Abstract
Interferon regulatory factor-1 (IRF-1) is a transcription factor that acts as a tumor suppressor and causes apoptosis in cancer cells. We evaluated IRF-1-induced apoptosis in gastric cancer cell lines. We established stable clones in AGS cells that have a tetracycline-inducible IRF-1 expression system. We used these clones and recombinant adenovirus expressing IRF-1 to explore the mechanism of IRF-1-induced apoptosis in gastric cancer. Expression of IRF-1 causes apoptosis in gastric cancer cell lines as shown by phosphatidylserine exposure and cleavage of caspase-8, caspase-3, and Bid with the mitochondrial release of cytochrome c. However, inhibition of caspase-8 and Bid did not inhibit apoptosis and did not decrease cleaved caspase-9 or mitochondrial release of cytochrome c. We then show that IRF-1 upregulates PUMA (p53 upregulated modulator of apoptosis), which is known to activate apoptosis by the intrinsic pathway; this can be p53-independent. IRF-1 binds to distinct sites in the promoter of PUMA and activates PUMA transcription. Moreover, molecular markers of mitochondrial apoptosis are eliminated in PUMA knockout and knockdown cells and phosphatidylserine exposure is decreased dramatically. Finally, we show that IFN-γ induces IRF-1-mediated upregulation of PUMA in cancer cells. We conclude that IRF-1 can induce apoptosis by the intrinsic pathway independent of the extrinsic pathway by upregulation of PUMA.
Similar content being viewed by others
Main
The interferon (IFN) regulatory factors (IRFs) have a vital role in various biological processes including growth regulation, pathogen responses, immune activation, and inflammation (reviewed by Tamura et al.1). IRF-1 is the first identified member of the IRF family; it can be effectively induced in most cell types after exposure to IFN-γ, IFN-α, IFN-β, tumor necrosis factor-α (TNF-α), retinoic acid, interleukin-1 (IL-1), and IL-6. IRF-1 binds to specific DNA sequences in promoters of genes and controls the transcription of these genes involved in mediating antiviral, immunomodulatory, and antiproliferative effects.
There is growing evidence that IRF-1 have an important role in apoptosis. IRF-1 not only induces apoptosis but is also a necessary mediator of apoptosis induced by DNA damage2 or other stimuli, including Faslodex3 and tamoxifen,4 as well as IFNs alone5 or in combination with retinoids,6 TRAIL,7 arsenic,8 cyclosporine A,9 and TNF-α10 in multiple cancer cell types. This regulation of apoptosis probably involves an IRF-1-mediated caspase cascade, as IRF-1 is known to upregulate and/or activate caspase-1,4 caspase-8,11 Fas ligand,12 and TRAIL.6 Our previous studies5, 13 indicated that overexpression of IRF-1 resulted in apoptosis in breast cancer cells in vitro and in vivo, which appears to involve both the extrinsic (death receptor) and intrinsic (mitochondrial) apoptotic pathways, but no death ligand nor any secreted ligand appears to be involved. However, the mechanism of IRF-1-induced apoptosis has been largely unstudied.
Gastric cancer is the second leading cause of cancer-related death in the world. There have been few studies indicating the role of IRF-1 in the pathology or treatment of gastric cancer. Frequent loss of heterozygosity at the IRF-1 locus has been found in gastric cancer patients.14 Furthermore, a point mutation in the second exon of the IRF-1 gene of the residual allele of a patient with gastric cancer has been reported, leading to the expression of functionally impaired IRF-1.15 These results suggest that IRF-1 has a critical role in the tumor suppression or surveillance of human gastric cancers.
In this study, we evaluated IRF-1-induced apoptosis in human gastric cancer cell lines. We established stable clones in AGS cells that have a tetracycline-inducible IRF-1 expression system. We used these cells and recombinant adenovirus expressing IRF-1 (Ad-IRF-1) to explore the mechanism of IRF-1-induced apoptosis in gastric cancer. Our data indicate that IRF-1 expression results in apoptosis in gastric cancer cells as assessed by FITC-Annexin V and propidium iodide (PI) staining, with the cleavage of caspase-8, caspase-3, and Bid, with the release of cytochrome c into the cytosol from mitochondria as assessed by immunoblotting. However, elimination of caspase-8 and Bid using RNA interference and the caspase-8 inhibitor did not inhibit apoptosis and did not decrease cleaved caspase-9 or the release of cytochrome c from the mitochondria. We then showed, for the first time, by both quantitative RT-PCR and immunoblotting analyses that IRF-1 upregulates PUMA (p53 upregulated modulator of apoptosis), which is known to activate apoptosis from the level of the mitochondria by the intrinsic pathway. IRF-1 binds to distinct sites in the promoter of PUMA and activates PUMA transcription as seen by luciferase reporter and chromatin immunoprecipitation (ChIP) analyses. Moreover, IRF-1-induced apoptosis is decreased dramatically in PUMA knockout and knockdown cells, and molecular markers of mitochondrial apoptosis appear to be eliminated. Finally, we analyzed that IFN-γ can upregulate PUMA in N87 cells that is mediated by IRF-1. Our findings show that IRF-1 can induce apoptosis by the intrinsic pathway independent of the extrinsic pathway involvement by upregulation of PUMA in cancer cells.
Results
IRF-1 induces apoptosis in gastric cancer cells
Our previous studies13, 16 as well as other studies2, 7 have showed that IRF-1 induces apoptosis in many cancer cell types, but this has not been analyzed in gastric cancer cells. The p53 wild-type gastric cancer cell line AGS was infected with Ad-IRF-1, the empty vector Ad-Null, or no infection (NI), and apoptosis was detected by flow cytometry analyses of Annexin V–FITC staining for phosphatidylserine (PS) exposure and PI staining for plasma membrane rupture at 24 or 48 h later. We found that Ad-IRF-1-induced apoptosis, but not Ad-Null or NI controls (Figure 1a). Cells that are Annexin V–FITC-positive and PI-negative are in early apoptosis as there is PS exposure but no plasma membrane rupture, and cells that are Annexin V–FITC-positive and PI-positive are in late apoptosis as plasma membrane rupture has occurred and confirms cell death. Figure 1b shows the total apoptotic (total Annexin V-positive) fraction in cells infected in three independent treatments in two different gastric cancer cell lines. There is a significant difference in apoptosis at 24 h and a markedly significant difference at 48 h. A similar effect was observed in N87 cells, and also in KATO III gastric cancer cells (Supplementary Figure 1a).
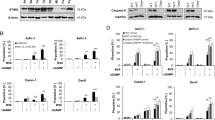
IRF-1 induces apoptosis in gastric cancer cells. (a) Cells were infected with Ad-IRF-1, the empty vector Ad-ψ5 (Ad-Null), or media only (no infection, NI) at 50 MOI for the indicated times. Apoptosis was detected by FACS analyses of Annexin V–FITC staining and propidium iodide (PI) staining as described in Materials and Methods, with representative bivariate data shown. (b) Samples including above were analyzed in three independently treated experiments (N=3) and the mean percentage apoptosis±S.D. is indicated graphically. (c–h) AGS cells were either treated with indicated amount of Ad-IRF-1 for 24 h or treated with 50 MOI for the indicated times; and tetracycline-inducible IRF-1 stably transfected AGS cells (AGSI) were treated with 2 μg/ml tetracycline at indicated times. Whole-cell lysates were harvested and cell extracts (20 μg) were immunoblotted for IRF-1, PARP (c–e), caspase-8, caspase-3, and caspase-9 (f–h). β-Actin was used as an internal loading control
Poly(ADP-ribose) polymerase (PARP) is known to be a key substrate associated with apoptosis at the level of the nucleus and is a more terminal hallmark of apoptosis in this context.17 To further confirm apoptosis, PARP cleavage was detected through time course and dose experiments. As seen in Figure 1c, the 85-kDa cleavage product of PARP is seen in cells infected with Ad-IRF-1, and the PARP cleavage is both dose (Figure 1c)- and time-dependent (Figure 1d). This specific 85 kDa band was not seen in the control cells (there is a nonspecific band smaller than 85 kDa seen throughout). PARP cleavage (both 85 and 31 kDa bands) was also seen in the tetracycline-inducible IRF-1 expression AGS cells as early as 24 h after tetracycline induction (Figure 1e).
It is well known that caspases have important roles in apoptosis. Caspases are cysteine–aspartyl proteases synthesized as inactive proenzymes. Once activated by cleavage, caspases then cleave their substrates and are responsible for most of the biochemical and morphological features of apoptotic cell death. Caspases are broadly categorized into initiators (caspase-2, -8, -9, and -10), effectors or executioners (caspase-3, -6, and -7), and inflammatory caspases (caspase-1, -4, and -5).17, 18 The activation of initiator caspases results from signaling through one of the two pathways: the extrinsic, death receptor pathway and the intrinsic, mitochondrial pathway.
We previously analyzed in breast cancer cells that IRF-1-induced apoptosis was caspase mediated and involves caspase-8 in the extrinsic pathway without death ligands. We did observe Bid cleavage and mitochondrial release of cytochrome c.5 We therefore examined the caspase cleavage pattern in AGS cells by dose and time course by western blot analysis. Figure 1f shows that caspase-3 and caspase-8 were cleaved into activated fragments at 25 MOI (multiplicity of infection), 24 h after infection, whereas the activated cleaved caspase-9 was able to be seen at 10 MOI, and the activation of these caspases correlated with IRF-1 expression. There was no activation of caspase-3, -8, or -9 in the controls, even at 100 MOI. Ad-IRF-1 results in IRF-1 induction (Figure 1g) by 12 h with the induction of caspase-3 cleavage by 24 h, which is increased at 30 h. Caspase-8 cleavage products were seen at 24 h and increased at 30 h, with decreased procaspase-8 at 30 h. However, the active caspase-9 product p35 was seen as early as 12 h, and increased at 24 and 30 h. The activation of caspase-3, -8, and -9 was also seen in tetracycline-inducible IRF-1 expression AGS clone cells after 24–72 h of tetracycline induction (Figure 1h).
The role of the intrinsic pathway in IRF-1-induced apoptosis
The activation of caspase-9 indicates an involvement of the intrinsic (mitochondrial) pathway in IRF-1-induced apoptosis. One of the hallmarks of the intrinsic pathway is the release of mitochondrial cytochrome c into the cytosol. Cytochrome c in the cytosol participates in the formation of the apoptosome, resulting in the activation of caspase-9 and downstream caspases.17 Using mitochondrial and cytosol fractionation, we detected a marked decrease in cytochrome c in the mitochondria and a marked increase in cytochrome c by 24 h in the cytosolic extracts prepared from AGS cells infected with Ad-IRF-1 compared with controls, which shows that IRF-1 induced cytochrome c release from the mitochondria (Figure 2a).
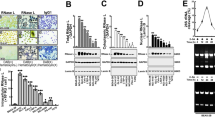
Intrinsic pathway in IRF-1-induced apoptosis. (a) Ad-IRF-1 induces mitochondrial release of cytochrome c. After infection (MOI=50), cells were harvested at 12, 24, and 30 h and fractionated. Cytosolic and mitochondrial fractions were then analyzed by western blot analysis for cytochrome c. VDAC is used as confirmation of successful and equivalent extraction of mitochondrial fraction. (b) AGS cells were transfected with nothing as control, non-targeting siRNA, or caspase-8 siRNA (25 nM) for 48 h, then the cells were infected with NI, Ad-Null, or Ad-IRF-1 (MOI=50) for 30 h. Apoptosis was detected by FACS analyses as described in Materials and Methods. (c) AGS cells were subjected to the indicated treatment as in panel b for 48 h, and caspase-8 and caspase-3 were analyzed by immunoblotting. C, control; N, non-targeting siRNA; and ‘8,’ caspase-8 siRNA. (d) The levels of caspase-9, Bid, and cytochrome c were detected by immunoblotting with the indicated treatments in AGS cells. (e) AGS cells were transfected with nothing as control, non-targeting siRNA, or Bid siRNA (25 nM) for 48 h, then the cells were infected with NI, Ad-Null, and Ad-IRF-1 (MOI=50) for 30 h. Apoptosis was detected by FACS analyses as described in Materials and Methods. (f) AGS cells were subjected to the indicated treatment as in panel e, and Bid, IRF-1, caspase-3, and caspase-9 were analyzed by western blotting. C, control; N, non-targeting siRNA; and B, Bid siRNA. (g) The levels of cytochrome c were assessed by western blot in cytosolic and mitochondrial fractions
As caspase-8-mediated activation of the pro-apoptotic BH3-only protein Bid may provide a link between extrinsic and intrinsic pathways,19 we investigated the role of caspase-8 and Bid on apoptosis upon IRF-1 induction in AGS cells using RNA interference. We found that caspase-8 siRNA did not inhibit IRF-1-induced apoptosis in AGS cells (Figure 2b), while completely eliminating the caspase-8 protein (Figure 2c). Immunoblot analysis showed that activated caspase-3 was detected irrespective of the elimination of caspase-8 (Figure 2c), and cleaved caspase-9 is not diminished (Figure 2d). We assessed for the cleavage of Bid by measuring the intact protein on immunoblot, which when proteolytically cleaved by caspase-8, acts to release cytochrome c from the mitochondria, and is reflected by decreased intact protein. We were unable to detect truncated Bid by immunoblot analysis, which can be difficult in certain cell types (XM Yin, personal communication), but is indirectly shown by the loss of intact Bid. There is no effect on Bid transcription by Ad-Null or Ad-IRF-1 based on RT-PCR (data not shown). The cleavage of Bid was inhibited by caspase-8 siRNA (Figure 2d). Nevertheless, cleaved caspase-9 was not diminished, indicating that IRF-1 can initiate the intrinsic pathway in AGS cells, which can be independent of Bid cleavage (Figure 2d). We also performed mitochondrial and cytosolic fractionation of treated cells and confirmed that cytochrome c release was unaffected by caspase-8 siRNA (Figure 2d). We also showed identical results using IETD, which is an irreversible peptide substrate inhibitor of caspase-8 (Supplementary Figure 2).
To further confirm that the intrinsic pathway of apoptosis is unaffected by Bid, we used RNA interference. As seen in Figure 2e–g, elimination of Bid by siRNA did not affect IRF-1-induced apoptosis by flow cytometry for PS exposure (Figure 2e), the presence of activated caspase-3 or caspase-9 (Figure 2f), or release of cytochrome c from the mitochondria (Figure 2g).
IRF-1 upregulates PUMA expression
Because BH3-only Bcl-2 family proteins are the initiators of mitochondrial apoptosis, we determined whether PUMA was involved in IRF-1-induced apoptosis. We evaluated the BH3-only proteins Noxa, Bad, and Bim in both AGS and N87 cells and did not find any increase in the expression by immunoblot analysis (Supplementary Figure 3). For PUMA expression, we evaluated dose and time course for Ad-IRF-1 in AGS cells. As shown in Figure 3a, IRF-1 induced PUMA protein in AGS cells in a dose- and time-dependent manner. PUMA protein began to increase at MOI of 10, and reached a maximum of 3.5-fold increase at MOI of 50, and increased at 12 h and further increased at 24 and 30 h after infection with Ad-IRF-1. To investigate whether IRF-1 induces PUMA transcription, we examined the effect of IRF-1 on PUMA mRNA. Quantitative real-time RT-PCR analysis showed a >2-fold increase in PUMA mRNA levels in AGS cells after Ad-IRF-1 infection at 24 h, compared with the Ad-Null control (Figure 3a). Moreover, we also examined the effect of IRF-1 on the expression of PUMA at protein and mRNA level using tetracycline-inducible clones. As shown in Figure 3b, IRF-1 dramatically increased the expression of PUMA not only at mRNA level but also at protein level. The alterations corresponded with changes in AGS cells infected with Ad-IRF-1. These data show that IRF-1 upregulates the expression of PUMA at the transcriptional level.
IRF-1 transcriptionally upregulates PUMA expression. (a) AGS cells were either treated with indicated amount of Ad-IRF-1 for 24 h or treated with 50 MOI for the indicated times. Cell extracts (20 μg) were assessed for IRF-1 and PUMA protein expression by western blot analysis. After infection with Ad-IRF-1 (MOI=50) for 24 h, expression levels of mRNA were also measured by quantitative real-time RT-PCR as described in Material and Methods and normalized to β-actin. N=3 for independently treated experiments±S.D. (b) Tetracycline-inducible AGS clone cells (AGSI) were treated with 2 μg/ml tetracycline at indicated times, and the expression of PUMA at both protein (left panel) and mRNA levels (right panel) were measured by western blot and quantitative real-time RT-PCR. N=3 for independently treated experiments±S.D. (c) N87 cells were infected with Ad-IRF-1 (MOI=50) for 24 h. The levels of PUMA were assessed by western blot and quantitative real-time RT-PCR. N=3 for independently treated experiments±S.D. (d) Tetracycline-inducible IRF-1 MDA468 (Pim) cells were treated with 1 μg/ml tetracycline for 24 h, and the expressions of PUMA protein and mRNA were measured as before. N=3 for independently treated experiments±S.D.
To determine the role of p53 in IRF-1-induced PUMA expression, N87 cells were used, which is a p53 mutant gastric cancer cell line. The results show that IRF-1 also induced PUMA expression in N87 cells (Figure 3c). Indeed, because there was virtually no expression of PUMA in control cells, presumably due to p53 mutation, there was a much greater fold increase in the expression as seen by quantitative RT-PCR (Figure 3c). We also used tetracycline-inducible IRF-1 MDA468 (designated Pim), a p53 mutated breast cancer cell line, to confirm the result (Figure 3d). Thus, we found that IRF-1 induced PUMA expression in both a p53 wild-type cell line (AGS) and p53 mutated cell lines (N87 and MDA468). These results confirm that IRF-1-induced PUMA expression can be p53-independent. We also evaluated the p53-null gastric cancer cell line KATO III and found identical results, with the activation of caspase-3 and caspase-9, as well as PARP cleavage (Supplementary Figure 1b).
IRF-1 increases PUMA promoter activity
We next addressed whether IRF-1 activates the transcriptional activity at the PUMA promoter by luciferase reporter. To identify potential DNA elements responsible for IRF-1-induced PUMA transcription, ∼2 kb fragment of the PUMA promoter (Frag E) and four smaller fragments in this region (Frags A–D, each containing ∼500 bp) were cloned into pBV-Luc (Figure 4a).20 Luciferase activity of these reporter constructs was examined in MDA468 and AGS cells after IRF-1 induction. As shown in Figure 4b, IRF-1 increased luciferase activity in the reporters that contain full-length Frag E or those that are more distant to the initiation site Frag D in MDA468 cells, but in Frags C, D, and E in AGS cells. These results indicate that the more distant regions of the PUMA promoter (Frags C and D, around −1 to −2 kb) are largely responsible for its transcription on IRF-1 induction.
IRF-1 increases PUMA promoter activity. (a) Schematic diagram of cloned, reporter constructs of PUMA promoter fragment E and four smaller fragments in the region. (b) The indicated PUMA reporter constructs were co-transfected into AGS and MDA468 cells with the expression construct of β-galactosidase, and then cells were infected the next day with indicated adenovirus at 25 MOI. The luciferase activities were analyzed 24 h later and normalized to β-gal expression. N=3 for independently treated experiments±S.D. (c) Schematic representation of the IRF-1-binding sites in the PUMA promoter. Putative tandem repeat IRF-1-binding sites (underlined and bold) in Frag C (left panel) and Frag D (right panel) were identified. (d) AGS cells were infected with Ad-IRF-1 (MOI=25) for 24 h, and then underwent chromatin immunoprecipitation (ChIP) assay to characterize the recruitment of IRF-1 to the PUMA promoter as described in Materials and Methods. IgG was used as a control. N=3 for independently treated experiments±S.D. (e) Tetracycline-inducible AGS clone cells (AGSI) treated with 2 μg/ml tetracycline for 24 h. ChIP was carried out to assess the binding of IRF-1 to the PUMA promoter. N=3 for independently treated experiments±S.D.
We then performed sequence analysis to determine putative IRF-1-binding sites in the PUMA gene promoter. A consensus IRF-1-binding sequence motif has been published to be 5′-G(A)AAAG/CT/CGAAAG/CT/C−3′ in previous reviews.21, 22 Two potential binding sites 5′-GAAAGC/T GAAAG-3′ were found in tandem in the Frag D of the PUMA promoter (Figure 4c), and a similar potential binding site 5′-GAAACTGAAAA-3′ was found in Frag C, with a possible tandem partner binding sequence 5′-GAATGGAAAGC-3′ (Figure 4c). This further suggests that PUMA is a transcriptional target of IRF-1.
Finally, ChIP assay was used to analyze the binding of IRF-1 to the PUMA promoter in AGS cells after IRF-1 induction. As seen in Figure 4d and e, using quantitative PCR as the readout for the ChIP analysis, immunoprecipitation of IRF-1 with fragmented chromatin shows increased PCR product using specific primers around the putative tandem binding sites in Frag C and Frag D in AGS cells, with a marked increase in PCR product with IRF-1 expression by adenovirus and the inducible model, indicative of increased binding of IRF-1 to the binding site.
The role of PUMA in IRF-1-induced apoptosis
To determine the role of PUMA in IRF-1-induced apoptosis, we infected HCT116 wild-type and PUMA−/− colon cancer cells with Ad-IRF-1, and also used RNA interference against PUMA in the AGS cells. The HCT116 cell lines expressed similar levels of IRF-1, but IRF-1 induced PUMA expression only in wild-type HCT116 cells, and PUMA siRNA completely eliminated PUMA protein in the AGS cells (Figure 5a). Although caspase-3 was activated in both HCT116 cell lines and all the treated AGS cells during infection of Ad-IRF-1, activated cleaved caspase-9 was only seen in the presence of PUMA. Furthermore, mitochondrial/cytosolic fractionation confirmed that cytochrome c was released from the mitochondria into the cytosol upon IRF-1 induction in HCT116 wild-type cells and control treated AGS cells only (Figure 5b), where PUMA was present. These results indicate that PUMA has an obligatory role in the mitochondrial pathway in IRF-1-induced apoptosis.
PUMA in IRF-1-induced apoptosis. (a) Left panel: HCT116 wild-type and HCT116 PUMA−/− cells were infected with indicated virus (MOI=100) for 48 h. Right panel: AGS cells were transfected with nothing as control (C), non-targeting siRNA (N), or PUMA siRNA (P, 25 nM) for 48 h, and then infected with NI, Ad-Null, and Ad-IRF-1 (MOI=50) for 30 h. Cell extracts (20 μg) were immunoblotted for IRF-1, PUMA, caspase-3, caspase-9, and β-actin. (b) HCT116 wild-type and HCT116 PUMA−/− cells, and AGS cells were treated as in panel a, and the levels of cytochrome c were assessed by western blot in cytosolic and mitochondrial fractions. (c) Left panel: The indicated HCT116 cells were infected with Ad-Null or Ad-IRF-1 (MOI=100) (or NI for control) and analyzed for apoptosis by flow cytometry 48 h later. N=3 for independently treated experiments±S.D. *P<0.0001. Right panel: AGS cells were treated as in panel a and then analyzed for apoptosis by flow cytometry. N=3 for independently treated experiments±S.D. *P=0.004 versus non-targeting control and no siRNA control
We then evaluated the effect on apoptosis by knockout of PUMA in HCT116 colon cancer cells and by knockdown in the PUMA siRNA-treated cells using Annexin V staining and flow cytometry. As shown in Figure 5c, IRF-1-induced apoptosis was significantly decreased in PUMA−/− cells compared with wild-type cells (P<0.0001), and in the AGS cells treated with PUMA siRNA (P=0.004), with >50% decreases in apoptosis in both cases.
IFN-γ induction of PUMA in N87 gastric cancer cells
IFN-γ is an essential mediator of the immune response to viruses and cancer, and is one of the strongest inducers of IRF-1 expression in cancer cells.23 We first confirmed that AGS cells are Stat1-deficient, which would prevent the induction of IRF-1 by IFN-γ in this cell line, as has already been established previously24 (Supplementary Figure 4). We then showed that IFN-γ increases IRF-1 expression in N87 cells and that PUMA protein is also found to be expressed in a dose-dependent manner, whereas in AGS cells, which do not express Stat1, IFN-γ does not appreciably increase IRF-1 expression and does not increase PUMA expression (Figure 6a). Furthermore, in N87 cells pretreated with siRNA against IRF-1 and then treated with IFN-γ, knockdown of IRF-1 expression by IRF-1 siRNA results in elimination of PUMA expression, confirming that IRF-1 mediates the increase in PUMA expression by IFN-γ (Figure 6b). As expected, IFN-γ induces both activated caspase-3 and caspase-9 at the doses that upregulate PUMA (500 and 1000 IU/ml, Figure 6c), and siRNA against PUMA eliminates activated caspase-9 in N87 cells (Figure 6d).
IFN-γ induction of PUMA in N87 gastric cancer cells through IRF-1. (a) N87 cells (left panel) and AGS cells (right panel) were treated with indicated amount of IFN-γ for 24 h, and then the expression of IRF-1 and PUMA were analyzed by western blot. (b) siRNA interference of IRF-1 significantly inhibits the expression of PUMA observed in IFN-γ-treated cells. N87 cells were treated with no siRNA (transfection agent alone), non-targeting control siRNA (Neg siRNA), or IRF-1-targeted siRNA (IRF-1 siRNA) and then cultured with IFN-γ (1000 IU/ml, 24 h) as indicated and underwent immunoblotting for IRF-1 and PUMA. (c) N87 cells were treated with indicated amount of IFN-γ for 48 h, and then caspase-3 and caspase-9 were analyzed by immunoblotting. (d) N87 cells were treated with no siRNA (transfection agent alone), 25 nM non-targeting siRNA (Neg siRNA) or 25 nM PUMA targeted siRNA (PUMA siRNA) and then cultured with IFN-γ (1000 IU/ml, 48 h) as indicated and underwent immunoblotting for PUMA, caspase-3, and caspase-9
Discussion
Our study is one of the few to investigate the mechanism of IRF-1-induced apoptosis in gastrointestinal malignancies. Previous studies have confirmed the importance of IRF-1 in tumor suppression of gastric cancer based on loss of heterozygosity or mutational analysis. Here, we show that IRF-1 induces apoptosis in gastric cancer cells in the mitochondrial pathway through upregulation of PUMA. Taken together, these observations support the important role of IRF-1 in gastric cancer and the use of strategies to augment IRF-1 expression or activity as a therapeutic option for gastric cancer.
To the best of our knowledge, this study is the first study to implicate PUMA as an IRF-1-regulated gene that can mediate apoptosis in cancer cells. PUMA has previously been found to be induced by infection and type I IFNs; in one study this appeared to be mediated by p53,25 and in another study, this appeared to be mediated by the Jak/Stat pathway,26 with IRF-1 being a signaling intermediate. To the best of our knowledge, this is the first study to show that the type II IFN IFN-γ can induce PUMA expression, and this can be independent of p53. IFN-γ stimulates PUMA expression in N87 cells, which is mediated by IRF-1 and activates caspase-3 and caspase-9 (Figure 6), and therefore further links the tumor immune response to apoptosis of cancer cells. This clearly may be a part of the mechanism of deletion of cancer cells or other cells as part of the immune surveillance against cancer or infection.
As seen in the AGS cell line in Figure 3, there is a low baseline expression of PUMA in the absence of IRF-1 expression presumably due to wild-type p53 expression. However, this is definitively enhanced in the setting of expression of IRF-1. In the p53 mutant cell lines N87 and MDA468, there is little or no baseline expression of PUMA, and with ectopic expression of IRF-1, there is a marked induction of PUMA. This confirms that p53 is not necessary for PUMA upregulation. It is now clear that PUMA can have a role in apoptosis independent of p53 status. For example, PUMA is necessary for glucocorticoid- and phorbol ester-induced apoptosis in thymocytes, which occur normally in p53−/− cells,27 and serum starvation-induced apoptosis appears to be mediated by PUMA also in p53-deficient cancer cells.20 In these circumstances, it seems clear that this occurs through transcriptional upregulation of PUMA without requiring p53. Further study is necessary to determine whether IRF-1 has a critical role in induction of PUMA independent of p53 in other settings.
It is notable that in the PUMA promoter, there appear to be possible tandem repeats of the IRF-1-binding site, which correlate with increased expression by luciferase reporter assay and the PCR fragments from the ChIP procedure (Figure 4). It is interesting that Kirchhoff et al.28 did report on the in vitro and in vivo formation of IRF-1 homodimers, for which it was difficult to reconcile the role of this homodimer in binding to a single putative IRF-1-binding site; perhaps, instead the homodimer has a more specific role in the promoters of genes that contain tandem repeat IRF-1-binding sites rather than single sites. This would require further study.
It is also notable that the gastric cancer cell line AGS is deficient in Stat1 expression, which prevents IFN-γ stimulation from upregulating IRF-1 and subsequently PUMA. In the setting of intact Stat1 expression in the N87 cell line, IFN-γ can induce PUMA expression. In AGS, this may be a mechanism of cancer cell evasion from tumor immunosurveillance, as has been shown in numerous cancer cell lines.29 Although gene therapy with IRF-1 would be one option for treatment, other modes of upregulation or activation of IRF-1 independent of Stat1 expression are possible. For example, retinoic acid is known to upregulate IRF-1 independent of Stat1,30 and we have shown in AGS cells that increased IRF-1 protein and reporter activity can be seen with retinoic acid (unpublished data).
Earlier we found that IRF-1 induced a caspase-mediated apoptosis in breast cancer cells that was mediated by the FADD/caspase-8 interaction but did not involve any soluble ligands.5 It is tempting to speculate that the transcriptional upregulation of PUMA, which remains intracellular, accounts for this phenomenon; however, because further level of caspase-3 cleavage and apoptosis can be seen with IRF-1 expression in the absence of PUMA, it is presumed that this occurs via the extrinsic pathway, and there is little precedent for PUMA interaction with the extrinsic pathway. We further proved this by knockdown of both proteins Bax and Bak in AGS cells using RNA interference, which should eliminate intrinsic pathway apoptosis and once again found that although apoptosis was decreased by ∼50% and molecular markers of intrinsic pathway apoptosis were eliminated, caspase-8 and caspase-3 activation as shown by cleavage on western blot was still seen (Supplementary Figure 5). Because IRF-1 has been found to upregulate caspase-8 and other extrinsic-pathway-related proteins, it is certainly possible that intracellular action of these upregulated factors account for this phenomenon. The fact that IRF-1 has transcriptional targets in both the extrinsic and intrinsic pathways of apoptosis further validates its important role in apoptosis.
In conclusion, we have shown that IRF-1 can directly mediate apoptosis through the intrinsic pathway by the upregulation of PUMA. We have linked PUMA with the immune response to cancer through IRF-1 and IFN-γ. This supports the role for IRF-1 as a mediator of apoptosis in cancer cells both in tumor immunosurveillance and in the treatments of cancer, where apoptosis is seen. Furthermore, PUMA (i.e., bbc3), has been found to have decreased expression in patients with poor prognosis in breast cancer,31 and can provide prognostic information for colon cancer patients receiving chemotherapy.32 Thus, this link may also implicate the role of IRF-1 and IFNs in these settings.
Materials and Methods
Cell lines and culture
The human gastric cancer cell lines, AGS, NCI-N87 (N87), and KATO III, and the breast adenocarcinoma cell line MDA-MB-468 (MDA468) were purchased from ATCC (Manassas, VA, USA). HCT116 wild-type and HCT116 PUMA−/−33 were provided by L Zhang with the permission of B Vogelstein. Gastric cancer cells were propagated in RPMI 1640 medium (BioWhittaker, Walkersville, MD, USA) with 10% fetal bovine serum (FBS, 20% for KATO III, FBS; Atlanta Biologicals, Norcross, GA, USA), 1% penicillin/streptomycin, and 1% L-glutamine (Invitrogen, Carlsbad, CA, USA). MDA468 tumor cells were propagated in 1 : 1 Dulbecco's modified Eagle's medium (DMEM; BioWhittaker) and Ham's F-12 (Invitrogen) with 10% FBS, 1% L-glutamine and 1% penicillin/streptomycin. HCT116 and HCT116 PUMA−/− tumor cells were propagated in DMEM with 10% FBS, 1% L-glutamine and 1% penicillin/streptomycin. Tetracycline-inducible IRF-1 stably transfected AGS cells (AGSI) were propagated with 10% Tet System Approved FBS from Clontech (Mountain View, CA, USA), 10 μg/ml Blasticidin and 100 μg/ml Zeocin. All cells were incubated at 37°C in a humidified atmosphere of 5% CO2.
Chemicals and reagents
Annexin V–FITC apoptosis detection kit and z-IETD-fmk (caspase-8 inhibitor) were purchased from BD Pharmingen (San Jose, CA, USA). Mitochondria Isolation Kit was purchased from Thermo Scientific (Rockford, IL, USA). Brilliant II SYBR Green QRT-PCR Master Mix Kit was purchased from Stratagene (La Jolla, CA, USA). Antibodies (Abs): mouse anti-PARP monoclonal antibody (BD Pharmingen); rabbit polyclonal Abs to caspase-3, cleaved caspase-9, and Bid; and monoclonal Abs (mAbs, clone if listed) to caspase-8 (1C12) (Cell Signaling Technology, Danvers, MA, USA); rabbit polyclonal Abs to IRF-1 (Santa Cruz Biotechnology, Santa Cruz, CA, USA); anti-cytochrome c mouse mAb (BioVision, Mountain View, CA, USA); and anti-PUMA rabbit Ab (Sigma, St Louis, MO, USA). Dimethylsulfoxide (DMSO) was purchased from ATCC.
Construction and infection of Ad-IRF-1
The construction and characterization of E1, E3-deleted non-replicating recombinant adenoviruses Ad-Ψ5 (empty vector control, Ad-Null), Ad-IRF-1, and Ad-EGFP (enhanced green fluorescent protein) were described previously.16 Optimal MOI was determined by using Ad-EGFP and by measuring the percentage of fluorescent cells 24 h later.
Construction and transfection of inducible IRF-1 expression plasmids
To construct an inducible IRF-1 expression system, we used the T-REx System from Invitrogen, which includes the regulatory vector (TetR expression plasmid) pcDNA6/TR and the expression vector pcDNA4/TO/myc-His. The IRF-1 gene was cloned into the expression vector pcDNA4/TO/myc-His under a strong CMV promoter with two copies of the Tet operator in tandem (TetO2) in the promoter. AGS cells were transfected with pcDNA 6/TR and stably transfected clones were selected against 10 μg/ml Blasticidin (Invitrogen). TetR expression was examined by western blot analysis using an anti-TetR antibody. One of the clones that expressed the strongest TetR was used for further transfection. The cells were transfected with the pcDNA 4-IRF-1 expression vector and the stably transfected clones were selected against 100 μg/ml Zeocin (Invitrogen). Tetracycline (0–10 μg/ml) was given to each clone for 24 h and the expression of IRF-1 in each clone was confirmed by western blot analysis. AGS clones were designated AGSI and MDA468 clones were designated Pim.
Apoptosis analysis by flow cytometry
Annexin V–FITC apoptosis analysis was carried out with Annexin V–FITC Apoptosis Detection Kit from BD Pharmingen according to the protocol. Apoptosis was assessed by flow cytometry using a Becton Dickinson FACSort (Franklin Lakes, NJ, USA).
Immunoblotting analysis
Western immunoblotting was carried out as described previously.16
Cellular fractionation
AGS cells were treated with no treatment, Ad-Null, or Ad-IRF-1 (MOI of 50) for 30 h. Cellular fractionation followed Mitochondria Isolation Kit (Thermo Scientific) instructions.
Quantitative real-time PCR analysis
Total RNA was isolated from treated cells using TRIzol Reagent (Invitrogen) according to the standard protocol. The qRT-PCRs were carried out using a Stratagene MX3000P System and BrilliantII SYBR Green QRT-PCR Master Mix Kit (Stratagene). The reactions contained 1 × Brilliant SYBR Green QPCR Master Mix, 30 nM ROX reference dye, each primer at 200 nM, 200 ng RNA, and 1 μl RT/RNase block enzyme mixture in a 25 μl reaction. All the reactions were carried out as the following conditions: 30 min at 50°C, 10 min at 95°C, and 40 cycles of 30 s at 95°C, 1 min at 55°C and 30 s at 72°C in 96-well optical reaction plates (Stratagene). The dissociation curve analysis was carried out at the end of amplification to confirm PCR product specificity. Fluorescence data were collected at the end of the extension step. The results were automatically determined using the MxPro software (Stratagene). The following primers were used:
PUMA Forward: 5′-ACGACCTCAACGCACAGTACGA-3′ and
Reverse: 5′-GCAGGAGTCCCATGATGAGATTGT-3′.
β-Actin forward : 5′-AGAAAATCTGGCACCACACC-3′ and
Reverse: 5′-CCATCTCTTGCTCGAAGTCC-3′.
On each experiment no-template control and no-RT control were also performed at the same time. No signals were detected in no-template controls and no-RT controls. β-Actin was used as a endogenous control.
Luciferase reporter gene assay
PUMA promoter luciferase constructs were constructed in the laboratory of J Yu20 MDA468 and AGS cells plated in 12-well plates were co-transfected with PUMA luciferase reporter constructs and β-gal plasmid using Lipofectamine 2000 Reagent (Invitrogen). Cells were infected the next day with indicated adenovirus at 25 MOI. After 24 h of infection, the cells were harvested, lysed, and the lysate was used for luciferase assay using the luciferase assay system (Promega, Madison, WI, USA) and readout of luciferase activity was recorded using a Autolumat LB953 luminometer (EG&G Flow Technology, Nashua, NH, USA). The luciferase activities were normalized to β-galactosidase to obtain relative luciferase activity.
ChIP assay
ChIP was carried out using the EZ-Magna ChIP A Chromatin Immunoprecipitation Kit Assay kit (Upstate Biotechnology, Lake Placid, NY, USA), according to manufacturer's instructions with minor modifications. Briefly, AGS cells were infected with indicated adenovirus at 25 MOI, and AGSI cells were treated with 2 μg/ml tetracycline. After 24 h, formaldehyde (1% final concentration) was directly added to the culture medium and incubated at 37°C for 10 min for cross-linking. Cross-linking was stopped by adding glycine to a final concentration of 0.125 M and incubating for 5 min at RT. Cells were washed twice with ice-cold PBS containing protease inhibitors, scraped, and resuspended in 0.5 ml cell lysis buffer with a protease inhibitor cocktail, then sonicated with four sets of 10 s pulses by a Model 100 Sonic Dismembrator (Fisher Scientific) to obtain DNA fragments in the range of 200–1000 bp. Extracts were incubated with 5 μg of control rabbit IgG (negative control) or anti-IRF-1 rabbit polyclonal Ab (Santa Cruz Biotechnology) and with protein A magnetic beads at 4°C overnight with rotation. Washed immunoprecipitates were digested with proteinase K followed by a 2 h incubation at 65°C to reverse the cross-linking. DNA was then purified and assessed using real-time quantitative PCR. PCR was carried out with primers surrounding the putative tandem repeat binding sites for IRF-1 from PUMA Fragment C (182 bp) 5′-CCACGTCGTGGAAGCAGC-3′ (forward) and 5′-ACAGGAATCCACGGCTTTG-3′ (reverse), and from Fragment D (161 bp) 5′-GGTTTACTCATGACCCACAG-3′ (forward) and 5′-GCAGGTAATCTATATCTCACG-3′ (reverse). Real-time PCR was carried out under the following conditions: 50 cycles of 30 s denaturation at 94°C and 1 min anneal and extension at 60°C.
Transient siRNA transfection
siRNA targeting IRF-1 (cat. no. 16708), caspase 8 (cat. no. 4392421), and negative control siRNA (cat. no. 4390844) were purchased from Ambion (Austin, TX, USA). siRNA targeting Bid (cat. no. L-004387-00), PUMA (cat. no. L-004380-00), and non-targeting control siRNA (cat. no. D-001810-01-05) were purchased from Dharmacon (Lafayette, CO, USA). Transfection was performed with Lipofectamine RNAiMAX (Invitrogen), according to the manufacturer's instructions. Cells were transfected at a concentration of 25 nM siRNA twice on sequential days to enhance knockdown. Reverse and forward transfections were carried out in each experiment.
Statistics
Data are presented as means±S.D. in each figure. Comparisons between values were analyzed using analysis of variance (ANOVA) or t-test. Differences were considered significant at P-values ⩽0.05.
Conflict of interest
The authors declare no conflict of interest.
Abbreviations
- IFN:
-
interferon
- IRF-1:
-
interferon regulatory factor-1
- PUMA:
-
p53 upregulated modulator of apoptosis
- PI:
-
propidium iodide
- TNF:
-
tumor necrosis factor
- IL:
-
interleukin
- ChIP:
-
chromatin immunoprecipitation
- PS:
-
phosphatidylserine
- PARP:
-
poly(ADP-ribose) polymerase
- MOI:
-
multiplicity of infection
References
Tamura T, Yanai H, Savitsky D, Taniguchi T . The IRF family transcription factors in immunity and oncogenesis. Annu Rev Immunol 2008; 26: 535–584.
Tamura T, Ishihara M, Lamphier MS, Tanaka N, Oishi I, Aizawa S et al. DNA damage-induced apoptosis and Ice gene induction in mitogenically activated T lymphocytes require IRF-1. Leukemia 1997; 11 (Suppl 3): 439–440.
Bouker KB, Skaar TC, Fernandez DR, O’Brien KA, Riggins RB, Cao D et al. Interferon regulatory factor-1 mediates the proapoptotic but not cell cycle arrest effects of the steroidal antiestrogen ICI 182,780 (Faslodex, Fulvestrant). Cancer Res 2004; 64: 4030–4039.
Bowie ML, Dietze EC, Delrow J, Bean GR, Troch MM, Marjoram RJ et al. Interferon-regulatory factor-1 is critical for tamoxifen-mediated apoptosis in human mammary epithelial cells. Oncogene 2004; 23: 8743–8755.
Stang MT, Armstrong MJ, Watson GA, Sung KY, Liu Y, Ren B et al. Interferon regulatory factor-1-induced apoptosis mediated by a ligand-independent fas-associated death domain pathway in breast cancer cells. Oncogene 2007; 26: 6420–6430.
Clarke N, Jimenez-Lara AM, Voltz E, Gronemeyer H . Tumor suppressor IRF-1 mediates retinoid and interferon anticancer signaling to death ligand TRAIL. EMBO J 2004; 23: 3051–3060.
Park SY, Seol JW, Lee YJ, Cho JH, Kang HS, Kim IS et al. IFN-gamma enhances TRAIL-induced apoptosis through IRF-1. Eur J Biochem 2004; 271: 4222–4228.
El Bougrini J, Pampin M, Chelbi-Alix MK . Arsenic enhances the apoptosis induced by interferon gamma: key role of IRF-1. Cell Mol Biol (Noisy-le-grand) 2006; 52: 9–15.
Beppu K, Morisaki T, Matsunaga H, Uchiyama A, Ihara E, Hirano K et al. Inhibition of interferon-gamma-activated nuclear factor-kappa B by cyclosporin A: a possible mechanism for synergistic induction of apoptosis by interferon-gamma and cyclosporin A in gastric carcinoma cells. Biochem Biophys Res Commun 2003; 305: 797–805.
Suk K, Chang I, Kim YH, Kim S, Kim JY, Kim H et al. Interferon gamma (IFNgamma) and tumor necrosis factor alpha synergism in ME-180 cervical cancer cell apoptosis and necrosis. IFNgamma inhibits cytoprotective NF-kappa B through STAT1/IRF-1 pathways. J Biol Chem 2001; 276: 13153–13159.
Ruiz-Ruiz C, Ruiz de Almodovar C, Rodriguez A, Ortiz-Ferron G, Redondo JM, Lopez-Rivas A . The up-regulation of human caspase-8 by interferon-gamma in breast tumor cells requires the induction and action of the transcription factor interferon regulatory factor-1. J Biol Chem 2004; 279: 19712–19720.
Chow WA, Fang JJ, Yee JK . The IFN regulatory factor family participates in regulation of Fas ligand gene expression in T cells. J Immunol 2000; 164: 3512–3518.
Pizzoferrato E, Liu Y, Gambotto A, Armstrong MJ, Stang MT, Gooding WE et al. Ectopic expression of interferon regulatory factor-1 promotes human breast cancer cell death and results in reduced expression of survivin. Cancer Res 2004; 64: 8381–8388.
Tamura G, Ogasawara S, Nishizuka S, Sakata K, Maesawa C, Suzuki Y et al. Two distinct regions of deletion on the long arm of chromosome 5 in differentiated adenocarcinomas of the stomach. Cancer Res 1996; 56: 612–615.
Nozawa H, Oda E, Ueda S, Tamura G, Maesawa C, Muto T et al. Functionally inactivating point mutation in the tumor-suppressor IRF-1 gene identified in human gastric cancer. Int J Cancer 1998; 77: 522–527.
Kim PK, Armstrong M, Liu Y, Yan P, Bucher B, Zuckerbraun BS et al. IRF-1 expression induces apoptosis and inhibits tumor growth in mouse mammary cancer cells in vitro and in vivo. Oncogene 2004; 23: 1125–1135.
Jin Z, El-Deiry WS . Overview of cell death signaling pathways. Cancer Biol Ther 2005; 4: 139–163.
Cohen GM . Caspases: the executioners of apoptosis. Biochem J 1997; 326 (Part 1): 1–16.
Yin XM . Signal transduction mediated by Bid, a pro-death Bcl-2 family proteins, connects the death receptor and mitochondria apoptosis pathways. Cell Res 2000; 10: 161–167.
Ming L, Sakaida T, Yue W, Jha A, Zhang L, Yu J . Sp1 and p73 activate PUMA following serum starvation. Carcinogenesis 2008; 29: 1878–1884.
Takaoka A, Tamura T, Taniguchi T . Interferon regulatory factor family of transcription factors and regulation of oncogenesis. Cancer Sci 2008; 99: 467–478.
Harada H, Taniguchi T, Tanaka N . The role of interferon regulatory factors in the interferon system and cell growth control. Biochimie 1998; 80: 641–650.
Yim JH, Ro SH, Lowney JK, Wu SJ, Connett J, Doherty GM . The role of interferon regulatory factor-1 and interferon regulatory factor-2 in IFN-gamma growth inhibition of human breast carcinoma cell lines. J Interferon Cytokine Res 2003; 23: 501–511.
Abril E, Real LM, Serrano A, Jimenez P, Garcia A, Canton J et al. Unresponsiveness to interferon associated with STAT1 protein deficiency in a gastric adenocarcinoma cell line. Cancer Immunol Immunother 1998; 47: 113–120.
Takaoka A, Hayakawa S, Yanai H, Stoiber D, Negishi H, Kikuchi H et al. Integration of interferon-alpha/beta signalling to p53 responses in tumour suppression and antiviral defence. Nature 2003; 424: 516–523.
Gomez-Benito M, Balsas P, Carvajal-Vergara X, Pandiella A, Anel A, Marzo I et al. Mechanism of apoptosis induced by IFN-alpha in human myeloma cells: role of Jak1 and Bim and potentiation by rapamycin. Cell Signal 2007; 19: 844–854.
Villunger A, Michalak EM, Coultas L, Mullauer F, Bock G, Ausserlechner MJ et al. p53- and drug-induced apoptotic responses mediated by BH3-only proteins puma and noxa. Science 2003; 302: 1036–1038.
Kirchhoff S, Schaper F, Oumard A, Hauser H . In vivo formation of IRF-1 homodimers. Biochimie 1998; 80: 659–664.
Dunn GP, Koebel CM, Schreiber RD . Interferons, immunity and cancer immunoediting. Nat Rev 2006; 6: 836–848.
Percario ZA, Giandomenico V, Fiorucci G, Chiantore MV, Vannucchi S, Hiscott J et al. Retinoic acid is able to induce interferon regulatory factor 1 in squamous carcinoma cells via a STAT-1 independent signalling pathway. Cell Growth Differ 1999; 10: 263–270.
van ‘t Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature 2002; 415: 530–536.
Sinicrope FA, Rego RL, Okumura K, Foster NR, O’Connell MJ, Sargent DJ et al. Prognostic impact of bim, puma, and noxa expression in human colon carcinomas. Clin Cancer Res 2008; 14: 5810–5818.
Yu J, Wang Z, Kinzler KW, Vogelstein B, Zhang L . PUMA mediates the apoptotic response to p53 in colorectal cancer cells. Proc Natl Acad Sci USA 2003; 100: 1931–1936.
Acknowledgements
J Gao was supported by the China Scholarship Council. JH Yim was supported by an NIH Grant K08CA098403 and the Susan G Komen Foundation for the Cure Grant BCTR0708040. L Zhang is supported by an NIH Grant R01CA106348. J Yu is supported by an NIH grant R01CA129829 and the FAMRI.
Author information
Authors and Affiliations
Corresponding author
Additional information
Edited by A Villunger
Supplementary Information accompanies the paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Supplementary information
Rights and permissions
About this article
Cite this article
Gao, J., Senthil, M., Ren, B. et al. IRF-1 transcriptionally upregulates PUMA, which mediates the mitochondrial apoptotic pathway in IRF-1-induced apoptosis in cancer cells. Cell Death Differ 17, 699–709 (2010). https://doi.org/10.1038/cdd.2009.156
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cdd.2009.156
Keywords
This article is cited by
-
Identification of STXBP6-IRF1 positive feedback loop in regulation of PD-L1 in cancer
Cancer Immunology, Immunotherapy (2021)
-
TNF-α synergises with IFN-γ to induce caspase-8-JAK1/2-STAT1-dependent death of intestinal epithelial cells
Cell Death & Disease (2021)
-
iNOS/NO is required for IRF1 activation in response to liver ischemia-reperfusion in mice
Molecular Medicine (2020)
-
Human pluripotent stem cell-derived cardiomyocytes as a target platform for paracrine protection by cardiac mesenchymal stromal cells
Scientific Reports (2020)
-
Anti-apoptotic effect by the suppression of IRF1 as a downstream of Wnt/β-catenin signaling in colorectal cancer cells
Oncogene (2019)