Abstract
In most multicellular organisms, the decision to undergo programmed cell death in response to cellular damage or developmental cues is typically transmitted through mitochondria. It has been suggested that an exception is the apoptotic pathway of Drosophila melanogaster, in which the role of mitochondria remains unclear. Although IAP antagonists in Drosophila such as Reaper, Hid and Grim may induce cell death without mitochondrial membrane permeabilization, it is surprising that all three localize to mitochondria. Moreover, induction of Reaper and Hid appears to result in mitochondrial fragmentation during Drosophila cell death. Most importantly, disruption of mitochondrial fission can inhibit Reaper and Hid-induced cell death, suggesting that alterations in mitochondrial dynamics can modulate cell death in fly cells. We report here that Drosophila Reaper can induce mitochondrial fragmentation by binding to and inhibiting the pro-fusion protein MFN2 and its Drosophila counterpart dMFN/Marf. Our in vitro and in vivo analyses reveal that dMFN overexpression can inhibit cell death induced by Reaper or γ-irradiation. In addition, knockdown of dMFN causes a striking loss of adult wing tissue and significant apoptosis in the developing wing discs. Our findings are consistent with a growing body of work describing a role for mitochondrial fission and fusion machinery in the decision of cells to die.
Similar content being viewed by others
Main
The role of mitochondria in apoptosis has been defined clearly in vertebrates, but has remained elusive in lower eukaryotes such as Caenorhabditis elegans and Drosophila melanogaster.1 Countless studies have shown that the release of cytochrome c from mitochondria is a critical step in vertebrate apoptosis and is regulated by the Bcl-2 family.2 In contrast, release of cytochrome c appears to be a dispensable step in Drosophila cell death,3, 4, 5, 6 with Bcl-2 family members having a secondary role at most.7, 8 Conversely, other studies have suggested that cytochrome c release has a role, although limited, in Drosophila cell death.9 In addition, other studies suggest that Drosophila mitochondria house apoptosis-inducing proteins that are critical for apoptosis initiation.10, 11, 12 It is therefore likely that mitochondria have at least a modulatory role in fly cell death, but it is unclear whether they are integrally involved in the decision of fly cells to undergo programmed cell death.13
Drosophila apoptosis is regulated primarily by inhibitor of apoptosis proteins (IAPs). Apoptosis in the fly is induced by inhibiting the Drosophila IAP, DIAP-1, which blocks the activity of caspases in virtually all somatic cells. In turn, DIAP-1 is inhibited by a family of antagonists that include Reaper, Hid, Grim, Sickle and Jafrac (collectively known as RHG proteins). These proteins prevent DIAP-1 from binding to caspases, thereby inducing cell death.14 Although RHG proteins can inhibit DIAP-1 in vitro through competitive inhibition,15 there is also evidence demonstrating that some members of this family of proteins can induce DIAP1 auto-ubiquitylation and degradation.16, 17, 18
The IAP antagonists Reaper, Hid and Grim localize to outer mitochondrial membranes.3, 19, 20, 21, 22, 23 We have shown that mitochondrial localization is essential for Reaper's ability to induce DIAP-1 ubiquitylation.23 Moreover, vertebrate mitochondria have been shown to release cytochrome c in response to Reaper.24 This induction of cytochrome c release is independent of caspases and can be inhibited with Bcl-2. In Drosophila cells, cytochrome c is released on Reaper expression, and this release requires both Reaper and caspase activity.2 Recent data also suggest that Drosophila Omi (dOmi), another IAP antagonist, localizes to the intermembrane space of mitochondria and has an important role in cell death.10, 11, 12 Importantly, proteins that regulate mitochondrial morphology can also regulate IAP-antagonist-induced apoptosis in the fly.3, 25 Although there has been some controversy regarding the requirement (or lack thereof) for mitochondrial fission in mitochondrial cytochrome c release, a number of reports have established a link between mitochondrial morphology and apoptosis initiation in vertebrates.26, 27 In particular, proteins that regulate mitochondrial dynamics, such as Mfn2 and Drp1 have been implicated in regulation of mitochondrial steps of apoptosis in mammalian cells. Conversely, Bax and Bak, the two principle multidomain proapoptotic Bcl-2 family members, have been shown to localize to mitochondrial fission sites and induce mitochondrial fragmentation; this is a hallmark of cell death occurring before cytochrome c release.27
Although the role of mitochondrial dynamics in programmed cell death has been studied a great deal in vertebrates, its importance has only recently been established in lower eukaryotes.3, 25, 28, 29 For instance, the yeast homolog of Drp1, Dmn1, induces mitochondrial fission during yeast cell death. Inhibition of mitochondrial fission in C. elegans leads to a decrease in the number of cells eliminated by cell death during worm development.29 In addition, two reports have demonstrated that disruption of mitochondrial dynamics in Drosophila has considerable effects on Drosophila cell death.3, 25
Given the likely importance of mitochondrial dynamics in the regulation of apoptosis, we assessed how mitochondrial RHG proteins in Drosophila might affect mitochondrial fusion/fission. In addition, we examined how members of the pro-fusion mitofusin family might affect cell death in the fly. Two key homologs of Mfn2 and Mfn1 can be found in Drosophila: Fuzzy Onions (Fzo) and dMFN (MARF).30 Disruption of the Fzo gene was shown to render flies sterile because of a defect in spermatid development. This phenotype was associated with severe alterations in spermatid mitochondrial morphology. In contrast to Fzo, which appears to be restricted primarily to the testes, dMFN is expressed ubiquitously.30 We report here that Drosophila Reaper can interact with MFN2 through its hydrophobic GH3 domain, which has been shown to be critical for dimerization and mitochondrial localization.21, 31 Moreover, we provide evidence to suggest that Reaper induces mitochondrial fragmentation by inhibiting pro-fusion proteins and that overexpression of dMFN to trigger hyperfusion of mitochondria inhibits apoptosis in the fly. Knockdown of dMFN leads to significant apoptosis induction and remarkable loss of adult wing tissue. These findings reveal that interplay between IAP antagonists and the mitochondrial fusion machinery help to determine whether Drosophila cells will live or die.
Results
Reaper induces mitochondrial fragmentation in Hela cells
Abdelwahid et al.3 and Goyal et al.25 recently reported that Reaper can induce mitochondrial fragmentation when overexpressed in Drosophila cells. However, given the small mitochondrial size, the non-adherent properties of commonly used Drosophila cell lines and the robust toxicity of Reaper in those cells, it was difficult to follow-up on these observations. Therefore, we wished to determine whether the effects of Reaper on mitochondrial dynamics could be examined in a more technically amenable system. Accordingly, we expressed Reaper in Hela cells in which mitochondrial morphology can be easily assessed using confocal microscopy. Hela cells are also relatively resistant to Reaper-induced cytochrome c release and apoptosis. Hela cells were transfected with an N-terminal Flag-tagged Reaper or a mutant variant of Reaper, lacking the GH3 domain, responsible for mitochondrial localization (ReaperΔGH3). Cells were subsequently stained with Flag antibody to visualize Reaper and cytochrome c antibody to visualize mitochondria. Mitochondria of transfected cells were scored for the amount of fragmented versus elongated mitochondria present in each cell. As shown in Figure 1a, mitochondria in Flag-Reaper-transfected cells appeared to be smaller and less interconnected, whereas cells transfected with Flag-ReaperΔGH3 showed elongated mitochondria similar to those seen in untransfected cells. Figure 1b shows quantitation of fragmentation in cells staining positive for Flag antibody.
Reaper induces mitochondrial fragmentation in Hela cells. (a) Hela cells were transfected with pCDNA3-Flag-Reaper and pCDNA3-Flag-ReaperΔGH3. Anti-Flag staining is shown on the left and anti-cytochrome c staining is shown on the right. Flag-Reaper has a punctate pattern (top), whereas ΔGH3 is more diffuse. (b) Quantitation of transfected cells with fragmented mitochondria. Data are presented as a percentage of total cells with fragmented mitochondria and is a representation of three experiments. Error bars are the S.E.M.
Reaper forms mitochondrial foci
Previous studies of Reaper localization have demonstrated that Reaper associates with the outer mitochondrial membrane in a GH3-dependent manner using biochemical fractionation in cell-free extract and fluorescence microscopy in Drosophila S2 cells.3, 21, 23 When visualized in Hela cells, Reaper appears to localize to foci associated with mitochondria rather than being distributed over the entire mitochondrial membrane. Figure 2a shows Hela cells transfected with pEBB-REAPER-Flag and stained with anti-cytochrome c and anti-Flag antibodies. The pattern of fluorescence indicates that Reaper accumulated at foci on mitochondria. It is important to note that for these experiments, Reaper was tagged on the C-terminus, as mitochondrial localization of the C-terminal fusion is more readily visualized than the N-terminally tagged protein, which stains prominently in the cytosol as well. The C-terminally tagged protein was not used in other experiments (e.g., to assess phenotypic consequences of expression) because its expression level was quite low, likely due to its interaction with IAPs and subsequent targeting for degradation. Figure 2b shows a similar pattern for GFP-Reaper when expressed in Drosophila S2 cells stained with Mitotracker red. These mitochondrial foci have a similar appearance to foci formed by mammalian Bax and Bak.27 Interestingly, in our original characterization of Reaper localization, we found that Reaper localized to only a subset of mitochondria.3, 21, 23 These better-resolved images suggest that the subsets were actually those in which foci were formed. In addition, these images show that Reaper is closely juxtaposed to, rather than superimposed on, the mitochondrial staining. This is consistent with Reaper foci on the outer mitochondrial membrane, adjacent to Mitotracker staining in the matrix. In addition, in Supplementary Figure 1, we show that Reaper foci do not appear to contain Bax. This was determined by co-staining Reaper-transfected cells with antibodies directed to Bax or directed to the active conformation of Bax. We found that Bax did not colocalize with Reaper and that active Bax could not be detected in cells displaying Reaper foci. Thus, although these foci are similar in appearance to those formed by Bax, they are likely distinct.
Reaper localizes to sub-mitochondrial foci. (a) Hela cells were transfected with pEBB-REAPER-Flag for 18 h. They were fixed and then stained with Flag antibody (shown in red) and cytochrome c antibody (shown in green). (b) S2 cells were transfected with pMT-ReaperGFP for 12 h and stained with MTKR. Cells were visualized live. Mitotracker is shown in red and Reaper-GFP is shown in green
Reaper interacts with MFN2
To begin to address the mechanism of Reaper-induced mitochondrial fragmentation, Reaper was co-expressed with regulators of mitochondrial fission and fusion. Flag-tagged Reaper was expressed with GFP-Drp1, GFP-MFN2 and a dominant-negative mutant of the pro-fission protein Drp1, GFP-Drp1K38A. Surprisingly, we found that co-expression with GFP-MFN2 markedly altered Reaper's distribution within the cell (Figure 3). When expressed with Mito-GFP, GFP-Drp1 or GFP-Drp1K38A, Reaper distributed throughout the cytosol and to mitochondrial foci. In contrast, when cells overexpressed GFP-MFN2 and Flag-Reaper, the two proteins colocalized in clusters, consistent with the hyperfused and condensed mitochondrial network often seen in cells overexpressing mitofusins. It appears that overexpression of GFP-MFN2 could promote a redistribution of Flag-Reaper from the cytosol and mitochondrial foci to a completely mitochondrial localization. This is not simply a result of mitochondrial condensation, as both GFP-DRP1K38A expression in Hela cells and Drp1 knockdown with RNAi in Drosophila S2 cells (RJK and KW, unpublished) results in mitochondrial condensation without markedly increasing Reaper localization to mitochondria. This provides strong evidence that Reaper and MFN2 interact in intact cells and that this interaction can occur at the mitochondria. Previous work has shown that Bax and Bak form foci that contain MFN2 and Drp1,32 suggesting that Reaper might potentially operate by a similar mechanism (though in molecularly distinct focal locations, as described above).
MFN2 overexpression enhances localization of reaper to mitochondria. Hela cells were co-transfected with pCDNA3-Flag-Reaper and Mito-GFP (a), GFP-Drp1 (b), GFP-Drp1K38A (c) or GFP-MFN2 (d). Reaper Flag is shown on the left column. Indicated GFP fusion proteins are shown in the middle column. Merged images are shown in the right column where Flag-Reaper is red and GFP fusion proteins are green. Colocalization between GFP-MFN2 and Flag-Reaper can be seen in 35% of co-transfected cells
We had previously reported that a synthetic peptide encompassing the majority of the Reaper sequence could target Reaper to mitochondria and induce mitochondrial cytochrome c release in a reconstitution system derived from Xenopus egg extract.21, 24 In these extracts, we also found that Reaper could interact physically with MFN2. In particular, a biotinylated peptide corresponding to amino acids 16–65 of Reaper was incubated in Xenopus egg extract and precipitated with streptavidin sepharose beads. Xenopus MFN2 was readily detected with an antibody raised to Human MFN2. As seen in Figure 4a, biotinylated Reaper peptide co-precipitated with endogenous Xenopus MFN2. We found that removal of the GH3 domain prevented Reaper from interacting with MFN2, which initially suggested simply that mitochondrial localization was critical for interaction. Interestingly, a peptide corresponding to the GH3 domain alone was able to precipitate MFN2, suggesting that the GH3 domain might be sufficient for binding of Reaper to MFN2 at the mitochondria. To confirm our binding experiments, His-tagged recombinant human MFN2 was incubated in Xenopus egg extract treated with biotinylated Reaper peptide (Figure 4c) or full-length Reaper peptide (data not shown). His-MFN2 was precipitated using nickel beads. The western blot in Figure 4c shows that Reaper is found in the His-MFN2 precipitate.
Reaper binds to MFN2. (a) Biotinylated Reaper-16-65, (b) 16-65ΔGH3 or GH3 domain were incubated in Xenopus egg extract and pulled down with streptavidin sepharose. The precipitate was then analyzed by western blotting using MFN2 antibody. Control (Ctrl) lanes show precipitations with free biotin bound to streptavidin beads. (c) Purified His-tagged MFN2 was bound to nickel beads and incubated in Xenopus egg extracts treated with Reaper-16–65. Control lanes show pull downs with nickel beads bound to free histidine
Interaction with MFN2 requires the GH3 domain of Reaper and is required for mitochondrial fragmentation
As seen in Figure 1, deletion of the GH3 domain prevented Reaper from inducing mitochondrial fragmentation. In addition, it is clear from Figure 4 that the GH3 domain was sufficient for interaction with MFN2. As shown in Figure 5, we also found that only Reaper molecules lacking the GH3 domain were unable to colocalize with MFN2 (Figures 5a–e and 6a). As seen in Figures 5b and c, the first 30 and final 23 amino acids of Reaper were not required for binding, as deletions either containing amino acids 31–65 or containing 1–42 readily colocalized with GFP-MFN2. In contrast, Figures 5d and e show that mutants either containing amino acids 39–65 or containing 1–33 (i.e., truncated at the N- and C-terminal ends of the GH3 domain) were unable to colocalize with GFP-MFN2. In Figure 6a, we summarize the results from these deletions and others, indicating that only mutations interfering with the GH3 domain lacked the ability to bind to MFN2. In addition, the ability of these deletion mutants to bind to MFN2 correlated well with their ability to induce mitochondrial fragmentation (Figure 6b). These findings suggest that the GH3 domain is required for both MFN2 binding and mitochondrial fragmentation.
The GH3 domain of Reaper is required for MFN2 interaction. (a–e) Colocalization of reaper deletion mutants with GFP-MFN2 is shown. Hela cells were cotransfected with GFP-MFN2 and (a) Flag-Reaper, (b) Flag-Reaper 31–65, (c) Flag-Reaper 1–42, (d) Flag-Reaper 39–65 or (e) Flag-Reaper–33 and stained with anti-Flag antibody and an Alexa 594 secondary. (f and g) Cells were transfected with (f) Flag-ReaperΔGH3-ActA or (g) Flag-Reaper-ActA and stained with cytochrome c antibody to visualize mitochondria
The GH3 domain of Reaper is required for mitochondrial fragmentation. (a) Summary of data of deletion mutants and their ability to colocalize with GFP-MFN2. The GH3 domain is highlighted in red. (b) Mitochondrial fragmentation induced by deletion mutants of Reaper. Mitochondrial morphology was assessed in cells expressing Flag-Reaper, Flag-Reaper 1–33, Flag-Reaper 1–42, Flag-Reaper 31–65, Flag-Reaper 39–65, Flag-ReaperΔGH3-ActA or Flag-RPR-ActA. Error bars represent the S.E.M.
One possibility is that the GH3 domain simply confers mitochondrial localization, which is required for both fragmentation and binding. To test this, we generated mutants of Reaper and ReaperΔGH3, in which mitochondrial localization was achieved using the 27 C-terminal amino acids from the Actin assembly-inducing protein from Listeria (ActA), which comprise its mitochondrial tail-anchoring domain.33 It is apparent from Figures 5f and g that both Reaper-ActA and ReaperΔGH3-ActA readily localize to mitochondria. From the images in Figures 5f and g and the quantitation in Figure 6b, it is clear that mitochondrial fragmentation occurred readily in cells transfected with Reaper-ActA, but not in cells transfected with ReaperΔGH3-ActA. Taken together, these data demonstrate that the GH3 domain is critical for binding to MFN2 and that this interaction is likely responsible for Reaper-induced mitochondrial fragmentation.
Reaper interacts with the Drosophila mitofusin dMFN
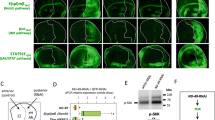
To demonstrate that this interaction occurs in Drosophila, we first cloned the ubiquitously expressed mitofusin, dMFN (FlyBase FBgn0029870 Marf). Although there are two Drosophila mitofusins, dMFN was used in for these experiments, as dFzo1 is expressed primarily in the male germ line and dMFN is ubiquitous.30 As seen in Figure 7a, GFP-dMFN largely colocalized with cytochrome c. GFP-dMFN is shown in green and cytochrome c is shown in red.
Reaper interacts with dMFN in S2 cells. (a) S2 cells were transfected with GFP-tagged dMFN. Cells were fixed and then stained with anti-cytochrome c antibody. Cytochrome c is shown in red and GFP-dMFN is shown in green. (b) dMFN overexpression enhances Reaper localization to mitochondria. S2 cells were transfected with Reaper-GFP and vector control or Myc-dMFN. Cells were fixed and then stained for cytochrome c. GFP and Reaper-GFP are shown in green and cytochrome c is shown in red in the merged image. (c) Biotinylated Reaper-16–65 was incubated in S2 cell extracts and precipitated with avidin sepharose. Proteins eluted from the Reaper-16–65-avidin beads were then analyzed by SDS-PAGE. The gels were then analyzed by western blotting with anti-hMFN2 antibody
The results shown in Figures 3 and 4 suggested that human MFN2 could interact with Drosophila Reaper. To confirm that this protein interaction occurs in Drosophila, we tested to see whether dMFN could also enhance localization of Reaper to mitochondria. Accordingly, Drosophila S2 cells were co-transfected with pAMW-dMFN and GFP-Reaper under the copper-inducible metallothionine promoter for 24 h. Cells were then treated with copper to induce GFP-Reaper expression. The cells were subsequently fixed and stained with cytochrome c antibody. It is important to note that previous work has shown that Reaper expression induces cytochrome c release.3 In these experiments, cells were fixed at an early time point when little cytochrome c release is seen. Figure 7b shows that expression of dMFN altered the distribution of GFP-Reaper. When co-transfected with a vector control, Reaper distributed to the cytosol and to a punctate pattern, whereas GFP-Reaper accumulated predominantly on mitochondria when co-expressed with dMFN. This morphology was seen in approximately 40% of the cells containing GFP-Reaper. On dMFN overexpression, cytochrome c staining and GFP-Reaper appeared as a concentrated mass in the center of the cell. The unusual mitochondrial morphology is a consequence of increased mitochondrial fusion, and is also seen on drp-1 knockdown.13 Cells have been outlined with a white line to indicate the approximate location of the plasma membrane. In addition to colocalization, biotinylated Reaper 16–65 peptide bound to streptavidin beads, as previously performed in Xenopus cell lysates, was able to retrieve dMFN from S2 cell lysates (Figure 7c). Thus, Drosophila Reaper interacts with dMFN as well as human and Xenopus MFN2.
Reaper inhibits mitochondrial fusion
To clarify the mechanism of mitochondrial fragmentation shown in Figure 1, we assessed Reaper's ability to promote mitochondrial fragmentation using a mitochondrial photoactivatable GFP (mito-PA-GFP).34 In theory, Reaper-induced changes in mitochondrial length could have resulted from enhanced fragmentation (e.g., stimulation of Drp1), inhibited fusion, or both. Given the interaction of Reaper with MFN2, we speculated that Reaper could function as an MFN2 inhibitor to prevent mitochondrial fusion. To test this, Hela cells were co-transfected with mito-PA-GFP and pCDNA3, Flag-Reaper or a known inhibitor of mitochondrial fusion, vMIA.35 A small number of mitochondria were then photoactivated in a region of interest (ROI) using the UV laser on a Leica SP5 confocal (Leica Microsystems, Inc., Buffalo Grove, IL, USA). The amount of activated GFP was then measured over time in the activated ROI. In cells with actively fusing mitochondria, the mito-PA-GFP diffuses throughout the cell, whereas non-fusing mitochondria will maintain their levels of mito-PA-GFP over time. Time-lapse videos are shown in Supplementary data. Figure 8a shows images of cells at the initial time point after photoactivation and 20 min post-activation. As shown in Figure 8b, mito-PA-GFP diffused quickly in cells co-transfected with pCDNA3, whereas mitochondria from cells transfected with Reaper or vMIA showed constant levels of GFP over time. These data strongly suggest that mitochondrial fusion is impaired by Reaper.
Reaper inhibits mitochondria fusion. Hela cells were cotransfected with mito-PA-GFP and pCDNA3, pCDNA3-Myc-vMIA or pCNA3-Flag-Reaper and incubated for 18 h. Cells were then visualized on a heated stage by confocal microscopy. To activate the mito-PA-GFP, regions of interest containing mitochondria were illuminated with a UV laser for 3 s. The UV illuminated region was then measured for GFP fluorescence every 30 s for 20 min. (a) Representative images from cells transfected with either pCDNA3 or pCDNA3-FL-Reaper at time 0 following UV illumination or at 20 min. (b) Time course of GFP intensity in initial region of interest following UV illumination in cells transfected with pCDNA3, pCDNA3-FL-Reaper or pCDNA3-Myc-vMIA. Data are presented as a percentage of the initial fluorescence in the UV illuminated region of interest. Data is representing mitochondria from two regions of interest in 10–15 cells per transfection. Error bars represent the S.E.M.
dMFN inhibits cell death in Drosophila
Given its biochemical interactions with Reaper, we wished to assess the potential for dMFN to modulate Reaper-induced cell death. Accordingly, S2 cells were co-transfected with pMT-Reaper (inducible with copper sulfate) and Myc-tagged dMFN under the control of an actin promoter (pAMW-dMFN). Following transfection, the cells were treated with copper sulfate for 3 or 6 h and assayed for apoptotic morphology. As seen in Figure 9a, S2 cells transfected with pAMW-dMFN were less susceptible to apoptosis induced by Reaper. This protection from Reaper-induced cell death was not because of any influence of dMFN on the ability of Reaper to induce DIAP1 degradation, as DIAP1 loss induced by etoposide occurred normally in the presence of dMFN, as assessed either by western blot of tagged DIAP1 (Supplementary Figure 2A), or by antibody staining of endogenous DIAP1 (data not shown). Moreover, the presence of dMFN did not appear to affect the interaction between DIAP1 and Reaper (Supplementary Figure 2A).
dMFN inhibits Drosophila apoptosis. (a) S2 cells were transfected with pMT-Reaper-HA and pAC5.1 or pAC5.1-dMFN. Reaper-HA expression was induced for 3 or 6 h with CuSO4. Cell death was analyzed by counting cells with apoptotic morphology. (b) Wing discs were dissected from untreated or irradiated larvae expressing en-gal4 (control) or en-gal4 with UAS-dMFN. Discs were stained with an anti-Engrailed antibody and active caspase 3 antibody. Discs were also dissected from untreated larvae expressing en-gal4 with UAS-dMFN dsRNA and stained for engrailed and active caspase 3. (c) Quantitation of cells with active caspase 3 (CP3+) from B. (d) Wings from adult flies expressing engrailed-gal4 UAS-dMFN-RNAi or driver control
To assess the ability of dMFN to modulate apoptosis in vivo, we generated fly strains with a UAS-Myc-dMFN transgene and obtained flies with a UAS-dMFN RNAi transgene. Myc-dMFN or RNAi expression was driven using an engrailed-Gal4 (en-gal4) factor to target expression to the posterior compartment of the developing wing. To induce apoptosis without overexpressing Reaper, Drosophia larvae were γ-irradiated, and cell death was assayed in the wing disc at 4 h later by staining with an anti-active caspase 3 antibody (CP3+). Engrailed staining indicated the region of dMFN expression. As shown in Figure 9b, irradiated wing discs from larvae expressing Myc-dMFN had decreased apoptosis in the engrailed domain. Conversely, cells expressing dMFN RNAi appeared to have increased caspase activity in the absence of radiation, suggesting that loss of dMFN leads to the induction of apoptosis. Interestingly, increased apoptosis could also be seen in the anterior compartment of the en-gal4 UAS-dMFN RNAi wing disc, indicating a non-cell autonomous effect of dMFN knockdown. The reason for this non-autonomous effect remains to be explored. Quantitation of these data is shown in Figure 9c. As shown in Figure 9d, wings from adult flies expressing engrailed gal4 and dMFN RNAi exhibited extensive loss of posterior wing tissue. This is consistent with the increased apoptosis detected in wing discs (Figure 9b). Taken together, these experiments indicate that dMFN protects cells from Reaper-induced and irradiation-induced apoptosis, and that decreased mitochondrial fusion can lead to apoptosis.
Discussion
Cell death in Drosophila is largely governed by IAP antagonists that trigger caspase activation by neutralizing IAPs. Although removal of caspase inhibition is sufficient to induce apoptosis, it appears that other cellular processes can be modulated by IAP antagonists to control cell death.36 Although mitochondrial outer membrane permeabilization appears to be dispensable for cell death in worms and flies,37 the ability of various mitochondrial regulators to impinge on the cell death pathway suggests that mitochondria are nevertheless involved in the death decision.3, 25, 29 We report here that in mammalian cells, like in fly cells, expression of Reaper can induce mitochondrial fragmentation; we propose that this occurs through an inhibitory interaction with mitofusins. We demonstrate this interaction by pull-down experiments (Figures 4 and 7) and by the ability of both human MFN2 and Drosophila dMFN to enhance the accumulation of Reaper on mitochondria (Figures 3, 5 and 7). Moreover, we show that Reaper inhibits mitochondrial fusion (Figure 8) and that dMFN overexpression can inhibit apoptosis in Drosophila (Figure 9).
Although previous work has shown that Reaper localizes to mitochondria,21, 23 we show that Reaper localizes to sub-mitochondrial foci. It is possible that these foci correlate with sites of stalled mitochondrial fusion. Inhibition of mitochondrial fission, using either dominant-negative or null alleles of Drp-13, 25 or overexpression of dMFN (this study) suppresses apoptosis in fly tissues and in S2 cells. The importance of Drp-1 function for cell death and the anti-apoptotic effect of dMFN indicate that mitochondrial morphology has a role in cell death in the fly. More specifically, it appears that the mitochondrial network must break down for apoptosis to be completed efficiently. We also show that inhibition of fusion can result in apoptosis, suggesting that mitochondrial status is sensed by the cell to activate the apoptotic program.
The role of mitochondria in Drosophila and C. elegans cell death remains elusive. There is evidence to suggest that cytochrome c may not have a critical role in apoptosis in each organism, but in both Drosophila and C. elegans, mitochondria appear to be involved in some manner. It is possible that mitochondria provide a scaffold for cell death signaling molecules, or that proapoptotic molecules other than cytochrome c contribute to apoptosis. Alterations in mitochondrial dynamics might alter the availability of apoptotic regulators or their ability to interact. This hypothesis is supported by previous findings that indicate that mitochondrial localization is required for Reaper-induced DIAP1 degradation;23 however, from the data shown in Supplementary Figure 2, it appears that the central interaction between DIAP1 and Reaper is not notably affected by the presence of dMFN. In some fly tissues, a role for cytochrome c in developmentally important caspase activation has been demonstrated.9, 38 However, in the majority of tissues, including the wing discs tested in this study, it may well be that mitochondria act as a platform of cell death signaling that cannot function properly in the hyperfused state. The ability of mitochondrial morphology to impinge on cell death pathways in evolutionarily distant organisms suggests that mitochondria have a fundamental role in programmed cell death that extends beyond cytochrome c release.
The diversity of apoptotic regulators located either at the surface of or within mitochondria provides strong prima facie evidence for a broad role of mitochondria in apoptosis. It is not yet clear whether particular mitochondrial morphologies are required simply to support the functioning of these molecules and/or changes in mitochondrial morphology are sensed to determine cell death or survival. Nevertheless, the results presented here add to the growing body of evidence that the interplay between apoptotic regulators and mitochondrial morphology critically determines the cellular fate. Indeed it is possible that the fundamental roles that mitochondria have in apoptotic signaling have yet to be fully elucidated.
Materials and Methods
Vectors
The pEBB-Reaper-Flag vector was described previously.23 pMT-GFP-Reaper was generated by subcloning the RPR cDNA into a pMT-GFP vector that was described previously.23 pCDNA3-Flag-Reaper and deletion mutants were generated by first cloning Reaper cDNA or mutants into pENTR-3C (Invitrogen Corp., Carlsbad, CA, USA). Reaper-ActA and ReaperΔGH3-ActA were fully synthesized with appropriate restriction sites (Integrated DNA Technologies, Inc., Coralville, IA, USA). Using LR Clonase II (Invitrogen) recombinase, Reaper was cloned into pCDNA3-Flag-Dest, which generates protein with an N-terminal Flag tag. GFP-Drp1K38A and GFP-MFN2 were generated by cloning hDrp1K38A and hMFN2 into pEGFP-C1 (Clontech, Mountain View, CA, USA), pCDNA3-MFN2 was a generous gift from Richard Youle (NINDS, NIH, Bethesda, MD, USA). pDEST-17-MFN2 was generated by first sub-cloning MFN2 into pENTR-3C and then performing a LR clonase II (Invitrogen) recombination into pDEST-17 (Invitrogen). pAMW-dMFN, pAGW-dMFN and pTMW-dMFN were generated by first cloning dMFN cDNA (DGRC-RE04414) into pENTR-3C and then performing a recombination reaction into pAMW (actin promoter with N-terminal Myc tag), pAGW (actin promoter with N-terminal GFP tag) and pTMW (pUAST vector with N-terminal Myc tag). pEGFP-mito was derived from pdsRED2-Mito vector (Clontech). pEGFP-mito-PA was a generous gift from Richard Youle (NIH).
Immunofluoresence
Imaging of Reaper-Flag constructs in Hela cells was performed by plating Hela cells at 50% confluency on MatTek (MatTek Corp., Ashland, MA, USA) coverslip four-well chamber slides. Cells were allowed to adhere and were then transfected with the indicated constructs and incubated for 18 h. Cells were fixed with 4% formaldehyde and permeabilized using 0.25% Triton X-100 (Sigma-Aldrich, St Louis, MO, USA), in phosphate-buffered saline (PBS). Cells were washed with PBS and then incubated with PBS with 5% goat serum (Invitrogen) for 30 min. For visualizing EGFP and Flag, cells were incubated with mouse-anti-Flag antibody (Agilent Technologies, Inc., Santa Clara, CA, USA) at a dilution of 1 : 10 000 in PBS with 5% goat serum. Slides were washed three times with PBS and were incubated in 5% BSA for 1 h to reduce nonspecific anti-Flag staining. The cells were then incubated with Goat Anti-Mouse Alexa-594 (Invitrogen) diluted by 1 : 1000 in PBS with 5% goat serum. For visualizing the Flag tag and cytochrome c, cells were incubated with Rabbit-anti-Flag (Sigma) at a 1 : 5000 dilution and mouse-anti-cytochrome c (6H2.B4; BD Biosciences, Franklin Lakes, NJ, USA) at a 1 : 1000 dilution. Goat-anti-rabbit Alexa 488 and goat-anti-mouse Alexa 594 were used as secondary antibodies at dilutions of 1 : 1000.
Mitochondrial fusion experiments
Hela cells were transfected with mito-PA-GFP34 and pCDNA3 or pCDNA3-Flag-Reaper at a 1 : 8 ratio (mito-PA-GFP:pCDNA3) for 18 h using Fugene 6 (Roche, Basel, Switzerland). The cells were then visualized on a Leica SP5 confocal microscope. A small number of mitochondria were illuminated with a UV laser (405 nM) for 3 s. Images were then taken every 30 s for 20 min.
Binding experiments
Crude Xenopus interphase extracts were prepared as previously described.24 To precipitate endogenous Xenopus Mfn2, biotinylated Reaper16-65, Reaper16-65ΔGH3 and GH3 peptides were conjugated to streptavidin-sepharose beads (GE Healthcare, Waukesha, WI, USA); control streptavidin-sepharose beads were blocked in free biotin (64 μ M) for 1 h at 4°C. Control and peptite-conjugated beads were washed in egg lysis buffer (ELB: 250 mM sucrose, 2.5 mM MgCl2, 10 mM Hepes pH 7.7, 50 mM KCl and 1 mM DTT), blocked in 1% milk in ELB for 1 h at 4°C, and incubated in Xenopus egg extract containing 0.6% digitonin for 1 h at 4°C with agitation. Following incubation, the beads were washed three times in ELB, solubilized with SDS sample buffer, resolved by SDS-PAGE, transferred and immunoblotted using anti-Mfn2 anti-serum.
His-Mfn2 protein was purified from bacteria expressing pDEST17-Mfn2 using Ni-affinity chromatography (Ni-NTA agarose, Qiagen Inc., Valencia, CA, USA). Control nickel beads were pre-blocked in 125 mM histidine, and then both sets of beads (control nickel and Ni-bound His-Mfn2) were blocked in 1% casamino acids and washed in ELB before incubation with Reaper peptide. Control Ni-beads and Ni-bound His-Mfn2 beads were incubated in Xenopus egg extracts treated with DMSO vehicle or Reaper peptide (16 μ M). Following 1 h incubation at 4°C, the beads were washed three times in ELB before SDS-PAGE and immunoblotting.
Drosophila S2 cells were resuspended in a hypotonic buffer (20 mM Hepes-KOH pH 7.5, 10 mM KCl, 1.5 mM MgCl2, 1 mM EDTA, 1 mM EGTA, 250 mM sucrose, 1 mM DTT and with Complete Protease Inhibitors (Roche)) and allowed to swell on ice 1 h before lysis by sonication. Reaper16-65-conjugated and control beads were prepared as described above before incubation with cell lysates. Cleared cell lysates were incubated with beads for 2 h at 4°C. Beads were then washed three times in PBS with 300 mM NaCl before loading onto SDS-PAGE gels for immunoblotting.
Reaper-DIAP1 co-immunoprecipitations were performed by transfecting Drosophila Kc cells with pAW, pAGW-dMFN and pAWF-DIAP1 or pMT-RPR-HA, pAW and pAWF-DIAP1 or pMT-RPR-HA, pAGW-DIAP1 and pAWF-DIAP1. Cells were collected and resuspended in lysis buffer (50 mM Tris 7.5, 150 mM NaCl, 1% NP40 and 0.5% Na-Deoxycholate with complete protease inhibitor tablets). Lysates were spun down at 15000 r.p.m. for 10 min at 4°C in an Eppendorf microcentrifuge. Supernatant was added to 25 μl HA affinity matrix beads (Roche) and incubated at 4°C for 1 h. The beads were washed twice in lysis buffer and then incubated in 25 μl SDS sample buffer for 5 min at 95°C. The samples were then loaded on a 10–20% Criterion gel (Bio-Rad Laboratories, Hercules, CA, USA), transferred onto PVDF and probed for anti-HA, anti-GFP and anti-Flag.
In vivo analysis of dMFN in cell death
The en-gal4 strain was crossed to UAS-dMFN or UAS-dMFN RNAi (VDRC, Vienna, Austria) at 22°C. Larvae were γ-irradiated and dissected for staining 4 h later.39 Discs were stained with anti-active caspase 3 (Cell Signaling Technology Inc., Danvers, MA, USA; 1 : 50) and anti-Engrailed antibody (Developmental Studies Hybridoma Bank, Iowa City, IA, USA; 1 : 2). Secondary antibodies were Alexa 488-conjugated donkey anti-mouse and Alexa 647-conjugated goat anti-rabbit IgG (Invitrogen; 1 : 200). Adult wings were dissected and mounted in Euparal (BioQuip, Rancho Dominguez, CA, USA). Three wing discs for each genotype were used for quantification of anti-active CP3 staining, which recognizes several cell death-related epitopes in flies.40 CP3+ cell numbers were obtained from four randomly chosen × 40, zoom 4, en+ fields from en-gal4 UAS-dMFN or en-gal4 alone. Means of the numbers of active caspase-3-positive cells per field in each disc were obtained and statistically analyzed, (mean±S.D.). Two different genomic insertions of UAS-dMFN produced similar results.
Analysis of dMFN on cell death in culture
Drosophila S2 cells were transfected with 3 μg pMT-Reaper-HA, 0.3 μg of pIE-Ds-Red and either 3 μg pAC 5.1 or pAC 5.1 dMFN vector using Cellfectin transfection reagent (Invitrogen). After incubation for 3 days, Reaper expression was induced by washing the cells with serum-free media and then adding fresh complete media with CuSO4 (210 μ M) and photographed after 3 or 6 h using fluorescence microscopy. The images were then scored for the percent of transfected cells with apoptotic morphology.
Abbreviations
- AP:
-
inhibitor of apoptosis protein
- RPR:
-
reaper
- DIAP1:
-
Drosophila inhibitor of apoptosis protein 1
- PA-GFP:
-
photoactivatable GFP
- ROI:
-
region of interest
- MOMP:
-
mitochondrial outer membrane permeabilization
- ELB:
-
egg lysis buffer
- FL:
-
flag
- Cp3+:
-
active caspase 3 positive
References
Abdelwahid E, Rolland S, Teng X, Conradt B, Hardwick JM, White K . Mitochondrial involvement in cell death of non-mammalian eukaryotes. Biochim Biophys Acta 2011; 1813: 597–607.
Cory S, Adams JM . The Bcl2 family: regulators of the cellular life-or-death switch. Nat Rev Cancer 2002; 2: 647–656.
Abdelwahid E, Yokokura T, Krieser RJ, Balasundaram S, Fowle WH, White K . Mitochondrial disruption in Drosophila apoptosis. Dev Cell 2007; 12: 793–806.
Dorstyn L, Mills K, Lazebnik Y, Kumar S . The two cytochrome c species, DC3 and DC4, are not required for caspase activation and apoptosis in Drosophila cells. J Cell Biol 2004; 167: 405–410.
Dorstyn L, Read S, Cakouros D, Huh JR, Hay BA, Kumar S . The role of cytochrome c in caspase activation in Drosophila melanogaster cells. J Cell Biol 2002; 156: 1089–1098.
Zimmermann KC, Ricci JE, Droin NM, Green DR . The role of ARK in stress-induced apoptosis in Drosophila cells. J Cell Biol 2002; 156: 1077–1087.
Sevrioukov EA, Burr J, Huang EW, Assi HH, Monserrate JP, Purves DC et al. Drosophila Bcl-2 proteins participate in stress-induced apoptosis, but are not required for normal development. Genesis 2007; 45: 184–193.
Galindo KA, Lu WJ, Park JH, Abrams JM . The Bax/Bak ortholog in Drosophila, Debcl, exerts limited control over programmed cell death. Development 2009; 136: 275–283.
Mendes CS, Arama E, Brown S, Scherr H, Srivastava M, Bergmann A et al. Cytochrome c-d regulates developmental apoptosis in the Drosophila retina. EMBO Rep 2006; 7: 933–939.
Khan FS, Fujioka M, Datta P, Fernandes-Alnemri T, Jaynes JB, Alnemri ES . The interaction of DIAP1 with dOmi/HtrA2 regulates cell death in Drosophila. Cell Death Differ 2008; 15: 1073–1083.
Igaki T, Suzuki Y, Tokushige N, Aonuma H, Takahashi R, Miura M . Evolution of mitochondrial cell death pathway: proapoptotic role of HtrA2/Omi in Drosophila. Biochem Biophys Res Commun 2007; 356: 993–997.
Challa M, Malladi S, Pellock BJ, Dresnek D, Varadarajan S, Yin YW et al. Drosophila Omi, a mitochondrial-localized IAP antagonist and proapoptotic serine protease. EMBO J 2007; 26: 3144–3156.
Krieser RJ, White K . Inside an enigma: do mitochondria contribute to cell death in Drosophila? Apoptosis 2009; 14: 961–968.
Steller H . Regulation of apoptosis in Drosophila. Cell Death Differ 2008; 15: 1132–1138.
Means JC, Muro I, Clem RJ . Lack of involvement of mitochondrial factors in caspase activation in a Drosophila cell-free system. Cell Death Differ 2006; 13: 1222–1234.
Holley CL, Olson MR, Colon-Ramos DA, Kornbluth S . Reaper eliminates IAP proteins through stimulated IAP degradation and generalized translational inhibition. Nat Cell Biol 2002; 4: 439–444.
Ryoo HD, Bergmann A, Gonen H, Ciechanover A, Steller H . Regulation of Drosophila IAP1 degradation and apoptosis by reaper and ubcD1. Nat Cell Biol 2002; 4: 432–438.
Yokokura T, Dresnek D, Huseinovic N, Lisi S, Abdelwahid E, Bangs P et al. Dissection of DIAP1 functional domains via a mutant replacement strategy. J Biol Chem 2004; 279: 52603–52612.
Claveria C, Albar JP, Serrano A, Buesa JM, Barbero JL, Martinez AC et al. Drosophila grim induces apoptosis in mammalian cells. Embo J 1998; 17: 7199–7208.
Claveria C, Caminero E, Martinez AC, Campuzano S, Torres M . GH3, a novel proapoptotic domain in Drosophila Grim, promotes a mitochondrial death pathway. EMBO J 2002; 21: 3327–3336.
Freel CD, Richardson DA, Thomenius MJ, Gan EC, Horn SR, Olson MR et al. Mitochondrial localization of Reaper to promote inhibitors of apoptosis protein degradation conferred by GH3 domain-lipid interactions. J Biol Chem 2008; 283: 367–379.
Haining WN, Carboy-Newcomb C, Wei CL, Steller H . The proapoptotic function of Drosophila Hid is conserved in mammalian cells. Proc Natl Acad Sci USA 1999; 96: 4936–4941.
Olson MR, Holley CL, Gan EC, Colon-Ramos DA, Kaplan B, Kornbluth S . A GH3-like domain in reaper is required for mitochondrial localization and induction of IAP degradation. J Biol Chem 2003; 278: 44758–44768.
Evans EK, Kuwana T, Strum SL, Smith JJ, Newmeyer DD, Kornbluth S . Reaper-induced apoptosis in a vertebrate system. EMBO J 1997; 16: 7372–7381.
Goyal G, Fell B, Sarin A, Youle RJ, Sriram V . Role of mitochondrial remodeling in programmed cell death in Drosophila melanogaster. Dev Cell 2007; 12: 807–816.
Sheridan C, Delivani P, Cullen SP, Martin SJ . Bax- or Bak-induced mitochondrial fission can be uncoupled from cytochrome c release. Mol Cell 2008; 31: 570–585.
Suen DF, Norris KL, Youle RJ . Mitochondrial dynamics and apoptosis. Genes Dev 2008; 22: 1577–1590.
Fannjiang Y, Cheng WC, Lee SJ, Qi B, Pevsner J, McCaffery JM et al. Mitochondrial fission proteins regulate programmed cell death in yeast. Genes Dev 2004; 18: 2785–2797.
Jagasia R, Grote P, Westermann B, Conradt B . DRP-1-mediated mitochondrial fragmentation during EGL-1-induced cell death in C. Nature 2005; 433: 754–760.
Hwa JJ, Hiller MA, Fuller MT, Santel A . Differential expression of the Drosophila mitofusin genes fuzzy onions (fzo) and dmfn. Mech Dev 2002; 116: 213–216.
Sandu C, Ryoo HD, Steller H . Drosophila IAP antagonists form multimeric complexes to promote cell death. J Cell Biol 2010; 190: 1039–1052.
Karbowski M, Lee YJ, Gaume B, Jeong SY, Frank S, Nechushtan A et al. Spatial and temporal association of Bax with mitochondrial fission sites, Drp1, and Mfn2 during apoptosis. J Cell Biol 2002; 159: 931–938.
Zhu W, Cowie A, Wasfy GW, Penn LZ, Leber B, Andrews DW . Bcl-2 mutants with restricted subcellular location reveal spatially distinct pathways for apoptosis in different cell types. EMBO J 1996; 15: 4130–4141.
Karbowski M, Arnoult D, Chen H, Chan DC, Smith CL, Youle RJ . Quantitation of mitochondrial dynamics by photolabeling of individual organelles shows that mitochondrial fusion is blocked during the Bax activation phase of apoptosis. J Cell Biol 2004; 164: 493–499.
Norris KL, Youle RJ . Cytomegalovirus proteins vMIA and m38.5 link mitochondrial morphogenesis to Bcl-2 family proteins. J Virol 2008; 82: 6232–6243.
Thomenius M, Kornbluth S . Multifunctional reaper: sixty-five amino acids of fury. Cell Death Differ 2006; 13: 1305–1309.
Oberst A, Bender C, Green DR . Living with death: the evolution of the mitochondrial pathway of apoptosis in animals. Cell Death Differ 2008; 15: 1139–1146.
Arama E, Bader M, Srivastava M, Bergmann A, Steller H . The two Drosophila cytochrome C proteins can function in both respiration and caspase activation. EMBO J 2006; 25: 232–243.
Moon NS, Frolov MV, Kwon EJ, Di Stefano L, Dimova DK, Morris EJ et al. Drosophila E2F1 has context-specific pro- and antiapoptotic properties during development. Dev Cell 2005; 9: 463–475.
Fan Y, Bergmann A . The cleaved-Caspase-3 antibody is a marker of Caspase-9-like DRONC activity in Drosophila. Cell Death Differ 2010; 17: 534–539.
Acknowledgements
This work was funded by NIH grants RO1 GM080333 (SK), GM55568 (KW), a supplement for collaborative research GM55568-S1 (KW and SAK), and an MGH Fund for Medical Discovery (EA).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by E Baehrecke
Supplementary Information accompanies the paper on Cell Death and Differentiation website
Rights and permissions
This work is licensed under the Creative Commons Attribution-NonCommercial-No Derivative Works 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Thomenius, M., Freel, C., Horn, S. et al. Mitochondrial fusion is regulated by Reaper to modulate Drosophila programmed cell death. Cell Death Differ 18, 1640–1650 (2011). https://doi.org/10.1038/cdd.2011.26
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cdd.2011.26
Keywords
This article is cited by
-
Mitochondrial function and nutrient sensing pathways in ageing: enhancing longevity through dietary interventions
Biogerontology (2022)
-
Apoptosis in Drosophila: which role for mitochondria?
Apoptosis (2016)
-
Screening of candidate proteins interacting with IE-2 of Bombyx mori nucleopolyhedrovirus
Molecular Biology Reports (2013)
-
Drosophila larvae lacking the bcl-2 gene, buffy, are sensitive to nutrient stress, maintain increased basal target of rapamycin (Tor) signaling and exhibit characteristics of altered basal energy metabolism
BMC Biology (2012)