Research, Lafayette, CO, USA); sihnRNP Q1, 5′-GAUCAGAAGAGGAAAGAAATT-3′ (Bioneer, Daejeon, Republic of Korea); sihnRNP Q2, 5′-CAACAGAGGUUAUGCGUUUTT-3′ (Bioneer). For siRNA transfection into NIH3T3 cell lines, a microporator (Digital-Bio, Seoul, Republic of Korea) was used, according to the manufacturer’s instructions.
Accession codes
Abbreviations
- rAANAT:
-
rat arylalkylamine N-acetyltransferase
- CHX:
-
cycloheximide
- Fluc:
-
firefly luciferase
- FACS:
-
fluorescence-activated cell sorting
- MEF:
-
mouse embryonic fibroblast
- IRES:
-
internal ribosomal entry site
- ITAF:
-
IRES trans-acting factor
- hnRNP:
-
heterogeneous nuclear ribonucleoprotein
- Rluc:
-
Renilla luciferase
- RPL32:
-
ribosomal protein L32
- UTR:
-
untranslated region
References
Zhan Q, Carrier F, Fornace AJ . Induction of cellular p53 activity by DNA-damaging agents and growth arrest. Mol Cell Biol 1993; 13: 4242–4250.
Maltzman W, Czyzyk L . UV irradiation stimulates levels of p53 cellular tumor antigen in nontransformed mouse cells. Mol Cell Biol 1984; 4: 1689–1694.
Lin D, Shields MT, Ullrich SJ, Appella E, Mercer WE . Growth arrest induced by wild-type p53 protein blocks cells prior to or near the restriction point in late G1 phase. Proc Natl Acad Sci USA 1992; 89: 9210–9214.
Yonish-Rouach E, Grunwald D, Wilder S, Kimchi A, May E, Lawrence JJ et al. p53-mediated cell death: relationship to cell cycle control. Mol Cell Biol 1993; 13: 1415–1423.
Maslon MM, Hupp TR . Drug discovery and mutant p53. Trends Cell Biol 2010; 20: 542–555.
Hastak K, Agarwal MK, Mukhtar H, Agarwal ML . Ablation of either p21 or Bax prevents p53-dependent apoptosis induced by green tea polyphenol epigallocatechin-3-gallate. FASEB J 2005; 19: 789–791.
Haupt Y, Maya R, Kazaz A, Oren M . Mdm2 promotes the rapid degradation of p53. Nature 1997; 387: 296–299.
Colman MS, Afshari CA, Barrett JC . Regulation of p53 stability and activity in response to genotoxic stress. Mutat Res 2000; 462: 179–188.
Tibbetts RS, Brumbaugh KM, Williams JM, Sarkaria JN, Cliby WA, Shieh SY et al. A role for ATR in the DNA damage-induced phosphorylation of p53. Genes Dev 1999; 13: 152–157.
Shieh SY, Ahn J, Tamai K, Taya Y, Prives C . The human homologs of checkpoint kinases Chk1 and Cds1 (Chk2) phosphorylate p53 at multiple DNA damage-inducible sites. Genes Dev 2000; 14: 289–300.
Zhao X, Wu N, Ding L, Liu M, Liu H, Lin X . Zebrafish p53 protein enhances the translation of its own mRNA in response to UV irradiation and CPT treatment. FEBS Lett 2012; 586: 1220–1225.
Mazan-Mamczarz K, Galban S, Lopez de Silanes I, Martindale JL, Atasoy U, Keene JD et al. RNA-binding protein HuR enhances p53 translation in response to ultraviolet light irradiation. Proc Natl Acad Sci USA 2003; 100: 8354–8359.
Gajjar M, Candeias MM, Malbert-Colas L, Mazars A, Fujita J, Olivares-Illana V et al. The p53 mRNA-Mdm2 interaction controls Mdm2 nuclear trafficking and is required for p53 activation following DNA damage. Cancer Cell 2012; 21: 25–35.
Takagi M, Absalon MJ, McLure KG, Kastan MB . Regulation of p53 translation and induction after DNA damage by ribosomal protein L26 and nucleolin. Cell 2005; 123: 49–63.
Clemens MJ, Bushell M, Jeffrey IW, Pain VM, Morley SJ . Translation initiation factor modifications and the regulation of protein synthesis in apoptotic cells. Cell Death Differ 2000; 7: 603–615.
Sherrill KW, Lloyd RE . Translation of cIAP2 mRNA is mediated exclusively by a stress-modulated ribosome shunt. Mol Cell Biol 2008; 28: 2011–2022.
Pelletier J, Sonenberg N . Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature 1988; 334: 320–325.
Kim TD, Woo KC, Cho S, Ha DC, Jang SK, Kim KT . Rhythmic control of AANAT translation by hnRNP Q in circadian melatonin production. Genes Dev 2007; 21: 797–810.
Kim DY, Woo KC, Lee KH, Kim TD, Kim KT . hnRNP Q and PTB modulate the circadian oscillation of mouse Rev-erb alpha via IRES-mediated translation. Nucleic Acids Res 2010; 38: 7068–7078.
Dobbyn HC, Hill K, Hamilton TL, Spriggs KA, Pickering BM, Coldwell MJ et al. Regulation of BAG-1 IRES-mediated translation following chemotoxic stress. Oncogene 2008; 27: 1167–1174.
Bushell M, Stoneley M, Kong YW, Hamilton TL, Spriggs KA, Dobbyn HC et al. Polypyrimidine tract binding protein regulates IRES-mediated gene expression during apoptosis. Mol Cell 2006; 23: 401–412.
Nevins TA, Harder ZM, Korneluk RG, Holcik M . Distinct regulation of internal ribosome entry site-mediated translation following cellular stress is mediated by apoptotic fragments of eIF4G translation initiation factor family members eIF4GI and p97/DAP5/NAT1. J Biol Chem 2003; 278: 3572–3579.
Ray PS, Grover R, Das S . Two internal ribosome entry sites mediate the translation of p53 isoforms. EMBO Rep 2006; 7: 404–410.
Decorsiere A, Cayrel A, Vagner S, Millevoi S . Essential role for the interaction between hnRNP H/F and a G quadruplex in maintaining p53 pre-mRNA 3′-end processing and function during DNA damage. Genes Dev 2011; 25: 220–225.
Rosenstierne MW, Vinther J, Mittler G, Larsen L, Mann M, Norrild B . Conserved CPEs in the p53 3′ untranslated region influence mRNA stability and protein synthesis. Anticancer Res 2008; 28: 2553–2559.
Christian KJ, Lang MA, Raffalli-Mathieu F . Interaction of heterogeneous nuclear ribonucleoprotein C1/C2 with a novel cis-regulatory element within p53 mRNA as a response to cytostatic drug treatment. Mol Pharmacol 2008; 73: 1558–1567.
Moreno SG, Dutrillaux B, Coffigny H . Status of p53, p21, mdm2, pRb proteins, and DNA methylation in gonocytes of control and gamma-irradiated rats during testicular development. Biol Reprod 2001; 64: 1422–1431.
Grover R, Ray PS, Das S . Polypyrimidine tract binding protein regulates IRES-mediated translation of p53 isoforms. Cell Cycle 2008; 7: 2189–2198.
Appella E, Anderson CW . Post-translational modifications and activation of p53 by genotoxic stresses. Eur J Biochem 2001; 268: 2764–2772.
Lee SJ, Kim DC, Choi BH, Ha H, Kim KT . Regulation of p53 by activated protein kinase C-delta during nitric oxide-induced dopaminergic cell death. J Biol Chem 2006; 281: 2215–2224.
Vagner S, Galy B, Pyronnet S . Irresistible IRES. Attracting the translation machinery to internal ribosome entry sites. EMBO Rep 2001; 2: 893–898.
Mizutani A, Fukuda M, Ibata K, Shiraishi Y, Mikoshiba K . SYNCRIP a cytoplasmic counterpart of heterogeneous nuclear ribonucleoprotein R, interacts with ubiquitous synaptotagmin isoforms. J Biol Chem 2000: 2759823–2759831.
Boggs K, Reisman D . Increased p53 transcription prior to DNA synthesis is regulated through a novel regulatory element within the p53 promoter. Oncogene 2006; 25: 555–565.
Reich NC, Levine AJ . Growth regulation of a cellular tumour antigen, p53, in nontransformed cells. Nature 1984; 308: 199–201.
Chen J, Kastan MB . 5′-3′-UTR interactions regulate p53 mRNA translation and provide a target for modulating p53 induction after DNA damage. Genes Dev 2010; 24: 2146–2156.
Ciriolo MR, De Martino A, Lafavia E, Rossi L, Carri MT, Rotilio G . Cu,Zn-superoxide dismutase-dependent apoptosis induced by nitric oxide in neuronal cells. J Biol Chem 2000; 275: 5065–5072.
Cardaci S, Filomeni G, Rotilio G, Ciriolo MR . Reactive oxygen species mediate p53 activation and apoptosis induced by sodium nitroprusside in SH-SY5Y cells. Mol Pharmacol 2008; 74: 1234–1245.
Yoo BC, Hong SH, Ku JL, Kim YH, Shin YK, Jang SG et al. Galectin-3 stabilizes heterogeneous nuclear ribonucleoprotein Q to maintain proliferation of human colon cancer cells. Cell Mol Life Sci 2009; 66: 350–364.
Woo KC, Kim TD, Lee KH, Kim DY, Kim W, Lee KY et al. Mouse period 2 mRNA circadian oscillation is modulated by PTB-mediated rhythmic mRNA degradation. Nucleic Acids Res 2009; 37: 26–37.
Kwak E, Kim TD, Kim KT . Essential role of 3′-untranslated region-mediated mRNA decay in circadian oscillations of mouse Period3 mRNA. J Biol Chem 2006; 281: 19100–19106.
Acknowledgements
This work was supported by grants from the National Research Foundation of Korea (NRF) (Nos. 20110027957, 20110031234, 20120005830, and 20110031517), the Brain Korea 21 program, and the World Class University program (R31-10105) funded by the Korean Ministry of Education, Science, and Technology (MEST).
D-YK and K-TK designed research; D-YK, WK, K-HL, S-HK, H-RL, H-JK, YJ, and J-HC performed experiments; D-YK, WK, K-HL, and K-TK analyzed data; and D-YK, WK, and K-TK wrote the paper.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by M Oren
Author contributions
D-YK and K-TK designed research; D-YK, WK, K-HL, S-HK, H-RL, H-JK, YJ, and J-HC performed experiments; D-YK, WK, K-HL, and K-TK analyzed data; and D-YK, WK, and K-TK wrote the paper.
Supplementary information accompanies the paper on Cell Death and Differentiation website
Supplementary information
Rights and permissions
About this article
Cite this article
Kim, DY., Kim, W., Lee, KH. et al. hnRNP Q regulates translation of p53 in normal and stress conditions. Cell Death Differ 20, 226–234 (2013). https://doi.org/10.1038/cdd.2012.109
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cdd.2012.109
Keywords
This article is cited by
-
DAP5 increases axonal outgrowth of hippocampal neurons by enhancing the cap-independent translation of DSCR1.4 mRNA
Cell Death & Disease (2019)
-
Genome-wide CRISPR screen identifies ELP5 as a determinant of gemcitabine sensitivity in gallbladder cancer
Nature Communications (2019)
-
The EGF/hnRNP Q1 axis is involved in tumorigenesis via the regulation of cell cycle-related genes
Experimental & Molecular Medicine (2018)
-
Translational upregulation of Aurora-A by hnRNP Q1 contributes to cell proliferation and tumorigenesis in colorectal cancer
Cell Death & Disease (2017)
-
The Six1 oncoprotein downregulates p53 via concomitant regulation of RPL26 and microRNA-27a-3p
Nature Communications (2015)