Abstract
Caspase-8, a cysteine-protease, initiates apoptosis when activated by death receptors. Caspase-8 is also essential for initiating T lymphocyte proliferation following T-cell antigen receptor (TCR) signaling. Given these disparate functions of caspase-8, we sought to determine whether this represented only a difference in the magnitude of caspase-8 activation, or different intracellular locations of active caspase-8. We demonstrate by high-resolution multicolor confocal laser scanning microscopy an aggregation of active caspase-8 within membrane lipid rafts in T cells stimulated with anti-CD3. This suggests that following TCR stimulation active caspase-8 physically interacts with lipid raft proteins, possibly to form a signaling platform. In contrast, Fas stimulation of T cells resulted in a much more profound activation of caspase-8 that was exclusively cytosolic. These confocal microscopic findings were confirmed using discontinuous sucrose gradient ultracentrifugation to isolate lipid raft versus cytosolic components. This sequestration model of caspase-8 activation was further supported by the observation that a classic caspase-8 substrate, BID, was not cleaved in CD3-stimulated T cells, but was cleaved after Fas engagement. Our data support a model that the location of active caspase-8 may profoundly influence its functional capacity as a regulator of either cell cycling or cell death.
Similar content being viewed by others
Main
Caspases are cysteine proteases with the ability to cleave proteins after certain aspartic acid residues.1 They are involved with a variety of cell processes, including cell death,2 inflammation,3 and in some instances cell growth.4 Caspases are present in the cytoplasm as inactive procaspases or zymogens.5 Caspase classification distinguishes between initiator (apical) caspases, activated by death receptor clustering, and effector (executioner) caspases. Initiator caspases (e.g., caspases-2, -8, -9, and -10) cleave inactive pro-forms of effector caspases, as well as substrates such as BID, which promote mitochondrial permeability.6 Effector caspases (e.g., caspases-3, -6, and -7) in turn cleave downstream protein substrates that promote the apoptotic process.7
The structural parameters of caspase-8 activation by engagement of death receptors, and the subsequent signaling cascade, have been well characterized.8 The commonly accepted ‘induced-proximity model’ proposes an oligomerization of death receptors upon triggering with appropriate ligands, which allows recruitment of the adapter molecule FADD by their mutual death domains. As a signaling scaffold FADD subsequently attracts procaspase-8 by their common death effector domains, which allows a clustering of the inactive procaspase-8 to form the death-inducing signaling complex (DISC).9 DISC formation leads to an autoproteolytic cleavage of procaspase-8 and a rearrangement of the cleaved domains to form the fully active, heterotetrameric caspase-8.10
Recent findings indicate an entirely separate function for caspase-8 in T lymphocyte activation. Humans lacking functional caspase-8 develop an immunodeficiency syndrome characterized by a decreased proliferative capacity of T cells.11 A deficiency of caspase-8 in murine T cells also diminishes their capacity to proliferate.12 This was accompanied by decreased NF-κB activity and IL-2 production following T-cell antigen receptor (TCR) ligation,11 thus making caspase-8 indispensable for lymphocyte proliferation.
Lipid rafts, cholesterol-, and sphingolipid-rich plasma membrane domains, orchestrate protein trafficking, and potentiate intracellular signaling.13, 14 In the context of lymphocyte signaling, lipid rafts have been suggested to assemble as platforms for the regulation and transduction of TCR signals at the plasma membrane.15 However, the concept of intracellular protein associations being regulated by membrane-microdomain-associated clusters constitutes a spatial model for the regulation of cell signaling.16
We recently reported, using discontinuous sucrose gradient ultracentrifugation analysis, that although essentially all caspase-8 in resting T cells was inactive and localized in the non-raft cytosol, within 30 min of T-cell activation, a small amount of total caspase-8 became active and appeared only in the lipid raft fraction.17 These findings suggested that spatially concentrating active caspase-8 in lipid rafts was not only important for T-cell activation, but also confining it to this region might be critical in preventing active caspase-8 from accessing substrates that might promote cell death. However, the definition of lipid rafts as a detergent-resistant membrane fraction remains controversial because the purification process has the possibility of creating detergent-induced artifacts, and therefore may not indicate the true state of proteins and lipids in native membranes.18 This biochemical approach also lacked the ability to analyze individual cells. Direct imaging of individual lipid rafts in their cellular context presents its own challenges. Lipid rafts lack clear morphological features, and their putative size ranges below the resolution of commonly used optical microscopes, hence necessitating the use of fluorescent-labeled proteins associated with lipid rafts, such as monosialotetra-hexosylgangliosid (GM1) and glycosylphosphatidyl-inositol (GPI)-anchored proteins.19
Given the seemingly paradoxical involvement of caspase-8 in T-cell growth and death, and the inherent technical concern with biochemical lipid raft fractionation, we therefore sought to investigate the kinetics and spatial patterns of caspase-8 activation following TCR stimulation versus Fas (CD95)-mediated apoptosis using confocal laser scanning microscopy (CLSM). To identify lipid rafts we used Jurkat T lymphocytes expressing enhanced GFP fused to the minimal membrane-anchoring sequence of the tyrosine kinase Lck (Jurkat L10-GFP).20 Here we demonstrate by high-resolution multicolor CLSM the rapid appearance of active caspase-8 in discrete small foci within membrane lipid rafts of T cells following CD3 stimulation. In contrast, Fas stimulation of T cells resulted in a much more profound activation of caspase-8 that was exclusively cytosolic. These results were substantiated using traditional discontinuous sucrose gradient ultracentrifugation to isolate lipid raft versus cytosolic components. The findings suggest that active caspase-8 is sequestered in different cellular compartments following activation for proliferation versus apoptosis, which may profoundly alter access to caspase-8 substrates.
Results
Active caspase-8 localizes to distinct cellular regions upon TCR versus Fas stimulation
Caspase-8 is activated and required in T cells following either TCR stimulation or Fas engagement, yet the outcome is proliferation in the former and cell death in the latter. The total amount of caspase-8 does not change during the transition of T lymphocytes from a resting to a proliferating state.21, 22 However, in contrast to freshly isolated T cells, a small fraction of total caspase-8 becomes active in effector T cells, and this is critical for their initiation of proliferation.17 These findings lead to the hypothesis that depending on the involved receptor, caspase-8 is activated to different degrees, in different cellular locations, and that after TCR engagement active caspase-8 interacts with lipid raft molecules to serve as a signal platform. We thus designed an approach to visualize caspase-8 activation within individual cells following TCR versus Fas stimulation.
Initial experiments to visualize active caspase-8 by CSLM in lymphocytes utilized the wild-type A3 subclone of the human Jurkat T lymphocyte cell line. This subclone is sensitive to Fas-induced apoptosis, which generates high levels of active caspase-8.23 The I9.2 cell line is a caspase-8-deficient mutant of the A3 Jurkat cell line,24 and was used in these studies as a negative control for caspase-8 staining. Active caspase-8 detection by CLSM employed an affinity label methodology, initially using the caspase-8-preferred substrate leucine-glutamic acid-threonine-aspartic acid (LETD)25 linked to a fluoromethylketone (fmk) moiety, which reacts covalently with the catalytic cysteine residue in the active enzymatic center.26 A 6-carboxyfluorescein (FAM) group linked to LETD-fmk was used as a reporter. Nuclear morphology was evaluated by staining with Hoechst 33342.
Figure 1 displays a stack of n optical slices (n=10–12, thickness 1 μm each) along the z axis (depth) in the sample. Unstimulated Jurkat T cells incubated with FAM-LETD-fmk generated no detectable fluorescence signal, indicating that levels of active caspase-8 were low to negligible in these cells (Figure 1a–d). By contrast, active caspase-8 was observed in both CD3-activated (Figure 1e–h) as well as Fas-stimulated cells (Figure 1i–l) 60 min after stimulation. The staining pattern and intensity after the two stimuli, however, were significantly different. Following CD3 stimulation active caspase-8 was confined to a peripheral region of the cell (Figure 1f). In contrast, lymphocytes stimulated by Fas featured active caspase-8 as an extensive intracellular accumulation, distributed throughout the cytoplasm (Figure 1j). Furthermore, CD3-activated cells displayed intact nuclei (Figure 1e), whereas Fas stimulation caused a nuclear condensation and blebs, reflecting the induction of cell death (Figure 1i).
Distinct spatial differences of active caspase-8 after T-cell antigen receptor (TCR) engagement versus Fas stimulation. Two-color confocal laser scanning microscopy (CLSM) of wild-type A3 Jurkat T cells either unstimulated (a–d), anti-CD3-stimulated (e–h), or anti-Fas-stimulated (i–l). Active caspase-8 was then labeled with 6-carboxyfluorescein (FAM)-leucine-glutamic acid-threonine-aspartic acid (LETD)-fluoromethylketone (fmk, green) and nuclear integrity was monitored by Hoechst 33342 counterstaining (blue). Orthogonal views (d, h, l) were derived from 10–12 optical slices each. Negative controls to verify the specificity of the active caspase-8 staining included the caspase-8-deficient I9.2 Jurkat cell line, stimulated either with anti-CD3 (m), or anti-Fas (n), and the blocking of caspases by benzyloxycarbonyl-valine-alanine-aspartic acid (zVAD)-fmk prior to stimulation with anti-CD3 (o), or anti-Fas (p) and FAM-LETD-fmk labeling. Bar represents 15 μm
The specificity of the FAM-LETD-fmk staining was verified by two methods. First, caspase-8-deficient I9.2 Jurkat cells were stimulated either by CD3 (Figure 1m) or Fas (Figure 1n). No staining with FAM-LETD-fmk could be observed following either stimulation in I9.2 cells. Second, wild-type Jurkat A3 cells were treated with the caspase blocker z-VAD-fmk, then stimulated either with anti-CD3 (Figure 1o) or anti-Fas (Figure 1p) and stained with FAM-LETD-fmk. Again no fluorescence was observed under these conditions.
To quantify the frequency of cells bearing active caspase-8 following CD3 versus Fas stimulation for 60 min, a minimum of 200 individual cells was counted from at least three independent experiments (Figure 2). Caspase-8 activity in unstimulated cells was rarely detected. By contrast 26% (±2.2) of CD3-stimulated cells exhibited detectable active caspase-8, whereas 65% (±4.3) of Fas-stimulated cells scored positive for caspase-8 activation (Figure 2a). As the microscopic samples were nuclear counterstained, a further evaluation of the levels of apoptosis was possible based on the amount of nuclear condensation and fragmentation. Anti-CD3-stimulated cells manifested no elevation of nuclear fragmentation. In contrast, 52% (±6.5) of anti-Fas-treated cells scored as apoptotic based on nuclear fragmentation (Figure 2b). The findings suggested that the location and level of active caspase-8 determines whether a cell initiates cell death.
T-cell antigen receptor (TCR) stimulation induces limited caspase-8 activity, but no nuclear fragmentation. A minimum of 200 cells per sample as shown in Figure 1 was evaluated microscopically for the proportion of active caspase-8 (a) versus nuclear fragmentation (b). Activities of caspase-8 and effector caspases 3 and 7 were measured, respectively, by an isoleucine-glutamic acid-threonine-aspartic acid (IETD)-luminescence (c) or rhodamine-DEVD release assay (d), using wild-type A3 Jurkat cells (open bars) or caspase-8-deficient I9.2 Jurkat cells (gray bars). Additionally, DNA strand breaks after CD3 or Fas stimulation were quantified by terminal-dUTP-nick-end-labeling (TUNEL) assay (e). (a.u.)=Arbitrary units
The results of caspase-8 activity defined by confocal microscopy in individual cells were confirmed by enzymatic measurement of the individual activities of caspase-8 (Figure 2c) versus the effector caspases-3 and -7 (Figure 2d) in unstimulated and CD3- or Fas-stimulated populations of Jurkat T cells. Whereas CD3 stimulation for 60 min caused only a slight increase of caspase-8 activity, Fas engagement for the same time generated high levels of activity of both the initiator as well as effector caspases. These results paralleled the microscopic observation that CD3 stimulation activated caspase-8, yet the level of caspase-8 activity was considerably lower than T cells undergoing Fas-mediated cell death.
To further correlate the level of caspase-8 activity with actual cell death, cells were stimulated under identical conditions as for confocal microscopy and then stained by terminal-dUTP-nick-end-labeling (TUNEL) assay, and apoptotic cells quantified by flow cytometry (Figure 2e). Both unstimulated, as well as CD3-stimulated Jurkat cells showed only marginal labeling, indicating a negligible level of DNA damage. In comparison 24% (±1.4) of Fas-stimulated lymphocytes were TUNEL-positive, consistent with the results by nuclear fragmentation.
Colocalization of active caspase-8 with CD3 in lipid rafts
As TCR proteins associate in lipid rafts upon stimulation,27 we examined whether the peripheral cellular pattern of active caspase-8 that we observed following TCR ligation represented localization to membrane lipid rafts. To define lipid rafts for CLSM we used Jurkat T cells stably expressing a fusion protein of enhanced green fluorescent protein (eGFP) and a 10 amino-acid sequence derived from the membrane-anchor domain of the tyrosine kinase Lck (Jurkat L10-GFP), consequently targeting eGFP to the lipid raft fraction of the cellular membrane.20, 28
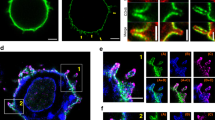
Jurkat L10-GFP cells were stimulated with either anti-CD3 or anti-Fas antibodies for up to 90 min, and subsequently stained with the caspase-8-specific substrate rhodamine-isoleucine-glutamic acid-threonine-aspartic acid (IETD)-fmk, and Hoechst 33342 nuclear counterstain (Figure 3). Active caspase-8, indicated now by red fluorescence, could be visualized in distinct patches in proximity to the GFP-labeled lipid rafts of the cellular membrane (Figure 3g and k). Active caspase-8 was detectable in discrete foci at 30 min after CD3 activation, and continued to be detectable in the same location after 90 min of stimulation (Figure 3e–l). However, following Fas stimulation for 30 min active caspase-8 staining was entirely cytoplasmic (Figure 3m–p), and at 90 min was still present in the cytoplasm (Figure 3q–t). Cellular alterations, indicating the initiation of apoptotic processes, could be distinguished as deterioration of the cellular membrane (fading green fluorescence of eGFP) and nuclear fragmentation. In some Fas-stimulated cells, in addition to the clearly central cytoplasmic staining for active caspase-8, there was also the appearance of an annular fluorescence of active caspase-8 (Figure 3o). However, more detailed analysis revealed that this area of active caspase-8 was just internal of eGFP-labeled membranes. First, there was no colocalization of active caspase-8 with lipid rafts based on merged images (Figure 3p), as well as calculated colocalization coefficients (Table 1). Furthermore, measurements of the fluorescence boundaries of red active caspase-8 were clearly within the border of green lipid raft fluorescence (Supplementary Figure S1). Finally, biochemical lipid raft separation did not reveal active caspase-8 in lipid rafts following Fas stimulation (see Figure 7).
Active caspase-8 is detectable in lipid rafts after stimulation with anti-CD3, but remains cytosolic after anti-Fas stimulation. Three-color confocal laser scanning microscopy (CLSM) of unstimulated (a–d), anti-CD3-stimulated (e–l), or anti-Fas-stimulated (m–t) wild-type Jurkat cells expressing L10-GFP (green) to identify lipid rafts. Cells were stimulated for either 30 or 90 min prior to staining with rhodamine-isoleucine-glutamic acid-threonine-aspartic acid (IETD)-fluoromethylketone (fmk) for active caspase-8 and Hoechst 33342 for nuclear integrity. Despite some annular fluorescence of active caspase-8 in (o), the GFP-fluorescence of the membrane did not merge with the red fluorescence of active caspase-8 (p), indicating no colocalization. Bar represents 10 μm
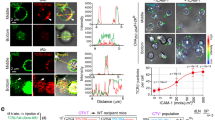
Colocalization in fluorescence imaging characterizes the overlap extent between two different fluorescent labels with different emission wavelengths. The detection of fluorescence signals from two differently labeled proteins within the same voxel (three-dimensional pixel) determines that these proteins are located in the same area or very near to each other. Two perfectly colocalized fluorescence signals, each displayed on separate x and y axes, will generate a scatter plot wherein the points fall in a line at 45° to either axis. In the situation of non-colocalized molecules, the resulting scatter plot reveals each color along its own axis, with no overlap at 45°. Figure 4 shows representative three-dimensional remodeling of Jurkat cells (based on an average of 50 z-stacked images) after CD3 (Figure 4a) or Fas (Figure 4b and c) stimulation, with the respective scatter plot generated from representative regions of interest (white rectangles) within these images. Following CD3 stimulation, the lipid raft fluorescence (green) and active caspase-8 fluorescence (red) were largely overlapping, as indicated in the scatter plot at 45° (Figure 4d). However, following Fas stimulation, active caspase-8 did not overlap with lipid raft fluorescence, but was entirely cytosolic (Figure 4e and f). Calculations of Pearson's correlation coefficient of colocalization are shown in Table 1 and demonstrate that a significant colocalization between active caspase-8 and lipid rafts exists only after CD3 stimulation.
Active caspase-8 colocalizes with lipid rafts after CD3 stimulation. Three-dimensional reconstruction of Jurkat cells expressing L10-GFP after anti-CD3 stimulation (a) or anti-Fas stimulation (b, c), on which is based the calculation for the colocalization scatter plots of active caspase-8 with lipid rafts after anti-CD3 stimulation (d) or anti-Fas stimulation (e, f). Colocalization is defined as the detection of one fluorescence location in both of two acquired fluorescence channels. Colocalized pixels fall on a 45° line between the axes of the two fluorophores (lower panels). Each axis represents the increasing fluorescence intensity present within analyzed voxels. Coefficients for colocalization of active caspase-8 with lipid rafts and stimulating antibodies are displayed in Table 1
Conceivably the strong cytosolic pattern of active caspase-8 observed with a particular dose of anti-Fas, examined at a particular time, may have obscured an additional membrane component of active caspase-8. We examined this possibility in two ways. First, the concentration of anti-Fas antibody was titered down and caspase-8 activation monitored at 90 min. Despite the fact that the proportion of cells bearing active caspase-8 decreased with lower anti-Fas concentrations, we were not able to observe a lipid raft colocalization of caspase-8 (Supplementary Figure S2). Second, the concentration of anti-Fas antibody was kept constant and cells were analyzed at several early time points. After 10 min of stimulation active caspase-8 became detectable; however, at this and all later time points it was present only in the cytoplasm (Supplementary Figure S3).
To examine the real-time kinetics of caspase-8 activation, live Jurkat T cells were observed at intervals of 1 min in an environmental chamber mounted on the CSLM (Figure 5). Active caspase-8 became detectable as distinct patches in the membrane as early as 26 min after CD3 stimulation (Figure 5b). This activation pattern remained stable for the 2 h duration of the recording (Figure 5d). Of interest was that following Fas stimulation active caspase-8 was observed in distinct foci in the cytoplasm as early as 8 min (Figure 5f), but at no time point did active caspase-8 colocalize with lipid rafts (see image sequences in Supplementary Figure S4 and S5). Unstimulated cells did not show active caspase-8 at any time during the observation (data not shown).
Kinetics and levels of active caspase-8 differ markedly after anti-CD3 versus anti-Fas stimulation. Live cells in culture medium were equilibrated with rhodamine-isoleucine-glutamic acid-threonine-aspartic acid (IETD)-fluoromethylketone (fmk) substrate prior to anti-CD3 stimulation (a–d) or anti-Fas stimulation (e–h). Red fluorescence indicates active caspase-8 and was recorded in real time by confocal laser scanning microscopy (CLSM). Representative time points are shown. Measurable active caspase-8 was detectable 26 min after addition of anti-CD3 antibody and remained confined to lipid rafts (arrows indicate loci of active caspase-8). By contrast, active caspase-8 was observed after 8 min following anti-Fas stimulation, and was localized solely in the cytoplasm. Bar represents 20 μm
In further studies, the site of the stimulating anti-CD3 or anti-Fas antibodies on the cell surface was determined by an additional staining using an Alexa633-conjugated secondary antibody (Figure 6). Active caspase-8 staining was detected in close proximity to CD3 (Figure 6e). Stimulating anti-Fas antibodies were also detectable on the cell surface, but due to the broadly cytoplasmic distribution of active caspase-8, Fas was not in proximity to active caspase-8 (Figure 6j). Table 1 summarizes calculated overlap coefficients for active caspase-8/lipid raft, active caspase-8/anti-CD3, or active caspase-8/anti-Fas colocalization, from representative experiments shown in Figures 4 and 6.
Active caspase-8 colocalizes with lipid rafts in the vicinity of CD3. Four-color confocal laser scanning microscopy (CLSM) of L10-GFP-expressing Jurkat cells after anti-CD3 (a–e) or anti-Fas (f–j) stimulation. Cells were stimulated for 90 min prior to staining with rhodamine-isoleucine-glutamic acid-threonine-aspartic acid (IETD)-fluoromethylketone (fmk) for active caspase-8, Hoechst 33342 for nuclear morphology, and goat-anti-mouse immunoglobulin conjugated to Alexa Fluor 633 for detection of stimulating anti-CD3 or anti-Fas antibodies. Bar represents 10 μm
Confirmation of the confocal microscopic localizations of active caspase-8 was assessed by biochemical lipid raft separation. Jurkat cells were stimulated with anti-CD3 or anti-Fas for 90 min and then further incubated with biotin-VAD-fmk for an additional 15 min to selective label active caspases. Cells were lysed, fractionated by a discontinuous sucrose gradient ultracentrifugation, and lysates were collected as 12 1 ml fractions. Immunoblots of lipid rafts (fractions 3–5) and cytoplasm (fractions 9–11) revealed that a small proportion of cleaved p43caspase-8 was present in the lipid raft fraction 5 of unstimulated Jurkat cells. However, a much larger proportion of uncleaved caspase-8 was present in the cytosolic fraction (Figure 7a). Following stimulation with anti-CD3, the amount of p43caspase-8 relative to full-length p55caspase-8 increased in the lipid raft fractions (Figure 7a and b). Intriguingly, p43caspase-8 disappeared from the lipid raft fractions after Fas stimulation, and became strongly present in cytosolic fraction 11, as was fully active pro-apoptotic p18caspase-8 (Figure 7a).
Active caspase-8 is present in lipid rafts of Jurkat cell lysates following CD3 stimulation. Jurkat wild-type cells were unstimulated, CD3-stimulated, or Fas-stimulated for 90 min, and further incubated with biotin-VAD-fluoromethylketone (10 μ M) during the final 15 min. Lysates were subjected to ultracentrifugation on a discontinuous sucrose gradient and then immunoblotted with anti-caspase-8 antibody (a). Lanes marked 3–5 represent lipid raft fractions; lanes marked 9–11 represent cytosolic fractions. After CD3 stimulation, the amount of p43caspase-8 increased in the lipid raft fraction, whereas it decreased after Fas stimulation and, instead, became the dominant form in the cytoplasm (a). Densitometry confirmed that the ratio of p43caspase-8 to p55caspase-8 increased in the lipid raft fraction 5 only after CD3 stimulation, whereas Fas stimulation led to an increased ratio only in the cytosolic fraction (b). The remainder of the same samples as used in (a) was subjected to precipitation of active caspase-8 using avidin-sepharose. Precipitates were then immunoblotted for caspase-8. This revealed that active caspase-8 appeared in the lipid raft fraction of CD3-stimulated cells, but in Fas-stimulated cells appeared only in the cytoplasm, including the pro-apoptotic p18 fragment (c). Lipid raft and cytosolic fractions were immunoblotted for the caspase substrates BID (d) and ICAD (e). NS=nonspecific band
Caspase-8 can also be active in its full-length form.30 Thus, by standard immunoblot it is not possible to determine whether any full-length caspase-8 is active. Hence, to more accurately define active caspase-8 in this system, the lipid raft and cytosolic fractions were individually precipitated from biotin-VAD-fmk-labeled lysates using streptavidin-sepharose beads, and then immunoblotted for active caspase-8 (Figure 7c). A small amount of active caspase-8 was detected exclusively in the cytosol of unstimulated cells. In contrast, CD3 stimulation resulted in the appearance of active p55caspase-8 and p43caspase-8 in the lipid raft fraction, whereas Fas-stimulated cells displayed very high levels of active caspase-8 in the p55 and p43 forms, as well as fully cleaved p18caspase-8, exclusively in the cytoplasm. Additionally, the traditional cytosolic caspase-8 substrate BID was not cleaved in any fraction following CD3 stimulation, but was cleaved in the cytosolic fraction after Fas stimulation (Figure 7d). A similar finding was also observed for another downstream caspase substrate, ICAD (Figure 7e). Thus, the findings using biochemical lipid raft separation closely parallel the results with the confocal imaging of active caspase-8.
Discussion
The current findings represent the first direct comparison of the distinct spatial differences of active caspase-8 in T cells following CD3 versus Fas stimulation using two independent methods. Extensive research on the plasma membrane of mammalian cells has demonstrated that it is not a uniform lipid bilayer, containing randomly distributed membrane, and associated proteins.31 Instead, the plasma membrane is believed to contain organized structural microdomains termed lipid rafts that are enriched in cholesterol and glycosphingolipid,13 and may serve as platforms that link activated receptors and signal transduction partners.32 Several lines of evidence indicate that TCR stimulation of T cells is accompanied by lipid membrane remodeling and recruitment into the immunologic synapse.27 The role of lipid rafts remains controversial, however, and it also has been suggested that they are randomly distributed during T-cell stimulation, and that the apparent enrichment of lipid rafts observed at the TCR contact site is the sole consequence of nonspecific membrane ruffling.33 Nonetheless, CD3 stimulation of T lymphocytes leads to a clustering of kinase-rich lipid raft microdomains in which Lck colocalizes with GPI-linked proteins.34 Consequently, we used two methods to investigate the spatial and kinetic differences of caspase-8 activation after TCR versus Fas stimulation.
By discontinuous sucrose gradient ultracentrifugation of lipid raft and cytosolic fractions, we previously observed in murine T cells evidence for limited activation of caspase-8 confined to lipid rafts, within 30 min of TCR stimulation.17 However, those experiments were not capable of visualizing the kinetics and spatial arrangement of active caspase-8 at the single-cell level. Furthermore, there is not uniform agreement about what precisely is defined anatomically by the detergent insoluble (lipid raft) fraction of cells. Yet, the independent methodology of confocal microscopy is in close agreement with our earlier biochemical analysis of caspase-8 activation following TCR ligation. The current findings further extend our previous observations by showing that Fas stimulation results in purely cytoplasmic active caspase-8. These results suggest a sequestration model in which TCR-induced caspase-8 activation occurs within a confined location of the cell membrane in proximity with the TCR/CD3 complex. This may serve to both limit caspase-8 activation as well as target it toward substrates associated with cell activation, and at the same time sequester active caspase-8 away from substrates that would promote cell death (e.g., BID).
The use of Jurkat cells stably expressing a fluorescent lipid raft marker (L10-GFP) enabled the direct examination of lipid raft dynamics during T-cell responses without the use of cholera toxin or specific antibodies to lipid raft components.35 Triggered TCRs associate within lipid rafts, and their crosslinking causes co-aggregation of raft-associated proteins, including the tyrosine-kinase Lck.36 L10-GFP is derived from the membrane-anchor sequence of Lck, and, although it may also label a small portion of the nonlipid raft membrane fraction, the fusion protein efficiently targets lipid rafts and concentrates in this membrane fraction.20, 34, 35, 37 Individual lipid rafts are too small to be defined by conventional microscopy, nevertheless this labeling has previously revealed that the basic raft units become large platforms that transport membrane proteins and signaling partners to specific cell sites during lymphocyte migration and activation.38 In our experiments using CLSM we were for the first time able to visually demonstrate the differences in the level and location of caspase-8 activation after CD3- or Fas stimulation at the single-cell level.
Caspase-8 accomplishes two apparently paradoxical cellular functions: full-length functional active caspase-8 is crucial for induction of T lymphocyte proliferation,39, 40 and yet caspase-8 is also an initiator of apoptosis. These seemingly opposite functions of caspase-8 demand a precise control mechanism. The sequestration model that caspase-8 is recruited to lipid rafts upon TCR activation is attractive, as the repertoire of caspase-8 substrates for growth versus death might be also spatially separated. The caspase-8 homologue and substrate, cFLIP, was also shown to be recruited to lipid rafts upon TCR ligation, and to be rapidly cleaved at its known caspase-8 cleavage site, Asp 376, to form p43FLIP only in lipid rafts.17 This supports the view that active caspase-8 is enriched in lipid rafts following TCR ligation. By contrast, the cytosolic caspase-8 substrate, BID, was not cleaved following TCR stimulation, but only after Fas activation.
At present, it is not known how active caspase-8 is recruited to membrane lipid rafts. CARMA1, MALT1, and Bcl-10 serve adapter functions in TCR signaling,41 and associate with each other to promote activation of the NF-κB pathway. These proteins also coprecipitated with active caspase-8 in proliferating murine effector T cells.17 CARMA1 has been reported to associate with TCRs in lipid rafts after stimulation, which makes it a potential candidate to engage active caspase-8 and translocate it to the cellular membrane.42
Materials and Methods
Cell culture
Jurkat wild-type (A3, ATCC, Manassas, VA, USA; CRL-2570) and Jurkat caspase-8-deficient (I9.2, ATCC CRL-2571) T lymphocyte cell lines were cultivated in RPMI 1640 medium (CellGro Mediatech, Herndon, VA, USA), supplemented with 10% fetal bovine serum (HyClone, Logan, UT, USA). Jurkat cell line L10-GFP, stably expressing a fusion protein of 10 amino acids of the minimal membrane anchor domain of Lck and enhanced GFP to label membrane lipid rafts, was produced as previously described.20
Antibodies and reagents used for cell staining
The following antibodies were used for short-term stimulation experiments: anti-CD3 (TR66)43 or anti-Fas (CH-11, Upstate, Temecula, CA, USA), at final concentrations, respectively, of 10 or 0.5 μg/ml, unless otherwise stated.
Antibodies used for western blot analysis included anti-caspase-8 (BD Pharmingen), anti-BID (R&D Systems, Minneapolis, MN, USA), anti-ICAD (Santa Cruz Biotechnology, Santa Cruz, CA, USA), and goat-anti-mouse, and donkey-anti-goat immunoglobulin conjugated to horseradish-peroxidase (Jackson Immunoresearch, West Grove, PA, USA).
Caspase-8-specific assay kits containing substrates for active caspase-8 were either FAM-LETD-fmk (Image-iT® LIVE Green Caspase-8 Detection Kit, Invitrogen, Carlsbad, CA, USA; λEx=488 nm, λEm=530 nm), or rhodamine-IETD-fmk (EMD, San Diego, CA, USA; λEx=540 nm, λEm=570 nm). The pan-caspase inhibitor benzyloxycarbonyl-valine-alanine-aspartic acid-fmk (MP Biomedicals, Aurora, OH, USA) was used as a competitive inhibitor for negative control staining.
Nuclear counterstaining was achieved by incubation with 1 μM Hoechst 33342 (λEx=350 nm, λEm=461 nm; Invitrogen). A secondary goat-anti-mouse Alexa Fluor 633 antibody (Invitrogen; λEx=632 nm, λEm=647 nm) was chosen for detection of surface-bound stimulating antibodies where indicated.
Caspase activity assays
A luminescence-based assay was used to determine the activity of caspase-8 (Caspase-Glo 8 Assay, Promega, Madison, WI, USA), whereas the activity of the effector caspases-3 and -7 were measured with a fluorescence-based assay (Apo-One Homogeneous Caspase-3/7 Assay, Promega). Assays were performed according to the manufacturer's recommendations.
DNA strand breaks by TUNEL assay
To assess the extent of apoptosis, TUNEL was performed using fluorescein isothiocyanate (FITC)-dUTP (Roche Diagnostics, Mannheim, Germany). Briefly, single-cell suspensions were first stimulated for 90 min with antibodies to CD3 or Fas, and then washed twice with cold PBS. Cells were then fixed with fresh 1% formaldehyde (Ted Pella, Redding, CA, USA), and after twice washing, permeabilized with ice-cold 70% ethanol. DNA strand break labeling was achieved by incubation at 37°C in labeling mix (1 × TdT buffer, 2.5 mM CoCl2, 1 U TdT (Roche Diagnostics), and 0.1 nM FITC-dUTP). Cells were washed twice in cold PBS/BSA and stored in PBS/BSA/1% formaldehyde until analyzed on a BD LSRII flow cytometer (BD Bioscience).
Lipid raft and cytosolic fraction isolation
Isolation of lipid raft and cytosolic fractions was achieved by ultracentrifugation over a discontinuous sucrose density gradient as previously described.17, 44 Before lysis, cells were incubated with biotin-VAD-fmk (MP Biomedicals) at 37°C for 15 min to label active caspases. After centrifugation, the lipid raft and cytosolic fractions were partly used for immunoblot analysis, and the remainder subjected to precipitation of the active caspases using avidin-sepharose (Zymed, San Francisco, CA, USA) followed by immunoblotting for caspase-8 as previously reported.17
Densitometry of immunoblots was performed using a Fluor-S MultiImager System with Quantity One software (Bio-Rad, Hercules, CA, USA).
Cell staining and CLSM
Jurkat cells were stimulated either with soluble anti-CD3 (TR66, 10 μg/ml) or anti-Fas (CH11, 0.5 μg/ml) antibodies for 30 and 90 min, unless stated otherwise in Supplementary Figures S1 and S2. At the indicated time points, cells were washed twice with cold PBS, and transferred to coverslips previously coated with poly-L-lysine (Sigma, St. Louis, MO, USA) to facilitate adherence, and fixed with 3.5% fresh paraformaldehyde. Finally, cells were mounted (three-color analysis) on glass slides with Aquamount (Polysciences, Warrington, PA, USA). In the case of four-color analysis, prior to mounting, cells were blocked with 5% BSA (Sigma) in PBS for 60 min, and then stained for 60 min with a secondary antibody to detect the stimulating anti-CD3 or anti-Fas antibodies bound to the cell surface.
Stained cell samples were imaged with a LSM 510 Meta confocal scanning laser microscope (Zeiss, Goettingen, Germany; 405 nm diode, 488 nm Argon, 543 and 633 nm Helium-Neon excitation wavelengths), attached to an Axiovert 200M inverted light microscope (Zeiss) using a 63 × oil immersion lens (numerical aperture 1.4). Individual images were analyzed either separately, or stacks of individual images were used for colocalization and three-dimensional reconstitution studies using LSM 5 (Zeiss) or Volocity software (Improvision, Waltham, MA, USA).
For two-color analysis, stacks of images with a total thickness of approximately 15 μm were acquired at 1 μm intervals, using a dynamic range of 8 bit per pixel (256 gray levels) to reduce data size. Orthogonal projections for all conditions were composed by overlaying 10–12 images, viewed along the z axis as indicated by the blue lines in the xz plane (small top panel in Figure 1d, h and l) and the yz plane (small right panel in Figure 1d, h and l).
Three- or four-color confocal images were acquired as an image stack of optical slices along the z axis. For these studies it was necessary to replace the previously used caspase-8 reporter FAM-LETD-fmk by the substrate rhodamine-IETD-fmk for two reasons. First, FAM as a fluorescein derivate is susceptible for photo bleaching, diminishing its value as a reporter for spatial resolutions when more complex stacks of cells are acquired, which requires multiple scans. Second and more importantly, due to their spectral similarities, FAM and GFP emissions cannot be discriminated by conventional CLSM. For multicolor microscopy the dynamic range was increased to 12 bit per pixel (4096 gray levels). Sequential frames were set to overlap 1/2 thicknesses, and slice thickness was reduced to 0.3 μm, resulting in approximately 50 individual images per stack. Reprocessing of images allowed three-dimensional reconstruction and colocalization studies of cells.
Live imaging was performed using a CTI Controller/Tempcontrol 37 Incubation System (PeCon GmbH, Erbach, Germany), which allowed monitoring of cells under environmental culture condition during the course of data acquisition.
Colocalization expressed as a correlation coefficient indicates the strength and direction of the linear relationship between two fluorescence channels. Pearson's linear correlation coefficient (rP) was used in this study to calculate fluorescence channel correlations:

where xi=intensity of voxel i in image (xi=0 if xi is outside threshold of detection)
yi=intensity of voxel i in image (yi=0 if yi is outside threshold of detection)
xaver and yaver represent averages of the x and y channel intensities.
The value of rP is between −1 and 1, where 0 indicates no correlation, and −1 indicates negative correlation. Values>0 indicate a positive correlation.
Colocalization in the context of fluorescence microscopy is defined as the signal detection of two separated fluorescence channels at the same voxel location (i.e., three-dimensional pixel). Threshold settings were generated automatically from regions of interest. Colocalization coefficients were calculated according to published methodology29 in which

and

where xi,coloc=xi if yi is within the intensity range defined by region of interest
xi,coloc=0 if yi is outside the intensity range and
yi,coloc=yi if xi is within the intensity range defined by region of interest
yi,coloc=0 if xi is outside the intensity range.
Values of colocalization coefficients range between 0 and 1. A value of 0 indicates that none of the signal within thresholds in that channel exists as co-localized with the other channel. A value of 1 indicates that the entire signal within thresholds in that channel exists as colocalized with the other channel. Two perfectly colocalized images will generate a scatter plot where the points fall in a line at 45° to either axis.
Abbreviations
- CLSM:
-
confocal laser scanning microscopy
- FAM:
-
6-carboxyfluorescein
- FITC:
-
fluorescein isothiocyanate
- fmk:
-
fluoromethylketone
- IETD:
-
isoleucine-glutamic acid-threonine-aspartic acid
- LETD:
-
leucine-glutamic acid-threonine-aspartic acid
- TCR:
-
T-cell antigen receptor
- TUNEL:
-
terminal-dUTP-nick-end-labeling
- zVAD:
-
benzyloxycarbonyl-valine-alanine-aspartic acid
References
Cohen GM . Caspases: the executioners of apoptosis. Biochem J 1997; 326 (Part 1): 1–16.
Kumar S . Caspase function in programmed cell death. Cell Death Differ 2007; 14: 32–43.
Martinon F, Tschopp J . Inflammatory caspases and inflammasomes: master switches of inflammation. Cell Death Differ 2007; 14: 10–22.
Lamkanfi M, Festjens N, Declercq W, Vanden Berghe T, Vandenabeele P . Caspases in cell survival, proliferation and differentiation. Cell Death Differ 2007; 14: 44–55.
Donepudi M, Grutter MG . Structure and zymogen activation of caspases. Biophys Chem 2002; 101–102: 145–153.
Ho PK, Hawkins CJ . Mammalian initiator apoptotic caspases. The FEBS Journal 2005; 272: 5436–5453.
Degterev A, Boyce M, Yuan J . A decade of caspases. Oncogene 2003; 22: 8543–8567.
Salvesen GS . Caspase 8: igniting the death machine. Structure 1999; 7: R225–R229.
Peter ME, Krammer PH . The CD95(APO-1/Fas) DISC and beyond. Cell Death Differ 2003; 10: 26–35.
Salvesen GS, Dixit VM . Caspase activation: the induced-proximity model. Proc Natl Acad Sci USA 1999; 96: 10964–10967.
Chun HJ, Zheng L, Ahmad M, Wang J, Speirs CK, Siegel RM et al. Pleiotropic defects in lymphocyte activation caused by caspase-8 mutations lead to human immunodeficiency. Nature 2002; 419: 395–399.
Salmena L, Lemmers B, Hakem A, Matysiak-Zablocki E, Murakami K, Au PY et al. Essential role for caspase 8 in T-cell homeostasis and T-cell-mediated immunity. Genes Dev 2003; 17: 883–895.
Simons K, Ikonen E . Functional rafts in cell membranes. Nature 1997; 387: 569–572.
Simons K, Toomre D . Lipid rafts and signal transduction. Nat Rev Mol Cell Biol 2000; 1: 31–39.
He HT, Lellouch A, Marguet D . Lipid rafts and the initiation of T cell receptor signaling. Semin Immunol 2005; 17: 23–33.
Russell S, Oliaro J . Compartmentalization in T-cell signalling: membrane microdomains and polarity orchestrate signalling and morphology. Immunol Cell Biol 2006; 84: 107–113.
Misra RS, Russell JQ, Koenig A, Hinshaw-Makepeace JA, Wen R, Wang D et al. Caspase-8 and c-FLIPL associate in lipid rafts with NF-kappa B adaptors during T cell activation. J Biol Chem 2007; 282: 19365–19374.
Edidin M . The state of lipid rafts: from model membranes to cells. Annu Rev Biophys Biomol Struct 2003; 32: 257–283.
Lagerholm BC, Weinreb GE, Jacobson K, Thompson NL . Detecting microdomains in intact cell membranes. Annu Rev Phys Chem 2005; 56: 309–336.
Rodgers W . Making membranes green: construction and characterization of GFP-fusion proteins targeted to discrete plasma membrane domains. Biotechniques 2002; 32: 1044–1046, 1048, 1050–1041.
Kennedy NJ, Kataoka T, Tschopp J, Budd RC . Caspase activation is required for T cell proliferation. J Exp Med 1999; 190: 1891–1896.
Misra RS, Jelley-Gibbs DM, Russell JQ, Huston G, Swain SL, Budd RC . Effector CD4+ T cells generate intermediate caspase activity and cleavage of caspase-8 substrates. J Immunol 2005; 174: 3999–4009.
Juo P, Kuo CJ, Reynolds SE, Konz RF, Raingeaud J, Davis RJ et al. Fas activation of the p38 mitogen-activated protein kinase signalling pathway requires ICE/CED-3 family proteases. Mol Cell Biol 1997; 17: 24–35.
Juo P, Kuo CJ, Yuan J, Blenis J . Essential requirement for caspase-8/FLICE in the initiation of the Fas-induced apoptotic cascade. Curr Biol 1998; 8: 1001–1008.
Earnshaw WC, Martins LM, Kaufmann SH . Mammalian caspases: structure, activation, substrates, and functions during apoptosis. Annu Rev Biochem 1999; 68: 383–424.
Slee EA, Zhu H, Chow SC, MacFarlane M, Nicholson DW, Cohen GM . Benzyloxycarbonyl-Val-Ala-Asp (OMe) fluoromethylketone (Z-VAD.FMK) inhibits apoptosis by blocking the processing of CPP32. Biochem J 1996; 315 (Part 1): 21–24.
Janes PW, Ley SC, Magee AI . Aggregation of lipid rafts accompanies signaling via the T cell antigen receptor. J Cell Biol 1999; 147: 447–461.
Duplay P, Thome M, Herve F, Acuto O . p56lck interacts via its src homology 2 domain with the ZAP-70 kinase. J Exp Med 1994; 179: 1163–1172.
Manders M, Verbeek PJ, Aten JA . Measurement of co-localization of objects in dual colour confocal images. J Microsc 1993; 169: 375–382.
Micheau O, Thome M, Schneider P, Holler N, Tschopp J, Nicholson DW et al. The long form of FLIP is an activator of caspase-8 at the Fas death-inducing signaling complex. J Biol Chem 2002; 277: 45162–45171.
Edidin M . Lipids on the frontier: a century of cell-membrane bilayers. Nat Rev Mol Cell Biol 2003; 4: 414–418.
Harder T . Lipid raft domains and protein networks in T-cell receptor signal transduction. Curr Opin Immunol 2004; 16: 353–359.
Glebov OO, Nichols BJ . Lipid raft proteins have a random distribution during localized activation of the T-cell receptor. Nat Cell Biol 2004; 6: 238–243.
Magee T, Pirinen N, Adler J, Pagakis SN, Parmryd I . Lipid rafts: cell surface platforms for T cell signaling. Biol Res 2002; 35: 127–131.
Zacharias DA, Violin JD, Newton AC, Tsien RY . Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science 2002; 296: 913–916.
Kabouridis PS . Lipid rafts in T cell receptor signalling. Mol Membr Biol 2006; 23: 49–57.
Chichili GR, Rodgers W . Clustering of membrane raft proteins by the actin cytoskeleton. J Biol Chem 2007; 282: 36682–36691.
Manes S, Viola A . Lipid rafts in lymphocyte activation and migration. Mol Membr Biol 2006; 23: 59–69.
Lemmers B, Salmena L, Bidere N, Su H, Matysiak-Zablocki E, Murakami K et al. Essential role for caspase-8 in Toll-like receptors and NF-kappaB signaling. J Biol Chem 2007; 282: 7416–7423.
Su H, Bidere N, Zheng L, Cubre A, Sakai K, Dale J et al. Requirement for caspase-8 in NF-kappaB activation by antigen receptor. Science 2005; 307: 1465–1468.
Egawa T, Albrecht B, Favier B, Sunshine MJ, Mirchandani K, O’Brien W et al. Requirement for CARMA1 in antigen receptor-induced NF-kappa B activation and lymphocyte proliferation. Curr Biol 2003; 13: 1252–1258.
Gaide O, Favier B, Legler DF, Bonnet D, Brissoni B, Valitutti S et al. CARMA1 is a critical lipid raft-associated regulator of TCR-induced NF-kappa B activation. Nat Immunol 2002; 3: 836–843.
Lanzavecchia A, Scheidegger D . The use of hybrid hybridomas to target human cytotoxic T lymphocytes. Eur J Immunol 1987; 17: 105–111.
Muppidi JR, Siegel RM . Ligand-independent redistribution of Fas (CD95) into lipid rafts mediates clonotypic T cell death. Nat Immunol 2004; 5: 182–189.
Acknowledgements
This work was supported by grants from the National Institutes of Health (AI36333, AI45666 and AR43520 to RCB, and R01GM070001 to WAR). We thank Dr. Douglas Taatjes and Marilyn Wadsworth for help and discussions concerning confocal laser scanning microscopy, and Colette Charland and Julie Wolfe for technical assistance with flow cytometry, and Tim Hunter and Scott Tighe for technical assistance with densitometry.
Author information
Authors and Affiliations
Corresponding author
Additional information
Edited by DR Green
Supplementary Information accompanies the paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Rights and permissions
About this article
Cite this article
Koenig, A., Russell, J., Rodgers, W. et al. Spatial differences in active caspase-8 defines its role in T-cell activation versus cell death. Cell Death Differ 15, 1701–1711 (2008). https://doi.org/10.1038/cdd.2008.100
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cdd.2008.100
Keywords
This article is cited by
-
Glycolysis promotes caspase-3 activation in lipid rafts in T cells
Cell Death & Disease (2018)
-
IL-15 maintains T-cell survival via S-nitrosylation-mediated inhibition of caspase-3
Cell Death & Differentiation (2014)
-
Caspase-8 cleaves its substrates from the plasma membrane upon CD95-induced apoptosis
Cell Death & Differentiation (2013)
-
Caspase blockade induces RIP3-mediated programmed necrosis in Toll-like receptor-activated microglia
Cell Death & Disease (2013)
-
Regulation of NF-κB signaling by caspases and MALT1 paracaspase
Cell Research (2011)