Abstract
Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) induces selective apoptotic death of human cancer cells while sparing normal human cells. Although TRAIL holds great promise as a potential anticancer agent, some tumors develop resistance to TRAIL. Previously, we have shown that the activator protein 1 (AP-1) family member, c-Fos, is an important modulator of apoptosis. Although F- box protein 10 (FBXL10) has been implicated to regulate an AP-1 family protein, c-Jun, its role in mediating apoptotic pathways has not been previously investigated. Here, we report that FBXL10 is a transcriptional repressor of c-Fos and a target gene of nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB)-p65 in human cancers. We demonstrate that FBXL10 is an important anti-apoptotic molecule, which directly binds and represses c-Fos promoter in order for cancer cells to resist TRAIL-induced apoptosis. FBXL10 indirectly regulates c-FLIP(L) levels via c-Fos-dependent pathways. Silencing of FBXL10 sensitizes resistant cells to TRAIL, while, overexpression of FBXL10 represses TRAIL-induced apoptosis. Moreover, our results indicate that expression of FBXL10 functions via an NF-κB-dependent pathway, and TRAIL or proteasome inhibitors downregulate FBXL10 via inhibiting NF-κB signaling. Taken together, we find a novel functional role for FBXL10 as an anti-apoptotic molecule, and describe a new apoptotic-related pathway that involves NF-κB/FBXL10/c-Fos/c-FLIP. Therefore, silencing FBXL10 can help overcome resistant cancer cells for pro-apoptotic therapies.
Similar content being viewed by others
Main
Apoptotic pathways are commonly altered in neoplastic initiation and progression, making tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) an attractive agent, particularly since TRAIL induces death in cancer cells, but not normal cells. In order to maximize the pro-apoptotic effects of TRAIL, we have been investigating the molecular pathways that differentiate between TRAIL-resistant and TRAIL-sensitive cancer cells. Previously, we have shown that c-Fos and nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) are important regulators of TRAIL-induced apoptosis.1, 2, 3 We have demonstrated that c-FLIP(L) is necessary and sufficient to cause resistance to TRAIL in prostate cancer cells.4 We have shown that c-FLIP(L) is repressed by c-Fos, demonstrating that c-Fos, in addition to its well-known proto-oncogenic activity, also possesses a pro-apoptotic function by priming cancer cells to undergo apoptosis.2
NF-κB5, 6 is another important regulator of TRAIL-induced apoptosis, which widely regulates genes responsible for cell proliferation and survival. To regulate apoptotic activities, NF-κB regulates downstream anti-apoptotic genes such as c-FLIP,7 Bcl-xL, IAPs and XIAP.8
In order to better understand the molecular mechanisms that regulate resistance to TRAIL-induced apoptosis, we evaluated the role of F-box protein 10 (FBXL10), which is a substrate-recognition component of SCF ubiquitin–ligase complexes. FBXL10 has been known as a nucleolar protein that represses transcription of ribosomal RNA genes9 and a transcriptional repressor of the activator protein 1 (AP-1) family member, c-Jun.10 As we have found that the AP-1 family member, c-Fos, regulates TRAIL-induced apoptosis,2 we wished to determine whether FBXL10, a transcriptional repressor of one of the AP-1 family members, has any role in mediating apoptotic pathways.
We demonstrate, for the first time, that FBXL10 is a transcriptional repressor of c-Fos and a target gene of the apoptotic mediator, NF-κB-p65, in human cancers. We show that FBXL10 directly binds to the promoter region of c-Fos, and functions as a transcriptional repressor. On the other hand, FBXL10 is activated by NF-κB-p65 subunit. More importantly, we demonstrate that FBXL10 has an anti-apoptotic role and describe a novel apoptotic-related NF-κB/FBXL10/c-Fos/c-FLIP pathway. We suggest that silencing FBXL10 can help overcome resistant cancer cells for pro-apoptotic therapies.
Results
FBXL10 is downregulated by TRAIL in TRAIL-sensitive cancer cells whereas c-Fos expression is upregulated
Cancer cells have different degrees of sensitivity to TRAIL. Prostate cancer PC3 cells,2 kidney cancer A498 cells11 and breast cancer MDA-MB-231 cells12 are TRAIL sensitive, whereas prostate PC3TR and LNCaP cells are TRAIL resistant.4 We have shown that c-Fos is a pro-apoptotic agent and reactivated after treatment with TRAIL, thus repressing the activity of anti-apoptotic molecule c-FLIP(L).2 Recent studies have shown that FBXL10 is a repressor of c-Jun,10 an AP-1 family protein. Therefore, we investigated whether FBXL10 has a role in TRAIL-mediated apoptosis through regulating AP-1 family members.
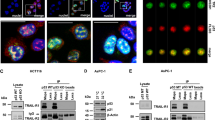
To test this hypothesis, first we treated the TRAIL-sensitive A498, PC3 and MDA-MB-231 cancer cells with TRAIL. Interestingly, TRAIL treatment led to a dramatic reduction in protein and mRNA levels of FBXL10 in all three different types of cancer cells, whereas c-Fos was upregulated, while expression of c-Jun was not affected (Figure 1a, Supplementary Figure S1A). However, in TRAIL-resistant PC3TR and LNCaP cancer cells, neither protein nor mRNA levels of FBXL10, c-Fos and c-Jun were affected by TRAIL (Figure 1b, Supplementary Figure S1B). Expression of FBXL11, a FBXL10 homologous protein, was not affected by TRAIL in either TRAIL-sensitive or TRAIL-resistant cells (Figure 1c). Immunofluorescence studies provided similar findings (Figure 1d). A498 kidney cancer cells treated with TRAIL showed a dose and time-dependent downregulation of FBXL10 and upregulation of c-Fos in protein (Figures 1e and f) and mRNA level (Supplementary Figure S1C, data not shown). These results suggest that downregulation of FBXL10 and upregulation of c-Fos are correlated.
Downregulation of FBXL10 and upregulation of c-Fos are correlated with TRAIL exposure in TRAIL-sensitive cells. (a–c) A498, PC3, MBA-MD-231 and LNCaP cells were treated with 50 ng/ml TRAIL for 8 h and immunoblot was performed (d) PC3 cells were treated as shown in (a–c) and immunofluorescence was assessed. (e) A498 cells were treated with TRAIL for 8 h (f) A498 cells were treated with 50 ng/ml TRAIL for the indicated times. (g and h) Representative fluorescent immunohistochemical analysis of orthotopically implanted human prostate cancer cells (PC3, LNCaP). Mice-bearing tumor xenografts were treated without (−) or with (+) Lexa, i.v. for 4 weeks (n=6). Original magnification, × 400. All in vitro experiments were repeated independently at least three times with similar results. All in vivo experiments were repeated independently twice. (Scale bar, 50 μm)
FBXL10 is inhibited in the orthotopic xenograft
To determine whether inhibition of FBXL10 that we observed in vitro is also found in in vivo models, we orthotopically implanted TRAIL-sensitive PC3 and TRAIL-resistant LNCaP cancer cells in the posterolateral lobe of the prostate of nude mice, respectively. Then, the mice-bearing PC3 or LNCaP tumors were treated with a TRAIL receptor 2 agonist antibody (lexatumumab (Lexa)). Immunohistochemistry showed that repression of FBXL10, concomitant with increased c-Fos was pronounced in PC3 xenografts after treatment, but not in LNCaP xenografts, while expression of c-Jun was not altered (Figures 1g and h). Altogether, these findings indicate that TRAIL inhibits FBXL10, yet upregulates c-Fos in TRAIL-sensitive tumors models, but not in the TRAIL-resistant models.
FBXL10 regulates transcription of c-Fos
Next, we evaluated whether FBXL10 regulates c-Fos-mediated transcription using reporter assays. Previously, we reported that c-Fos promoter activity is stimulated in response to TRAIL.2 Here, we found that the human c-Fos promoter was repressed when PC3 cells were co-transfected with FBXL10. TRAIL treatment potentiated c-Fos promoter activity, which was repressed with increasing FBXL10 concentration (Figure 2a). The specificity of c-Fos promoter repression was confirmed by using FBXL11 and FBXL10 CxxC mutant (Figure 2b).
FBXL10 represses c-Fos-mediated transcription. (a) PC3 cells were co-transfected with pc-Fos luciferase plasmids and FBXL10 plasmids with the weight ratio indicated for 24 h and then were treated with or without TRAIL for additional 8 h. Renilla luciferase was used as a positive control. Results are shown as the mean (bar) ± S.D. of at least three independent experiments. (b) FBXL10, FBXL11 or FBXL10CxxC mutation plasmids were co-transfected with pc-Fos luciferase plasmids with the weight ratio of 1 : 4 into PC3 cells for 24 h and then were treated with or without TRAIL for additional 8 h. (c) Schematic representation showing the human c-Fos promoter. FBXL10-BS, FBXL10 BS mutation (BSm) and their corresponding sequences are shown. (d) The pc-Fos luciferase plasmids and its mutant promoter constructs (c-Fos-promoter FBXL10 BSm) were co-transfected with FBXL10 plasmids into PC3 cells for 24 h. (e) In all, 8 μg of FBXL10, FBXL11 and control empty plasmids were co-transfected with 2 μg of the pc-Fos promoter luciferase reporter constructs into mice for 16 h using the hydrodynamic procedure. In vivo bioluminescence imaging was performed as described in ‘Materials and Methods section’. Data are representative of three independent experiments. All experiments were repeated independently at least three times with similar results
FBXL10 has been shown to target AP-1-binding sites (BSs),10 therefore, we used a bioinformatics program (TRANSFAC 4.0, BIOBASE, Wolfenbuttel, Germany) to predict possible FBXL10 BSs in the c-Fos promoter. We identified one putative FBXL10-BS, −300bp upstream of the c-Fos promoter transcription initiation site (Figure 2c). To further determine whether the site is functionally required for FBXL10 binding, we mutated the FBXL10-BS. The mutant showed reduced transcriptional activity and was not inhibited by FBXL10 (Figure 2d).
Hydrodynamic transfection is highly efficient in its ability to deliver foreign naked DNA into hepatocytes of live mice and mimic in vivo cellular conditions.13 Here, we introduced c-Fos promoter luciferase construct and FBXL10 expression construct into mice with hydrodynamic transfection, and found that only FBXL10 dramatically reduced c-Fos promoter activity in vivo (Figure 2e). Taken together, these results suggest that FBXL10 is a repressor of c-Fos in vitro and in vivo.
FBXL10 binds directly to c-Fos promoter
As mutation of the FBXL10-BS compromised the FBXL10-mediated inhibition (Figure 2d), we decided to determine their functional relevance. Using chromatin immunoprecipitation assay (ChIP) assay, we found that FBXL10 was associated with c-Fos promoter in both TRAIL-sensitive PC3 and -resistant PC3TR cells, and TRAIL treatment dramatically reduced the binding of FBXL10 to c-Fos promoter in PC3 cells, but not in PC3TR cells (Figure 3a). Next, we performed electrophoretic mobility shift assay (EMSA) assay using c-Fos promoter DNA (−322 to −261) containing the FBXL10-BS as a probe. We observed a super-shift in both PC3 and LNCaP cells in the presence of FBXL10 antibody. But TRAIL treatment abolished FBXL10 interaction with c-Fos promoter in PC3 cells, but not in LNCaP cells (Figure 3b). Finally, we observed that in vivo c-Fos promoter activity was reduced by FBXL10, not FBXL10 CxxCm with hydrodynamic transfection (Figure 3c). Collectively, we show that FBXL10 binds and represses the expression of c-Fos promoter activity.
FBXL10 binds directly to c-Fos promoter. (a) PC3 and PC3TR cells treated with TRAIL (50 ng/ml) for 8 h were subjected to ChIP assay using a FBXL10 antibody or IgG. (b) Nuclear extracts from PC3 or LNCaP cells treated with or without TRAIL were incubated with FBXL10BS probe in the presence or absence of FBXL10 antibody. Arrows indicate the super-shifted FBXL10 protein–DNA complexes. (c) In all, 8 μg of FBXL10 or FBXL10 CxxC mutation plasmids were co-transfected with 2 μg of the pc-Fos promoter reporter constructs into mice for 16 h using the hydrodynamic procedure. Data are representative of three independent experiments. All experiments were repeated independently at least three times with similar results
FBXL10 regulates sensitivity of TRAIL-mediated apoptosis
To assay the importance of FBXL10 in regulating sensitivity to TRAIL, we either knocked-down FBXL10 in TRAIL-resistant LNCaP cells with RNAi or stably overexpressed FBXL10 in TRAIL-sensitive PC3 cells and tested for changes in TRAIL sensitivity. Downregulation of FBXL10, accompanied a concomitant increase in c-Fos-sensitized LNCaP cells to TRAIL (Figures 4a–c, Supplementary Figure S2A). Of note, reduced FBXL10 did not promote cell death and merely primed TRAIL-resistant cells to undergo apoptosis after TRAIL treatment, which is consistent with earlier studies.2, 14 In contrast, overexpression of FBXL10 in PC3 cells compromised reactivation of c-Fos and significantly protected PC3 cells from TRAIL (Figures 4d–f, Supplementary Figure S2B). These results suggest that FBXL10 may have an important anti-apoptotic role.
FBXL10 regulates sensitivity/resistance of cancer cells to TRAIL. (a–c) LNCaP cells transfected with FBXL10 or control siRNA for 40 h were treated with TRAIL (50 ng/ml) for additional 8 h. (a) immunoblot, (b) MTT cell viability and (c) Annexin V assays. Results are shown as the mean (bar) ± S.D. of at least three independent experiments. (d–f) PC3 cells stably transfected with FBXL10 or control plasmids were treated with TRAIL (50 ng/ml) for 8 h: (d) immunoblot, (e) MTT cell viability and (f) Annexin V assay. (g and h) LNCaP cells transfected with FBXL10, c-Fos or control siRNA were treated with TRAIL (50 ng/ml): (g) immunoblot and (h) MTT cell viability assay. (i and j) PC3TR cells were stably transfected with shFBXL10 or control plasmids to generate stable cells. The stable cells transfected with c-Fos siRNA or control siRNA for 40 h were treated with TRAIL (50 ng/ml) for additional 8 h: (i) immunoblot and (j) MTT cell viability. (k and l) Stable PC3/FBXL10 cells transfected with FLIP siRNA or control siRNA for 40 h were treated with TRAIL (50 ng/ml) for additional 8 h: (k) immunoblot and (l) MTT cell viability assay. (m and n) Stable PC3TR/shFBXL10 cells transfected with pcDNA3-c-FLIP or control pcDNA3 for 40 h were treated with TRAIL (50 ng/ml) for additional 8 h: (m) Immunoblot and (n) MTT cell viability assay. The data represent means of average determinants ± S.E.M. **P<0.01; ***P<0.001
Similarly, PC3 xenografts that were treated with Lexa resulted in decreased tumor size and higher apoptotic activities as compared with PC3/FBXL10 xenografts (Figures 5a–d). We also found reduced c-Fos levels in the resistant PC3/FBXL10 xenografts (Figure 5e). In contrast, repression of FBXL10 in PC3TR/shFBXL10 xenografts after Lexa treatment led to decreased tumor size, increased apoptotic activities and increased c-Fos levels as compared with controls (Figures 5f–j). In other xenograft studies we had similar findings. We found that expression of FBXL10 was reduced and c-Fos level was increased in the TRAIL-sensitive A-498 renal caner xenografts, while in the TRAIL-resistant LNCaP xenografts the levels of c-Fos and FBXL10 were unchanged (Supplementary Figure S3). These results demonstrate that loss of FBXL10, which is accompanied by upregulation of c-Fos, is important to prime resistant cancer cells for TRAIL-mediated apoptosis.
FBXL10 has an anti-apoptotic role and regulates sensitivity of TRAIL-mediated apoptosis in vivo. (a) Representative images of PC3/FBXL10 and PC3/control tumor xenografts without (−) or with (+) Lexa treatment (n=6) and (b) tumor growth rates. Mice harboring PC3/FBXL10 or PC3/control cancer cells were treated with or without Lexa for 4 weeks, and tumor growth was measured every 4 days. Solid line, untreated group; dotted line, treated group. (c and d) Apoptosis in the tumor was detected by measuring cleavage of PARP and cleaved caspase 3. Original magnification, × 400. (e) Representative immunohistochemical analysis of implanted tumor. Mice-bearing tumor xenografts were treated without (−) or with (+) Lexa for 4 weeks. Original magnification, × 400. (f) Representative images of PC3TR/shFBXL10 and PC3TR/sh control tumor xenografts without (−) or with (+) Lexa treatment (n=6) and representative (g) tumor growth rates. Mice harboring PC3TR/shFBXL10 or PC3TR/sh control cancer cells were treated as shown in (b). Solid line, untreated group; dotted line, treated group. (h and i) Apoptosis was detected by measuring cleavage of PARP and cleaved caspase 3. Original magnification, × 400. (j) Representative immunohistochemical analysis of implanted tumor. Mice-bearing tumor xenografts were treated without (−) or with (+) Lexa for 4 weeks. Original magnification, × 400. All in vivo experiments were repeated independently twice. (Scale bar, 50 μm). The data represent means of average determinants ± S.E.M. *P<0.05; **P<0.01; ***P<0.001
FBXL10 regulates c-FLIP(L) via a c-Fos-dependent pathway
To determine whether FBXL10 can function as a regulator of c-FLIP(L), we knocked-down FBXL10 by siRNA in LNCaP cells. Repression of FBXL10 reactivated c-Fos, repressed c-FLIP(L) and induced cleavage of caspase 8 (Figure 4g), leading to sensitization of cells to TRAIL (Figure 4h). However, concomitant repression of FBXL10 and c-Fos protected the cells from TRAIL (Figures 4g and h). These results indicate that FBXL10 regulates c-FLIP(L) through a c-Fos-dependent pathway. Similar results were observed in the PC3TR cells that were stably transfected with the shFBXL10 construct (PC3TR/shFBXL10). Silencing FBXL10 reactivated c-Fos, leading to sensitization of cells to TRAIL. However, double knock-down of FBXL10 and c-Fos protected PC3TR/shFBXL10 cells from TRAIL (Figures 4i and j).
Furthermore, to strengthen the point that c-FLIP(L) contributes to FBXL10-mediated apoptotic pathway, we transfected c-FLIP siRNA into PC3/FBXL10 cells. FBXL10 overexpression protected PC3 cells from TRAIL. But this phenomenon was reversed by inhibition of c-FLIP (Figures 4k and l). Conversely, c-FLIP(L) cDNA was transfected into PC3TR/shFBXL10 cells. Silencing FBXL10 reactivated c-Fos and altered the phenotype of PC3TR/shFBXL10 from a TRAIL-resistant to a more sensitive phenotype. We found that ectopic expression of c-FLIP(L) provided a significant protection of PC3TR/shFBXL10 cells from TRAIL (Figures 4m and n). These findings indicate that FBXL10 has an anti-apoptotic role in TRAIL-mediated apoptosis, to a great extent, via the c-FLIP(L) signaling pathway.
MG132 sensitizes TRAIL-resistant cells by inhibiting FBXL10 and activating c-Fos
In this study, we have shown that FBXL10 has an anti-apoptotic role in TRAIL-sensitive cells through repressing c-Fos. Next, we wished to investigate the role of FBXL10 in TRAIL-resistant cells. Previously, we have shown that MG132 sensitizes resistant cells to TRAIL by promoting expression of c-Fos to prime resistant cancer cells to undergo apoptosis.14 Here, we found that protein and mRNA levels of FBXL10 were dramatically reduced, concomitant with increased levels of c-Fos in the presence of MG132, a condition that did not inhibit the anti-apoptotic molecule, c-FLIP(L) or promote cell death in TRAIL-resistant cells. However, TRAIL+MG132 led to dramatic reduction of c-FLIP(L) in resistant cells. The expression of FBXL11 was not affected (Figures 6a and b, Supplementary Figure S4). In addition, LNCaP cells treated with TRAIL+MG132 showed time-dependent downregulation of FBXL10 and upregulation of c-Fos (Figure 6c).
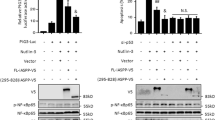
MG132 sensitizes TRAIL-resistant cells by inhibiting FBXL10 and activating c-Fos. (a) LNCaP cells were treated with 50 ng/ml TRAIL or/and 2 μM MG132 for 8 h and immunoblot was performed. (b) PC3TR cells were treated as shown in (a) and RT-PCR was performed. (c) LNCaP cells were treated with 50 ng/ml TRAIL and 2 μM MG132 for the times shown and immunoblot was performed. (d) PC3 and LNCaP cells transfected with pFBXL10 reporter plasmids and were treated as shown in a. Renilla luciferase was used as a positive control. Results are shown as the mean (bar) ± S.D. of at least three independent experiments. (e) PC3TR cells treated cells as shown in a were subjected to ChIP assay using a FBXL10 antibody or IgG. (f) Nuclear extracts from LNCaP cells treated as shown in a were incubated with FBXL10BS probe in the presence or absence of FBXL10 antibody. Arrows indicate the super-shifted FBXL10 protein–DNA complexes. All experiments were repeated independently at least three times with similar results
Our studies also revealed that transcriptional activity of FBXL10 was reduced in TRAIL-sensitive PC3 cells when the cells were exposed to either MG132 or TRAIL alone. In contrast, FBXL10 transcriptional activity in LNCaP cells was reduced only in the presence of MG132; and TRAIL alone did not reduce FBXL10 promoter activity. Of note, repression of FBXL10 promoter activity was more pronounced in both PC3 and LNCaP cells when treated with combination of MG132+TRAIL (Figure 6d). Collectively, in combination with our previous work,14 we show that MG132 primes resistant cells to undergo apoptosis by inhibiting FBXL10 promoter activity and reactivating c-Fos promoter activity, which only in the presence of TRAIL will lead to repression of c-FLIP(L) and induction of cell death.
Moreover, we found that FBXL10 was associated with c-Fos promoter in TRAIL-resistant PC3TR cells, but the binding of FBXL10 to c-Fos promoter was significantly reduced in the presence of MG132 (Figure 6e). EMSA assay provided a similar finding and the addition of an antibody to FBXL10 super-shifted the protein–DNA complex only in the absence of MG132 (Figure 6f), demonstrating that the reduced binding of FBXL10 to c-Fos promoter is specific and abrogated by MG132. Taken together, these results suggest that FBXL10 is a novel anti-apoptotic molecule, which directly binds and represses c-Fos promoter to make cancer cells resistant to TRAIL.
FBXL10 activity is NF-κB dependent
We have shown that TRAIL represses FBXL10 in sensitive cells. As NF-κB is an important regulator of apoptosis,1, 15, 16 we wished to determine whether NF-κB may have a role in regulating FBXL10.
First, we evaluated the relative expression of the constitutively active NF-κB-p65 in TRAIL-sensitive PC3 and -resistant LNCaP cells. We observed that PC3 cells expressed relatively lower levels of p65 than LNCaP cells (Figure 7a). As we have shown before,15 TRAIL significantly reduced nuclear p65 and levels in PC3 cells, whereas, in LNCaP cells, nuclear p65 levels were increased. Furthermore, MG132, which primes resistant cancer cells to TRAIL,14 repressed nuclear levels of p65 (Figure 7a). Concomitant with repression of p65, FBXL10 levels were reduced in the presence of MG132. Similar findings were reached with the downstream target gene of p65, Bcl-xL (Figure 7a). As a result of the correlative changes in levels of p65 and FBXL10 in the presence of TRAIL and MG132, next we wished to determine if NF-κB may directly regulate FBXL10.
FBXL10 activity is NF-κB dependent. (a) Nuclear extracts from PC3 or LNCaP cells treated with 50 ng/ml TRAIL or/and 2 μM MG132 as indicated for 8 h and immunoblot was performed. (b) PC3 or LNCaP cells were treated with MG132 (2 μM), Bay 11–7085 (5 μM) or bortezomib (20 nM) for 16 h and immunoblot was performed. (c) PC3 or LNCaP cells transfected with p65 siRNA or control siRNA for 40 h and immunoblot was performed. (d) LNCaP cells transfected with p65 siRNA or control siRNA for 40 h were treated with TRAIL (50 ng/ml) for additional 8 h. Cell viability was determined by MTT. (e and f) PC3 cells were stably transfected with p65 expression plasmid or control empty plasmid, respectively. The stable cells were treated with TRAIL (50 ng/ml): (e) immunoblot (f) MTT cell viability assay. (g and h) The PC3/p65 stable cells transfected by FBXL10 siRNA or control siRNA were treated with TRAIL (50 ng/ml): (g) immunoblot and (h) MTT cell viability assay. (i) PC3 or LNCaP cells transfected with wild-type pFBXL10 or NF-κB-BS mutated pFBXL10 reporter plasmids, respectively, were treated with or without 50 ng/ml TRAIL or/and 2 μM MG132 as indicated for 8 h. Renilla luciferase was used as a positive control. Results are shown as the mean (bar) ± S.D. of at least three independent experiments. (j and k) In all, 2 μg of pFBXL10 reporter plasmids were transfected into mice for 16 h using the hydrodynamic procedure. In vivo bioluminescence imaging was performed before and after treatment with Bay 11-7085 or vehicle (j) or before and after treatment with vehicle, LPS or Bortezomib (k). Data are representative of three independent experiments. All experiments were repeated independently at least three times with similar results
Treatment with the highly specific NF-κB inhibitor, Bay 11–7085, and clinically approved proteasome inhibitor, bortezomib, reduced FBXL10 expression dramatically in both PC3 and LNCaP cells (Figure 7b). Repression of p65 by siRNA decreased FBXL10 (Figure 7c, Supplementary Figure S5). TRAIL treatment led to a dramatic reduction in cell viability in p65-knockout-LNCaP cells (Figure 7d), which is consistent with a previous study.17 Overexpression of p65 protected c-FLIP(L) from TRAIL-mediated downregulation in PC3 cells (Figure 7e) and protected PC3 cells from TRAIL (Figure 7f). Moreover, suppression of FBXL10 in PC3/p65 stable cells reversed the p65 overexpression-mediated resistance (Figure 7g and h).
Subsequently, we identified one putative NF-κB-BS (−230 to −219 bp) in the FBXL10 promoter using bioinformatics tools. To further determine whether the site is functionally required, we mutated the NF-κB-BS and evaluated the relative FBXL10 promoter activity in PC3 and LNCaP cells. In both sensitive and resistant cell lines, treatment led to reduction in wt-FBXL10 promoter activity, whereas repression of FBXL10 promoter activity was abrogated in the mutant (Figure 7i). Taken together, these results suggest that NF-κB functions as an upstream regulator of FBXL10.
Using hydrodynamic transfection, we found that Bay11-7085 dramatically reduced the FBXL10 promoter activity, yet treatment with lipopolysaccharides (LPS) increased the FBXL10 promoter activity 1.4 times. Moreover, bortezomib led to a dramatic reduction in FBXL10 promoter activity (Figure 7j and k). Taken together, these findings suggest that existence of NF-κB signaling is critical for the transcription of FBXL10 not only in vitro, but also in vivo.
NF-κB directly binds to FBXL10 promoter
Next, we wished to determine if NF-κB directly binds to the FBXL10 promoter. We found that p65 associated with FBXL10 promoter in both TRAIL-sensitive PC3 and -resistant LNCaP cells (Figure 8a). Addition of TRAIL reduced the binding of p65 to FBXL10 promoter in PC3 cells, whereas in LNCaP cells, only TRAIL+MG132 reduced binding of p65 to FBXL10 promoter, whereas TRAIL alone did not dramatically affect the interaction (Figure 8a). In addition, using a fragment of the FBXL10 promoter that includes the NF-κB-BS demonstrated that the binding of NF-κB to FBXL10 promoter is specific and abrogated by addition of TRAIL in PC3 cells (Figure 8b). Addition of TRAIL to LNCaP cells enhanced interaction between NF-κB and FBXL10 promoter, while presence of MG132, abolished the interaction (Figure 8b). These results suggest that interaction between NF-κB and the FBXL10 promoter are specific, and because the promoter of the FBXL10 harbors one active NF-κB-BS, it is of considerable interest to evaluate its in vivo potential as transcriptional enhancers. To this end, we studied whether oligonucleotides corresponding to the putative NF-κB-BS could be used as decoys for repressing FBXL10 promoter activity. We introduced the FBXL10 promoter luciferase construct together with the oligonucleotides into mice with hydrodynamic transfection. We found that FBXL10 promoter was severely inhibited by the decoy oligonucleotides corresponding to the NF-κB-BS but not by the mutant oligonucleotide (Figure 8c).
NF-κB directly binds to FBXL10 promoter. (a) PC3 or LNCaP cells treated as shown in Figure 7a were subjected to ChIP assay using a p65 antibody or IgG. (b) An NF-κB BS probe was incubated without (lane 1, negative control) or with nuclear extracts from PC3 or LNCaP cells treated as shown in a. In all, 100-fold molar excess of unlabeled oligonucleotide (cold probe) was used as competitor. The probe was replaced by the mutant probe in the last lane of each panel. (c) Mice were transfected with 2 μg pFBXL10 reporter plasmids using the hydrodynamic procedure. In addition, the mouse to the left received 1 nmol oligonucleotide decoys (details in ‘Materials and Methods section’). The mouse to the right received 1 nmol mutant oligonucleotides. Data are representative of three independent experiments. (d) PC3 cells were co-transfected with pFBXL10 or NF-κB-BS mutated pFBXL10 reporter plasmids and p65 plasmids with the weight ratio as indicated for 24 h, respectively. Renilla luciferase was used as a positive control. Results are shown as the mean (bar) ± S.D. of at least three independent experiments
NF-κB signaling positively regulates FBXL10 transcription
We found that upregulation of NF-κB signaling in cells and in vivo only induced a slight or modest increase in FBXL10 expression, whereas NF-κB inhibitors always dramatically reduced FBXL10 expression. Therefore, we investigated whether NF-κB itself could have a positive regulatory role in FBXL10 promoter activity. We co-transfected the wt-FBXL10 promoter and NF-κB-BS mutated FBXL10 promoter together with increasing concentration of an expression vector p65/RelA into PC3 cells, respectively. We observed that the induction of FBXL10 promoter activity by 0.5–1 molar excess of p65 was limited; however, there was a pronounced induction of FBXL10 promoter activity (up to 2.5-fold) in the cells overexpressing p65/RelA (four molar excess). In contrast, mutant FBXL10 promoter could not be activated by the p65 plasmid (Figure 8d). This result suggests that NF-κB still maintains a positive regulatory role in transcription of FBXL10, but robust activity of NF-κB could be required to induce observable transcription of FBXL10.
Discussion
Here, we report that FBXL10 is a transcriptional repressor of c-Fos and a target gene of NF-κB-p65 in human cancers and describe, for the first time, the role of FBXL10 as an anti-apoptotic molecule. FBXL10 functions as an anti-apoptotic molecule by direct binding and repressing of the c-Fos promoter to make cancer cells resistant to TRAIL. Repression of FBXL10 in resistant tumors improves efficacy of TRAIL's pro-apoptotic activities. In addition, we present evidence that FBXL10 regulates c-FLIP(L) indirectly through a c-Fos-dependent pathway, while FBXL10 is regulated by NF-κB-p65 (Figure 9).
Schematic diagram of TRAIL or TRAIL+MG132 action in sensitive or resistant cells, respectively. Following TRAIL stimulation in sensitive cells, or TRAIL+MG132 stimulation in resistant cells, first, NF-κB activity is inhibited. Reduced activity of NF-κB leads to downregulation of FBXL10 transcription which increases c-Fos transcriptional activity. Second, reduced activity of NF-κB and increased c-Fos directly result in inhibition of c-FLIP(L), thus inducing apoptosis in TRAIL-sensitive and -resistant cells. Third, FBXL10 regulates c-FLIP(L) through a c-Fos-dependent pathway
TRAIL delivers potent antitumor activity, however, some tumors develop resistance. Various mechanisms of resistance have been proposed some of which include regulation of pro-apoptotic receptor transport to the cell membrane,18 Bcl-2-mediated downregulation of a mitochondrial machinery,19 bax mutations,20 XIAP inhibition21 and peroxiredoxin 6 interference,22 or downregulation of Mcl-1 or STAT3 when sorfenib is used in combination with TRAIL.23, 24 Previously, we have demonstrated that c-Fos-mediated inhibition of c-FLIP(L) is an important pathway in regulating resistance to TRAIL.2
FBXL10 has been implicated in the demethylation of trimethylated histone H3 lysine 4,9 a negative regulator of c-Jun10 and a new reference gene for gene expression studies in adipose tissue,25 while others have found that FBXL10 regulates cell proliferation and senescence through p15.26 However, FBXL10′s role in apoptosis has not been investigated. To investigate the role of FBXL10 in apoptosis, we used our previous model in which we demonstrated that c-Fos has a pro-apoptotic role and primes cancer cells to TRAIL-mediated apoptosis through repressing c-FLIP(L).2 Interestingly, in the TRAIL-mediated apoptosis model, the previously suggested concept that FBXL10 is a repressor of c-Jun10 could not be observed. Rather, both in vitro and in vivo studies indicated that only the expression of c-Fos, not c-Jun, were inversely correlated with FBXL10 levels (Figure 1 and Supplementary Figure S1). Moreover, our data support our hypothesis that FBXL10 functions as an anti-apoptotic molecule in TRAIL-mediated apoptosis.
Many pro-apoptotic stimuli including chemotherapy,27 UV radiation28, 29 and TNF-α exposure30can induce c-Fos. We2 and others31, 32 have shown that c-Fos levels increase after TRAIL treatment. Although transcriptional regulation of c-Fos has been extensively investigated, the interaction between FBXL10 and c-Fos signaling has not been previously described. In this study, we demonstrate that FBXL10 is a transcriptional repressor of c-Fos by direct binding to the c-Fos promoter (Figures 2 and 3), thus defining the role of FBXL10 as an anti-apoptotic agent (Figures 4 and 5, Supplementary Figure S2 and S3).
In order to determine whether FBXL10 has any role in TRAIL-resistant cancer cells, we utilized the proteasome inhibitor, MG132, which primes resistant cancer cells to TRAIL.14, 33 Concomitant with activation of c-Fos, FBXL10 was repressed and direct interactions between c-Fos promoter and FBXL10 were abrogated (Figure 6). Therefore, demonstrating that FBXL10 has an anti-apoptotic role in TRAIL-mediated apoptosis and repressing FBXL10 in resistant cells can improve efficacy of TRAIL pro-apoptotic therapies.
As FBXL10 regulates TRAIL-induced apoptosis, we next determined whether NF-κB, another important regulator of TRAIL-mediated apoptosis, may affect FBXL10. Previous studies have shown that NF-κB activity is decreased or maintained at low levels in TRAIL-sensitive cells.15 Whereas in TRAIL-resistant cells, TRAIL can activate NF-κB signaling at variable levels.16, 34 Downregulation of NF-κB sensitizes tumors to TRAIL.1, 35 Here, we found that MG132, bortezomib, Bay11-7085 and p65 siRNA all significantly downregulated FBXL10 expression (Figure 7 and Supplementary Figure S5). Also, silencing of p65 sensitized TRAIL-resistant cells (Figure 7c and d), and overexpression of p65 in sensitive cells protected the cells from TRAIL (Figure 7e and f), however, this finding was reversed by suppression of FBXL10 (Figure 7g and h). Further, we demonstrated that p65 directly associated with FBXL10 promoter and TRAIL treatment reduced the interaction between p65 and FBXL10 promoter in TRAIL-sensitive cells (Figure 8a–c).
In conclusion, we demonstrate, for the first time, that FBXL10 is a transcriptional repressor of c-Fos and a target gene of NF-κB-p65 in human cancers. We show that FBXL10 is an important anti-apoptotic molecule, which directly binds and represses c-Fos promoter in order for cancer cells to resist TRAIL-induced apoptosis. FBXL10 indirectly regulates c-FLIP(L) levels via c-Fos-dependent pathways. Silencing of FBXL10 sensitizes resistant cells to TRAIL, while overexpression of FBXL10 represses TRAIL-induced apoptosis. Moreover, our results indicate that expression of FBXL10 functions via a NF-κB-dependent pathway, thus describing a novel apoptotic-related pathway that involves NF-κB/FBXL10/c-Fos/c-FLIP (Figure 9). Therefore, silencing FBXL10 can help overcome resistant cancer cells for pro-apoptotic therapies.
Materials and Methods
Tissue culture, transfection
PC3, A498, MDA-MB-231, PC3TR and LNCaP cell lines were maintained in RPMI 1640 containing 10% FCS at 37 °C with 5% CO2. PC3TR (a TRAIL-resistant sub-population of PC3) was generated as previously described.4 Stable PC3/control and PC3/FBXL10 cells were established using 1 mg/ml G418 (Invitrogen, Carlsbad, CA, USA) after transfection of wild-type FBXL10 expression vector or control empty vector. Transfection of plasmid, shRNA and siRNA were performed using Fugene 6 or XtremeGENE according to the manufacturer's instructions (Roche, Indianapolis, IN, USA). Stable PC3TR/shcontrol and PC3TR/shFBXL10 cells were established using 0.8 μg/ml puromycin (Sigma, St Louis, MO, USA) after transfection of shFBXL10 or control shRNA.
Reagents
Antibodies to FBXL10 (ab5199, ab64920), FBXL11 (ab27867), TBP (ab51841), c-Fos (4384), c-Jun (9165), c-FLIPL (06-864), BCL-xL (2764), Caspase 8 (9746), GAPDH (2118), p65 (4764), c-Myc (9E10), PARP (9542) cleaved caspase 3 (9661) and normal rabbit IgG (2729) were purchased from Abcam (Cambridge, MA, USA), Millipore (Billerica, MA, USA), Cell Signaling (Danvers, MA, USA) or Santa Cruz (Santa Cruz, CA, USA). MG-132, Bay11-7085 and LPS were purchased from Sigma. Bortezomib was purchased from LC Laboratory (Woburn, MA, USA) and Lexa was provided as a generous gift from Human Genome Science, Inc. (Rockville, MD, USA) (Dr. Robin Humphreys). Lexa is a TRAIL receptor 2 agonist antibody, which is currently in clinical trials for advanced malignancies.36 Lexa has equivalent sensitivity and resistance profiles and induces similar downstream signaling molecules as recombinant TRAIL.37 Recombinant human TRAIL was purchased from R&D (Minneapolis, MN, USA). FBXL10 siRNA (M-014930-01-0005) FLIP siRNA (M-003772-06-0005) and non-targeting siRNA 1 (D-001210-01-05) were purchased from Dharmacon (Lafayette, CO, USA), c-Fos siRNA was from Santa Cruz (sc-29221), NF-κB p65 siRNA II was from Cell Signaling (6534). shFBXL10 plasmids (sc-75005-SH) and control shRNA plasmids (sc-108060) were purchased from Santa Cruz.
Plasmids constructs and luciferase assay
The wild-type FBXL10, FBXL11 and FBXL10CxxC mutant expression vectors were kindly provided by Ryo Koyama-Nasu (New York University, New York, NY, USA). The c-Fos promoter-luciferase construct was provided by Akihiko Yoshimura (Keio University of Medicine, Tokyo, Japan). pcDNA1-p65/Rel A was a gift from Abdalla Jama (Karolinska Institute, Stockholm, Sweden). pSV40-Renilla luciferase expression plasmid was from Promega (Madison, WI, USA). pcDNA3-c-FLIP expression plasmid was constructed as described previously.4 Human wild-type FBXL10 promoter (–1120, 0) was cloned from genomic DNA of PC3 cells with PCR. Sense primer: 5′-AATCTCGGTACCACCCGCAAGATCAGGCCTAG-3′, and antisense primer: 5′-CTACACAAGCTTTGTTTTGTTGTTGGGTCACGAGT-3′, which contain a KpnI site and HindIII site, respectively. The PCR products were inserted into pGL3-enhancer luciferase reporter vector (Promega). Site-directed mutagenesis was used to generate mutant versions of the c-Fos promoter and FBXL10 promoter luciferase construct. FBXL10-BS-mutated c-Fos promoter was made by changing the FBXL10-binding sequence 5′-TGCGTCAGCA-3′ to 5′-TAATTGCAAA-3′. NF-κB-BS-mutated FBXL10 promoter was made by changing sequence 5′-GGGGCTTTCA-3′ to 5′-TTTGCTAAAA-3′. All clones were verified by sequencing. At 24–48 h after transfection with the appropriate plasmids, cells were lysed and luciferase activity was measured in a Synergy Luminescence Microplate Reader (BioTek, Winooski, VT, USA) using the Dual-Luciferase reporter assay systems (Promega).
Semi-quantitative RT-PCR and real-time RT-PCR
Total RNA was isolated using RNeasy Mini Kit (Qiagen, Valencia, CA, USA) according to the manufacturer's instructions. The reverse transcription (RT) and PCR were performed using Supercript one step RT-PCR kit (Invitrogen). The following oligonucleotides were used: FBXL10 sense, 5′-ACAACGTCATCAGCCTAGAGTTCAGCCACA-3′, antisense, 5′-TGGTGGGTGAAGTGGAGCCATCGGT-3′; c-Fos sense, 5′-ATGATGTTCTCGGGCTTCAACGCAGA-3′, antisense 5′-TCACAGGGCCAGCAGCGTGGGTGA-3′; c-Jun sense, 5′-CTGAAACAGAGCATGACCCTGAACCTG-3′, antisense, 5′-TTGGAGGCAGCGATGCGGTT-3′; c-FLIP sense, 5′-TTGAAGATGGACAGAAAAGCTGTGGAGACC-3′, antisense, 5′-CACACAAAGCTGTCGTAGTCTCGGTGCTC-3′; GAPDH sense, 5′-TGATGACATCAAGAAGGTGGTGAAG-3′, antisense, 5′-TCCTTGGAGGCCATGTGGGCCAT-3′. The PCR products were subjected to electrophoresis on 1% agarose gels, and the signal was visualized with Fluor-S Multiimager (Bio-Rad, Hercules, CA, USA). Quantitative data were obtained using the Quantity One software (Bio-Rad). Real-time RT-PCR was performed using Power SYBR Green RNA-to-CT 1-Step Kit and done in 7300 Real-Time PCR System (Applied Biosystems, Foster City, CA, USA) according to the manufacturer's instructions. The reaction for each sample was done in triplicate.
Nuclear extraction, western blotting and immunofluorescence staining
Nuclear extraction was prepared according to the manufacturer's instruction (Pierce Biotechnology, Rockford, IL, USA). Immunoblot was performed as previously described.4 TRAIL-treated PC3 cells and controls were fixed in 4% paraformaldehyde, permeabilized with 0.2% Triton X-100/PBS and incubated with anti-FBXL10, c-Fos and c-Jun antibodies (1 : 1000–2000), respectively, followed by incubation with Cy3-conjugated Affinipure goat anti rabbit IgG (111-167-003, Jackson Immunoresearch Lab, West Grove, PA, USA). The sections were counterstained with DAPI and mounted with mounting medium (Vector Lab, Burlingame, CA, USA).
Cell viability and apoptosis assay
Cell viability was determined by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) method (Roche) as previously described.4 Cells were then treated with TRAIL. The percentage of apoptotic cells was determined by analysis of Annexin V-positive cells according to the manufacturer's instruction (Invitrogen). Briefly, cells were washed with PBS three times, and resuspended in 100 μl of 1 × binding buffer. After adding 5 μl of Annexin-V– Alexa Fluo 647 and 2 μl of propidium iodide, the cells were incubated at room temperature in the dark. Fifteen minutes later, cells were quantified by flow cytometry (BD FACSAria I cell sorter, BD Bioscience, Franklin Lakes, NJ, USA; FACSDiva 6.0 Software, BD Bioscience).
Electrophoretic mobility shift assay
EMSA was performed using LightShift Chemiluminescent EMSA Kit (Pierce Biotechnology). DNA–protein binding was performed at room temperature for 20 min in a final volume of 20 μl containing binding buffer (10 mM Tris, pH 7.5, 50 mM KCl, 1 mM dithiothreitol), 2.5% (vol/vol) glycerol, 5 mM MgCl2, 1 μg poly(dI-dC), 0.05% (vol/vol) Nonidet P-40, 8 pmol double-stranded biotinylated probe and 10 μg nuclear extract. The DNA–protein complexes were separated by 6% DNA retardation gel (Invitrogen) in 0.5 × TBE at 100 V at 4 °C for 1.5 h. DNA–protein complexes in the gel were transferred to Niodyne B nylon membrane (Pierce) by electroblotting with 0.5 × TBE at 100 V for 40 min. DNA–protein complexes were fixed to the membrane by a UV cross-linker and detected by a nonradioactive nucleic acid detection kit (Pierce). For the super-shift assay, anti-FBXL10 antibody (2 μl) was preincubated in the binding reaction for 30 min before the addition of probe. The oligonucleotide probes (FBXL10BS): 5′-CAGGATGTCCATATTAGGACATCTGCGTCAGCAGGTTTCCACGGCCTTTCCCTGTAGCCCT-3′. The oligonucleotide probes (NF-κBBS) 5′-GGTGCCAGGCCCCGGGGGCTTTCACGCCAGCATGCTCGGCTTGCATTTC-3′.
Chromatin immunoprecipitation assay
ChIP was performed using the SimpleChIP enzymatic chromatin IP kit as described by the manufacturer (Cell Signaling). In brief, cross-linking of protein–DNA complex in vivo was achieved by incubating cells in 1% formaldehyde solution for 10 min at room temperature. The cross-linking reaction was quenched by 125 mM glycine (final concentration) for 5 min at room temperature. Chromatin was prepared and digested with micrococcal nuclease for 20 min at 37°C. The lysate was subjected to sonication at 4°C using an ultrasonic processor (W-385, Heat System-Ultrasonic Inc., Farmingdale, NY, USA) The sonicated lysates were first pre-cleared by adding 50 μl protein A-agarose beads and rotated for 1 h at 4 °C. Subsequently, lysates were subjected to immunoprecipitation by adding 5 μl polyclonal anti-FBXL10, anti-p65 or normal rabbit normal Ig, respectively. Samples were rotated overnight at 4 °C; 50 μl protein A-agarose beads were added to each sample followed by rotation for another 2 h. The proteins were removed from DNA by digesting with 10 μg/ml proteinase K at 65 °C for 2 h. The DNA was further purified by spin column and eluted in 50 μl elution buffer. Eluted DNA (2 μl) was used for the PCR assay to detect DNA physically associated with the immunoprecipitated proteins. Values obtained from immunoprecipitated samples were normalized to input samples. The primers for the FBXL10-binding sequence were forward primer, 5′-CGCACTGCACCCTCGGTGTTGGCTG-3′, and reverse primer, 5′-TTCCTGTCTCAGAGGTCTCGTGGGC-3′. Primers for control were forward primer, 5′-GTCCCAGCAGTCGAGGTATT-3′, and reverse 5′-CCAGGAAGCTGTGCTTACG-3′. Primers for NF-κB-binding sequence were forward primer, 5′-GAGTCGGCCACACAGGTAATGTTGG-3′, and reverse primer, 5′-AGGGAGGTGCAGATTTGCACGGC-3′.
Hydrodynamic transfection
Hydrodynamic transfection of plasmids in DPBS was carried out as previously described.38 Briefly, oligonucleotide and plasmids as indicated were added to final volume of 1.8 ml of DPBS and steadily injected into tail vein of mice over a period of 5 s (N=4–6 animals per group). Live, anesthetized mice were imaged for 30 s to 2 min using an intensified CCD camera (IVIS imaging system, Xenogen Corp., Alameda, CA, USA). Image data were analyzed using LivingImage software (Xenogen Corp.). The 22–25 g female CD-1 mice were obtained from Charles River Laboratories (Wilmington, MA, USA). Bortezomib (1 g per g body weight, i.p.), LPS (4 g per g body weight, i.v.).39 Bay 11–7085 (20 μg per g body weight, i.p.).
Tumor xenograft experiments
PC3/FBXL10 and PC3TR/shFBXL10 xenografts
The stable PC3/FBXL10 and PC3/control cells were established as described in ‘Transfection section of SI Materials and Methods’. The 1 × 107 PC3/FBXL10 and PC3/control cells were suspended in 50% Matrigel (BD Biosciences) and injected subcutaneously into two sites per mouse (Female NCr homozygous nude mice, Taconic Farms, Germantown, NY, USA), respectively. Tumor growth was monitored by palpation, and the onset when tumors were detectable was noted. Ten mice harboring PC3/FBXL10 cells and 10 mice harboring PC3/control cells were randomly divided into two groups, respectively (each subgroup contained five mice): control groups were injected with PBS and treatment groups were treated with Lexa (Lexa 10 μg/g Lexa in PBS administered intravenously twice a week as previously described2, 37). Mouse body weight and tumor size were measured every 4 days. Tumor volume was quantified (volume=width2 × length × 0.52). Four weeks after treatment, all animals were euthanized, and xenografts were harvested and assessed for immunohistochemistry and apoptosis. Tumor tissues were fixed in 10% formalin and embedded in paraffin routinely. Immunohistochemistry was carried out as previously described.40 Tissue samples for western blot analysis were preserved in liquid nitrogen and then prepared in radioimmunoprecipitation assay buffer with 2% SDS. The stable PC3TR/shFBXL10 and PC3TR/shcontrol cells were established as described in ‘Transfection section of SI Materials and Methods’. Experimental protocol is as described in ‘PC3/FBXL10 xenografts’.
Xenograft orthotopic implantations and immunofluorescence analysis
Prostate and renal orthotopic implantations were carried out by injection of 1 × 106 cells in either the posterolateral lobe of the prostate or beneath the kidney capsule of athymic nude mice (Charles River Laboratories) at 6–8 weeks of age. Mice were implanted with PC3, LNCaP and A498 cells, respectively. Ten weeks after implantation of the xenografts, the athymic nude mice were randomly divided into treated and untreated groups and treated with Lexa via tail vein twice a week (10 mg/kg), as previously described.37 Four weeks after treatment, all animals were euthanized and xenografts were harvested, and assessed for immunostaining. Tumor tissues were fixed in 10% formalin and embedded in paraffin routinely, and cut as 5 μm cross-section. After deparaffinization, the tissues were then stained with anti-FBXL10, c-Fos and c-Jun antibodies (1 :100–200), respectively. Positive signal was developed with Cy3-conjugated Affinipure goat anti rabbit IgG (Jackson Immunoresearch Lab) and photographed under a fluorescent microscope (Nikon E800 microscope with epifluorescence illumination, Melville, NY, USA). Nuclear staining was done as described in cellular immunostaining section.
Ethics permission
The mice were housed and maintained in laminar flow cabinets under specific pathogen-free conditions. All animal experiments were approved by the Institutional Animal Care and Use Committee at Massachusetts General Hospital, and performed in accordance with ethical guidelines.
Abbreviations
- FBXL10:
-
F- box protein 10
- TRAIL:
-
tumor necrosis factor-related apoptosis-inducing ligand
- AP-1:
-
activator protein 1
- BS:
-
binding site
- NF-κB:
-
nuclear factor kappa-light-chain-enhancer of activated B cells
- Lexa:
-
lexatumumab
- LPS:
-
lipopolysaccharides
References
Roue G, Perez-Galan P, Lopez-Guerra M, Villamor N, Campo E, Colomer D . Selective inhibition of IkappaB kinase sensitizes mantle cell lymphoma B cells to TRAIL by decreasing cellular FLIP level. J Immunol 2007; 178: 1923–1930.
Zhang X, Zhang L, Yang H, Huang X, Otu H, Libermann TA et al. c-Fos as a proapoptotic agent in TRAIL-induced apoptosis in prostate cancer cells. Cancer Res 2007; 67: 9425–9434.
Ashkenazi A . Targeting death and decoy receptors of the tumour-necrosis factor superfamily. Nat Rev Cancer 2002; 2: 420–430.
Zhang X, Jin TG, Yang H, DeWolf WC, Khosravi-Far R, Olumi AF . Persistent c-FLIP(L) expression is necessary and sufficient to maintain resistance to tumor necrosis factor-related apoptosis-inducing ligand-mediated apoptosis in prostate cancer. Cancer Res 2004; 64: 7086–7091.
Johnstone RW, Frew AJ, Smyth MJ . The TRAIL apoptotic pathway in cancer onset, progression and therapy. Nat Rev Cancer 2008; 8: 782–798.
Gonzalvez F, Ashkenazi A . New insights into apoptosis signaling by Apo2L/TRAIL. Oncogene 2010; 29: 4752–4765.
Kreuz S, Siegmund D, Scheurich P, Wajant H . NF-kappaB inducers upregulate cFLIP, a cycloheximide-sensitive inhibitor of death receptor signaling. Mol Cell Biol 2001; 21: 3964–3973.
LaCasse EC, Baird S, Korneluk RG, MacKenzie AE . The inhibitors of apoptosis (IAPs) and their emerging role in cancer. Oncogene 1998; 17: 3247–3259.
Frescas D, Guardavaccaro D, Bassermann F, Koyama-Nasu R, Pagano M . JHDM1B/FBXL10 is a nucleolar protein that represses transcription of ribosomal RNA genes. Nature 2007; 450: 309–313.
Koyama-Nasu R, David G, Tanese N . The F-box protein Fbl10 is a novel transcriptional repressor of c-Jun. Nat Cell Biol 2007; 9: 1074–1080.
Asakuma J, Sumitomo M, Asano T, Asano T, Hayakawa M . Selective Akt inactivation and tumor necrosis actor-related apoptosis-inducing ligand sensitization of renal cancer cells by low concentrations of paclitaxel. Cancer Res 2003; 63: 1365–1370.
Shao R, Lee DF, Wen Y, Ding Y, Xia W, Ping B et al. E1A sensitizes cancer cells to TRAIL-induced apoptosis through enhancement of caspase activation. Mol Cancer Res 2005; 3: 219–226.
Yu L, Mohamed AJ, Simonson OE, Vargas L, Blomberg KE, Bjorkstrand B et al. Proteasome-dependent autoregulation of Bruton tyrosine kinase (Btk) promoter via NF-kappaB. Blood 2008; 111: 4617–4626.
Li W, Zhang X, Olumi AF . MG-132 sensitizes TRAIL-resistant prostate cancer cells by activating c-Fos/c-Jun heterodimers and repressing c-FLIP(L). Cancer Res 2007; 67: 2247–2255.
Zhang X, Huang X, Olumi AF . Repression of NF-kappaB and activation of AP-1 enhance apoptosis in prostate cancer cells. Int J Cancer 2009; 124: 1980–1989.
Falschlehner C, Emmerich CH, Gerlach B, Walczak H . TRAIL signalling: decisions between life and death. Int J Biochem Cell Biol 2007; 39: 1462–1475.
Khanbolooki S, Nawrocki ST, Arumugam T, Andtbacka R, Pino MS, Kurzrock R et al. Nuclear factor-kappaB maintains TRAIL resistance in human pancreatic cancer cells. Mol Cancer Ther 2006; 5: 2251–2260.
Jin Z, McDonald III ER, Dicker DT, El-Deiry WS . Deficient tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) death receptor transport to the cell surface in human colon cancer cells selected for resistance to TRAIL-induced apoptosis. J Biol Chem 2004; 279: 35829–35839.
Sinicrope FA, Penington RC, Tang XM . Tumor necrosis factor-related apoptosis-inducing ligand-induced apoptosis is inhibited by Bcl-2 but restored by the small molecule Bcl-2 inhibitor, HA 14-1, in human colon cancer cells. Clin Cancer Res 2004; 10: 8284–8292.
LeBlanc H, Lawrence D, Varfolomeev E, Totpal K, Morlan J, Schow P et al. Tumor-cell resistance to death receptor-induced apoptosis through mutational inactivation of the proapoptotic Bcl-2 homolog Bax. Nat Med 2002; 8: 274–281.
Stadel D, Mohr A, Ref C, Macfarlane M, Zhou S, Humphreys R et al. TRAIL-induced apoptosis is preferentially mediated via TRAIL receptor 1 in pancreatic carcinoma cells and profoundly enhanced by XIAP inhibitors. Clin Cancer Res 2010; 16: 5734–5749.
Choi H, Chang JW, Jung YK . Peroxiredoxin 6 interferes with TRAIL-induced death-inducing signaling complex formation by binding to death effector domain caspase. Cell Death Differ 2010 (e-pub ahead of print 10 September 2010; doi:10.1038/cdd.2010.113).
Chen KF, Tai WT, Liu TH, Huang HP, Lin YC, Shiau CW et al. Sorafenib overcomes TRAIL resistance of Hepatocellular carcinoma cells through the inhibition of STAT3. Clin Cancer Res 2010; 16: 5189–5199.
Ricci MS, Kim SH, Ogi K, Plastaras JP, Ling J, Wang W et al. Reduction of TRAIL-induced Mcl-1 and cIAP2 by c-Myc or sorafenib sensitizes resistant human cancer cells to TRAIL-induced death. Cancer Cell 2007; 12: 66–80.
Hurtado del Pozo C, Calvo RM, Vesperinas-Garcia G, Gomez-Ambrosi J, Fruhbeck G, Corripio-Sanchez R et al. IPO8 and FBXL10: new reference genes for gene expression studies in human adipose tissue. Obesity (Silver Spring) 2010; 18: 897–903.
He J, Kallin EM, Tsukada Y, Zhang Y . The H3K36 demethylase Jhdm1b/Kdm2b regulates cell proliferation and senescence through p15(Ink4b). Nat Struct Mol Biol 2008; 15: 1169–1175.
Jazirehi AR, Bonavida B . Cellular and molecular signal transduction pathways modulated by rituximab (rituxan, anti-CD20 mAb) in non-Hodgkin's lymphoma: implications in chemosensitization and therapeutic intervention. Oncogene 2005; 24: 2121–2143.
Quan T, He T, Voorhees JJ, Fisher GJ . Ultraviolet irradiation induces Smad7 via induction of transcription factor AP-1 in human skin fibroblasts. J Biol Chem 2005; 280: 8079–8085.
Tanos T, Marinissen MJ, Leskow FC, Hochbaum D, Martinetto H, Gutkind JS et al. Phosphorylation of c-Fos by members of the p38 MAPK family. Role in the AP-1 response to UV light. J Biol Chem 2005; 280: 18842–18852.
Manchester KM, Heston WD, Donner DB . Tumour necrosis factor-induced cytotoxicity is accompanied by intracellular mitogenic signals in ME-180 human cervical carcinoma cells. Biochem J 1993; 290 (Part 1): 185–190.
Drosopoulos KG, Roberts ML, Cermak L, Sasazuki T, Shirasawa S, Andera L et al. Transformation by oncogenic RAS sensitizes human colon cells to TRAIL-induced apoptosis by up-regulating death receptor 4 and death receptor 5 through a MEK-dependent pathway. J Biol Chem 2005; 280: 22856–22867.
Siegmund D, Mauri D, Peters N, Juo P, Thome M, Reichwein M et al. Fas-associated death domain protein (FADD) and caspase-8 mediate up-regulation of c-Fos by Fas ligand and tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) via a FLICE inhibitory protein (FLIP)-regulated pathway. J Biol Chem 2001; 276: 32585–32590.
He Q, Huang Y, Sheikh MS . Proteasome inhibitor MG132 upregulates death receptor 5 and cooperates with Apo2L/TRAIL to induce apoptosis in Bax-proficient and -deficient cells. Oncogene 2004; 23: 2554–2558.
Chen X, Kandasamy K, Srivastava RK . Differential roles of RelA (p65) and c-Rel subunits of nuclear factor kappa B in tumor necrosis factor-related apoptosis-inducing ligand signaling. Cancer Res 2003; 63: 1059–1066.
Ravi R, Bedi GC, Engstrom LW, Zeng Q, Mookerjee B, Gelinas C et al. Regulation of death receptor expression and TRAIL/Apo2L-induced apoptosis by NF-kappaB. Nat Cell Biol 2001; 3: 409–416.
Kelley SK, Ashkenazi A . Targeting death receptors in cancer with Apo2L/TRAIL. Curr Opin Pharmacol 2004; 4: 333–339.
Zhang L, Zhang X, Barrisford GW, Olumi AF . Lexatumumab (TRAIL-receptor 2 mAb) induces expression of DR5 and promotes apoptosis in primary and metastatic renal cell carcinoma in a mouse orthotopic model. Cancer Lett 2007; 251: 146–157.
Zhang G, Budker V, Wolff JA . High levels of foreign gene expression in hepatocytes after tail vein injections of naked plasmid DNA. Hum Gene Ther 1999; 10: 1735–1737.
Gross S, Piwnica-Worms D . Real-time imaging of ligand-induced IKK activation in intact cells and in living mice. Nat Methods 2005; 2: 607–614.
Olumi AF, Grossfeld GD, Hayward SW, Carroll PR, Tlsty TD, Cunha GR . Carcinoma-associated fibroblasts direct tumor progression of initiated human prostatic epithelium. Cancer Res 1999; 59: 5002–5011.
Acknowledgements
This work was partially funded by support from the Edwin Beer Fellowship–New York Academy of Medicine to AFO.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited V Dixit
Supplementary Information accompanies the paper on Cell Death and Differentiation website
Supplementary information
Rights and permissions
About this article
Cite this article
Ge, R., Wang, Z., Zeng, Q. et al. F-box protein 10, an NF-κB-dependent anti-apoptotic protein, regulates TRAIL-induced apoptosis through modulating c-Fos/c-FLIP pathway. Cell Death Differ 18, 1184–1195 (2011). https://doi.org/10.1038/cdd.2010.185
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cdd.2010.185
Keywords
This article is cited by
-
FBXL10 promotes EMT and metastasis of breast cancer cells via regulating the acetylation and transcriptional activity of SNAI1
Cell Death Discovery (2021)
-
Identification of Structural Elements of the Lysine Specific Demethylase 2B CxxC Domain Associated with Replicative Senescence Bypass in Primary Mouse Cells
The Protein Journal (2020)
-
FBXL10 contributes to the development of diffuse large B-cell lymphoma by epigenetically enhancing ERK1/2 signaling pathway
Cell Death & Disease (2018)
-
KDM2B, an H3K36-specific demethylase, regulates apoptotic response of GBM cells to TRAIL
Cell Death & Disease (2017)
-
Cyclodextrin mediated delivery of NF-κB and SRF siRNA reduces the invasion potential of prostate cancer cells in vitro
Gene Therapy (2015)