Abstract
Background:
Hypoxia imaging is a promising tool for targeted therapy but the links between imaging features and underlying molecular characteristics of the tumour have not been investigated. The aim of this study was to compare hypoxia biomarkers and gene expression in oropharyngeal squamous cell carcinoma (OPSCC) diagnostic biopsies with hypoxia imaged with 64Cu-ATSM PET/CT.
Methods:
64Cu-ATSM imaging, molecular and clinical data were obtained for 15 patients. Primary tumour SUVmax, tumour to muscle ratio (TMR) and hypoxic volume were tested for association with reported hypoxia gene signatures in diagnostic biopsies. A putative gene signature for hypoxia in OPSCCs (hypoxic volume-associated gene signature (HVS)) was derived.
Results:
Hypoxic volume was significantly associated with a reported hypoxia gene signature (rho=0.57, P=0.045), but SUVmax and TMR were not. Immunohistochemical staining with the hypoxia marker carbonic anhydrase 9 (CA9) was associated with a gene expression hypoxia response (rho=0.63, P=0.01). Sixteen genes were positively and five genes negatively associated with hypoxic volume (adjusted P<0.1; eight genes had adjusted P<0.05; HVS). This signature was associated with inferior 3-year progression-free survival (HR=1.5 (1.0–2.2), P=0.047) in an independent patient cohort.
Conclusions:
64Cu-ATSM-defined hypoxic volume was associated with underlying hypoxia gene expression response. A 21-gene signature derived from hypoxic volume from patients with OPSCCs in our study may be linked to progression-free survival.
Similar content being viewed by others
Main
Hypoxia has been extensively investigated in head and neck squamous cell carcinoma (HNSCC) and the association with poor outcome is well known (Nordsmark et al, 2005). A number of therapeutic interventions can target global or focal hypoxia in tumours (Rischin et al, 2010; Hendrickson et al, 2011); however, the selection of patients with hypoxic tumours who would benefit from these interventions has been the major limiting factor for translation into clinical practice.
Accurate detection and quantification of hypoxic tumours is essential to identify patients who have aggressive, treatment-resistant disease. Various methods have been investigated but with inconsistent and sometimes conflicting results, lacking the sensitivity and specificity needed for clinical utility (Aebersold et al, 2001; Overgaard et al, 2005). Hypoxic gene expression signatures from biopsy samples could have predictive value (Winter et al, 2007; Toustrup et al, 2011) but are not able to quantify hypoxia or provide information on its spatial distribution within tumours. Combining hypoxic gene signatures and 64Cu-ATSM PET imaging biomarkers may enable a more comprehensive assessment of the hypoxic status of tumours.
64Cu-ATSM is a PET radiotracer that has been shown to accumulate in hypoxic and other conditions of mitochondrial dysfunction (Lewis et al, 1999; Donnelly et al, 2012). Although the exact mechanism has not been elucidated and uptake may be dependent on cell or tumour type, clinical studies have clearly demonstrated the potential as an imaging biomarker that warrants further investigation, especially in HNSCC, with clear advantages over the nitroimidazole-based hypoxia tracers (Minagawa et al, 2011; Grassi et al, 2014; Sato et al, 2014).
Our study hypothesis was that hypoxia gene signatures from diagnostic FFPE biopsy samples would be associated with uptake of 64Cu-ATSM in patients with oropharyngeal squamous cell carcinoma (OPSCC), and a signature could be developed to identify patients with OPSCC who would benefit from further investigation with hypoxia imaging. This would, in turn, provide more information on the level and distribution of hypoxic regions, which could potentially guide treatment.
Materials and methods
Patients
Fifteen patients with newly diagnosed histologically proven stages III–IV squamous cell carcinoma of the oropharynx to be treated with standard radical concomitant chemoradiation were prospectively recruited as part of an initial pilot phase of the study. HPV status was determined by p16 IHC and by in situ hybridisation for high-risk subtype DNA. Patients received a static 64Cu-ATSM PET/CT scan of the head and neck approximately 1 week prior to the start of their treatment. Time between biopsy and imaging was recorded (Table 1, Supplementary Figure S7). On inspection of clinical follow-up, there was one reported death (not disease-specific) and all other patients so far have no recurrence (median time to follow-up, 24 months). Research Ethics Committee approval was obtained for the study (reference 12/LO/1123) and all patients gave written and voluntary consent.
Image acquisition and analysis
Detailed methods for image acquisition and analysis are provided as Supplementary Information. In summary, patients were injected with 545±27 (range 486–577) MBq of 64Cu-ATSM followed by an uptake period of 60 min. The first patient acquisition was performed on a GE Discovery VCT PET/CT scanner (General Electric Medical Systems, Waukesha, WI, USA). All subsequent patients were imaged on the GE Discovery 710 PET/CT scanner. The 64Cu-ATSM PET/CT images were interpreted by a nuclear medicine physician and clinical oncologist using Hermes Hybrid Viewer version 2.2C (Hermes Medical Solutions, Stockholm, Sweden). PET uptake was assessed semi-quantitatively using standardised uptake values (SUV) normalised to patient body weight determined using the following Equation:
SUVbw=64Cu-ATSM activity concentration measured in the tumour (Bq cc−1) × (patient body weight (kg)/injected activity of 64Cu-ATSM (Bq) decay corrected to the time of injection) × 1000 g cc−1.
Visible lesions on PET with uptake higher than background muscle uptake were considered hypoxic. Background uptake was evaluated by placing fixed 2.5 cm spheres over bilateral posterior neck muscles on the CT images to guide correct positioning, copying the spheres onto the PET images and calculating the average SUVmean. Regions of interest (ROIs) were outlined in multiple planes using a set zoom, SUV scaling and colour scale. An initial region was drawn using an automatic segmentation seeded region growing tool on each scan slice and manually edited. The seeded region growing tool starts with a seed pixel within the tumour and then adds pixels to the region in all directions. The operator determined the extent of pixel expansion and final region outlined. ROIs were then summed to create a volume or the hypoxic volume. For each primary tumour SUVmax, tumour to muscle ratio (TMR), tumour SUVmean and hypoxic volume were measured. TMR was determined by the ratio of tumour SUVmax to the average SUVmean of the posterior neck muscles.
Immunohistochemistry (IHC), RNA extraction, sequencing and analysis
Diagnostic biopsy slides were reviewed by a head and neck pathologist to confirm the diagnosis, and the tumour outline was marked on the slide without further selection. Sections from FFPE tumour blocks were placed onto slides for IHC staining for carbonic anhydrase 9 (CA9), an endogenous marker for hypoxia, as previously described (Watson et al, 2003). Blocks were then scored with a surgical blade to correspond with the tumour regions on the slides. Ten-micrometer-thick sections of the tumour area were cut on a microtome (minimum 10 sections, minimum tumour area 100 mm2) and sent to BGI (BGI TechSolutions, Wuhan, China). BGI performed RNA extraction, RNA-sequencing (seq) and small RNA-seq (Illumina HiSeq 2000, Illumina Inc., San Diego, CA, USA) and filtered the reads to remove adaptors and low-quality reads. RNA-seq reads aligned to reference genome (hg19; BWA v0.7.10-r789) were obtained from BGI and read counts per gene were enumerated using htseq-count (HTSeq v0.6.1p1 (Anders et al, 2015); union of exons). Small RNA reads were obtained from BGI and aligned to microRNA (miRNA) mature sequences (MirBase v21 (Kozomara and Griffiths-Jones, 2011) using bowtie2 v2.2.5 (Langmead and Salzberg, 2012) with local alignment (—local –a). Read counts per miRNA were enumerated using reads that were uniquely mapped among reported alignments. Differential expression analyses from count data and read count normalisation (rlog-transform) for visualisation and clustering were performed using DESeq2 v1.6.3 (Love et al, 2014). Differential expression analysis of HPV status was perfomed using DESeq2 with HPV status as the co-variable. Genes associated with hypoxic volume were identified using DESeq2 with hypoxic volume as a continuous co-variable. Gene set enrichment and leading edge analysis (GSEA v2.2.2 (Subramanian et al, 2005); preranked tool; minimum set size=5; 1000 permutations) was performed on genes preranked by fold-change (per unit increase of hypoxic volume) and filtered for coverage (baseMean >200). Gene sets with names containing the phrase ‘hypoxia’, ‘HIF1’ or ‘HIF2’ were preselected from all MSigDB curated gene sets (c2.all.v5.1.symbols.gmt (Subramanian et al, 2005)). Gene signature scores were estimated from normalised (rlog-transformed) read counts using a weighted sum of Z-scores for each gene in the respective gene list, with weights (+1, −1) according to the direction of expression in the original gene signature.
Analysis of external gene expression data sets
The following series were retrieved from the Gene Expression Omnibus (an international public genomics data repository): GSE686 (Chung et al, 2004) and GSE65858 (Wichmann et al, 2015). Follow-up information about progression-free survival (GSE65858) was obtained from GEO, and follow-up information about recurrence-free survival (GSE65858) was obtained from the original publication (Chung et al, 2004). A set of predefined exploratory analyses of follow-up were performed. Kaplan–Meier plots were used to inspect the hypoxic volume signature score by upper quartile, interquartile and lower quartile of the score values, samples in the upper quartile vs all other samples and samples in the lower quartile vs all other samples. After inspection of the Kaplan–Meier plots, exploratory log-rank tests were performed for 3-year follow-up and for the whole follow-up period, and hazard ratios were fitted to the same time periods using univariable Cox proportional hazards models.
Statistical analysis
Mann–Whitney U-tests, Kaplan–Meier plots, log-rank tests and Cox proportional hazard model fits were performed in the R environment, v3.1.2 and ‘survival’ package v2.38 (R Core Team, 2014). Exact Wilcoxon–Mann–Whitney tests were performed using the ‘coin’ package v1.1-2. Cox model P-values report the Wald test. Unless otherwise stated, two-tailed tests are reported. P<0.05 was considered to be statistically significant.
Results
Patient characteristics are summarised in Table 1. There were no immediate or late adverse reactions after tracer injection. Uptake was detected in all 15 primary tumours on 64Cu-ATSM PET scans (Figure 1A). Two patients had diagnostic tonsillectomies at diagnosis but uptake was seen in the residual disease. The mean SUVmax for all 15 patients was 3.32±0.85 (range 2.12–4.85) and the mean TMR was 4.51±1.82 (range 1.84–9.00; Table 1). SUVmax and TMR were not significantly different between HPV-positive and HPV-negative patients. Tonsillectomy patients were excluded from analyses using hypoxic volume and SUVmean.
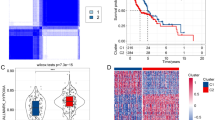
64Cu-ATSM PET imaging and association with CA9 immunohistochemical staining and a hypoxia gene expression response in disgnostic biopsies.(A–B) Example images of 64Cu-ATSM PET (left) and fused PET/CT (right) scans. (A) Patient with T2N2b left base of tongue SCC, with Cu-ATSM uptake in the primary but no uptake in left level II neck node. (B) Patient with T3N1 left tonsil SCC with high uptake in primary. (C) Immunohistochemistry staining of hypoxia biomarker CA9 vs each image feature. ‘High’ is defined as values greater than the median value. (D) Scatterplots of 64Cu-ATSM imaging parameters vs the previously reported Sorensen2015 hypoxia classifier. Hypoxic volume, TMR and SUVmax plotted against the 15-gene hypoxia classifier. A significant positive correlation is observed between the hypoxia score and hypoxic volume but not TMR or SUVmax. Red points indicate samples with low hypoxia score but high SUVmax. Samples from patients who had diagnostic tonsillectomies (Study IDs 9 and 11) were excluded as a hypoxic volume could not be determined.
The mean hypoxic volume in 13 primary tumours was 12.54 cm3±11.53 (range 0.68–36.89) and the mean SUVmean was 2.34±0.45 (range 1.71–3.11; Table 1). Supplementary Figure S1 demonstrates the examples of hypoxic volume delineation. Increased uptake was detected in 22 lymph nodes on 64Cu-ATSM PET scans out of 31 abnormal lymph nodes reported on CT imaging. Lymph node uptake appeared visually to be more heterogeneous than uptake in the primary tumour, especially in large nodes, which showed uptake in the periphery with central regions of no uptake, likely secondary to necrosis (Supplementary Figure S2). In general, the uptake in the nodes was lower than the primary. The mean SUVmax of the nodes was 2.43±0.93 (range 1.39–5.80) and the mean TMR 3.29±2.41 (range 1.35–13.18) (Supplementary Table S1).
The differential gene expression between HPV-positive and HPV-negative samples were compared with previously published signatures (Pyeon et al, 2007; Keck et al, 2015) as an initial screen to assess the RNA sequencing from FFPE samples (Supplementary Figure S3). Overall, the pattern of gene expression profiles in our HPV-positive vs HPV-negative samples were readily distinguishable and consistent with previous studies. There was no difference in hypoxic volume, TMR or SUVmax between HPV-positive and HPV-negative samples.
To establish whether RNA-seq data from the diagnostic biosies could report gene expression responses to hypoxia, we initially compared the gene expression of our samples with two different published hypoxia-associated signatures that have been used to assess hypoxia: a signature comprising genes regulated by hypoxia in head and neck cancers (Winter et al, 2007) and the 15-gene hypoxia classifier (Sorensen et al, 2015). There was high correlation between the two different signatures in our samples (Supplementary Figure S4C). Hypoxic volume was significantly associated with both the 15-gene hypoxia classifier (P=0.045; Spearman’s rho=0.57, two-tailed test, Figure 1D) and the gene list by Winter et al (P=0.05; Spearman’s rho=0.56, two-tailed test). SUVmax and TMR were not significantly associated with these hypoxia signatures; on inspection, four patient samples exhibited low scores for the 15-gene hypoxia signature but had among the highest values of SUVmax and TMR (Figure 1D). There were no significant differences in baseline characteristics between this ‘subgroup’ and the other samples, and these patients were among the oldest in the study (Mann–Whitney U, W=3, P=0.02).
To verify that hypoxic imaging volume and RNAseq-derived hypoxia gene expression response were associated with hypoxia in patient tumours, CA9 protein IHC, a marker for hypoxia, was performed on biopsy samples. Among the image features, hypoxic volume showed a positive trend with CA9 IHC scores (Mann–Whitney U one-tailed, threshold median of hypoxic volume, P=0.06 (Figure 1B and Supplementary Figure S4A); Spearman’s rho=0.43, P=0.15 (Supplementary Figure S4A)). SUVmax and TMR were not associated with CA9 (Figure 1B and Supplementary Figure S4A). Furthermore, the 15-gene hypoxia classifier score was significantly associated with CA9 IHC scores (Spearman’s rho=0.63, P=0.01 (Supplementary Figure S4B)). Genes ranked according to association with hypoxic volume were found to be enriched for previously curated hypoxia-associated gene sets with expression response to hypoxia or downstream of hypoxia-inducible factor 1 (HIF-1; Supplementary Table S2, Supplementary Figure S5).
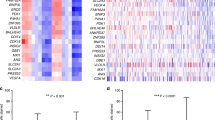
The 15-gene hypoxia classifier (Sorensen et al, 2015) was originally derived and tested in multiple cancer types and may represent hypoxia gene expression responses that are commonly found across differing tumour types (Winter et al, 2007; Sorensen et al, 2015). We hypothesised that alternative gene signatures could accurately evaluate the hypoxic phenotypes in OPSCCs. We used the 64Cu-ATSM hypoxic volume together with mRNA expression from biopsy samples to identify a gene signature for hypoxic imaging phenotypes in OPSCCs. Hypoxic volume was significantly associated with increased expression of 16 genes and decreased expression of 5 genes (hypoxic volume-associated gene signature (HVS)) (Figure 2A, Supplementary Table S3). Two genes overlapped with the 15-gene hypoxia classifier, LOX and PFKFB3. On inspection, there was concordance between the hypoxic volume-associated gene signature and expression of the 15-gene hypoxia classifier (Figure 2A). The HVS was more highly correlated with each of the image features (hypoxic volume, TMR and SUVmax) than the 15-gene hypoxia classifier, but only the hypoxic volume was significantly correlated with HVS (hypoxic volume: rho=0.89, P<10−4; TMR: rho=0.37, P=0.17; SUVmax: rho=0.45, P=0.09; Spearman’s correlation, two-tailed). Furthermore, the HVS was significantly associated with CA9 IHC staining (Mann–Whitney U one-tailed, threshold median of HVS score, P=0.03, Figure 2B).
HVS associated with hypoxic volume.(A) The heatmap displays the HVS, comprising 16 genes positively and 5 genes negatively associated with hypoxic volume. Samples are ordered by increasing hypoxic volume (base mean >200; adjusted P<0.05 or (italicised) adjusted P<0.1). Two genes that overlapped with the 15-gene hypoxia classifier gene list are LOX and PFKFB3. Yellow and blue colour bar shows the hypoxia 15-gene classifier score by rank within samples, for comparison (Sorensen2015). (B) CA9 IHC is shown compared with ‘low’ or ‘high’ HVS signature scores for all samples (n=15). ‘High’ is defined as HVS scores greater than the median of HVS scores.
To gain some initial insights into potential modes of gene regulation in tumours with a 64Cu-ATSM imaging hypoxic phenotype, miRNAs were ranked according to the association of expression with increasing 64Cu-ATSM hypoxic volume. The top-ranked genes for association with the hypoxia volume signature were miR-21-5p (P=0.004, adjusted P=0.99) and miR-10b-5p (P=0.005, adjusted P=0.99; Figure 2A, Supplementary Table S4). The study-derived HVS was then inspected in two publicly available data sets from head and neck cancers with long-term follow-up and derived from alternative gene expression platforms: GEO series GSE686 (Chung et al, 2004) and GSE65858 (Wichmann et al, 2015). The genes comprising the HVS were concordantly expressed within each data set (Supplementary Figure S6), which indicates that this signature may be widely relevant to head and neck tumour series. Finally, in exploratory analyses of each independent series, the HVS was found to be associated with poorer progression-free survival during a 3-year follow-up period (GSE65858; HR=1.5 (1.0–2.2), P=0.047), and there was a suggestive association with worse 3-year recurrence-free survival (GSE686; HR=3.5 (0.8–16), P=0.1; Supplementary Figure S6).
Discussion
This study demonstrates that 64Cu-ATSM-defined hypoxic volume correlates with a previously reported hypoxia gene expression response and may be an important imaging parameter to consider for assessment of hypoxia in OPSCCs using PET. In addition, to describe the gene expression differences that underlie the PET imaging features, we identified a refined gene signature associated with 64Cu-ATSM hypoxic volume in OPSCCs.
64Cu-ATSM PET is well tolerated and demonstrates a spectrum of hypoxic imaging phenotypes in OPSCCs. Cu-ATSM has been investigated as a promising radiotracer for hypoxia imaging, which is a challenging area of research as hypoxia is spatially and temporally heterogeneous. Small clinical studies in HNSCC have reported that different imaging parameters correlate with progression or outcome, but there is currently no consensus, and the molecular mechanisms underlying these associations are not well understood. In addition, SUVmax, while widely reported, is a single voxel measure and may be sensitive to technical factors affecting PET (Adams et al, 2010). Minagawa et al (2011) found that 62Cu-ATSM SUVmax, but not TMR, was significantly different in 17 locally advanced head and neck cancer patients with and without residual or recurrent tumours. A study of 25 head and neck cancer patients indicated that both increasing SUVmax and TMR were associated with worse progression-free survival (Sato et al, 2014). Grassi et al (2014) used 64Cu-ATSM to define a biological target volume in 11 patients with HNSCC and found this parameter, along with SUVmax, to have high sensitivity but low specificity in predicting complete response to therapy. The same group reported that hypoxic tumour volume and hypoxic burden (hypoxic tumour volume × SUVmean) in 18 patients with lung cancer or HNSCC were more robust prognostic parameters for progression-free survival after a median follow-up of 14.6 months (Lopci et al, 2015), which is in agreement with our data suggesting that hypoxic volume is an informative feature in 64Cu-ATSM PET scans. Our data suggest that hypoxic volume might be a more reliable correlate with gene expression patterns associated with hypoxia.
The optimal method for volume delineation is uncertain and requires further investigation. Grassi et al, 2014 used a cutoff of 42% of the lesion SUVmax, but their subsequent study did not apply a fixed threshold for contouring (Lopci et al, 2015). The limited spatial resolution of PET makes defining the edge of tumour uptake difficult and also means that PET imaging does not necessarily show the microregional, true heterogeneity of hypoxia within the tumour (Horsman et al, 2012). The mechanism of pO2 dependence and factors that can affect cellular uptake and retention in both normoxic and hypoxic tissues is also unclear. Cu-ATSM may be an indirect marker for hypoxia, correlating with levels of the biological reductants NADH and NADPH (Yoshii et al, 2012). Hypoxia is one of a number of conditions leading to NADH accumulation, providing the cellular reduction potential that leads to tracer retention. Other factors such as multidrug resistance protein 1 expression (Liu et al, 2009) and CD133+ expression (Yoshii et al, 2012) have also been shown to influence Cu-ATSM uptake and retention, as well as cellular copper metabolism and processing (Hueting et al, 2014). Cu-ATSM is a promising radiotracer to detect hypoxia, but other possible sources of uptake need to be taken into consideration when interpreting the images and determining clinical utility.
We therefore inspected the genes comprising our proposed gene signature associated with hypoxic volume in OPSCCs. There were a number of upregulated genes relating to the development of hypoxia in tumours. For example, SOD2 is an antioxidant enzyme, which prevents redox-mediated damage of mitochondrial proteins, and is associated with aggressive cancers with enhanced cell migration and metastases. Stress such as hypoxia leads to increased reactive oxygen species (ROS) and tumours may increase their expression of SOD2 to prevent ROS-mediated DNA damage (Connor et al, 2007). Also among the upregulated genes, NAMPT tissue expression has also been found to be upregulated in tumours and shown to induce cell proliferation and angiogenesis (Shackelford et al, 2013). It is the rate-limiting enzyme for the biosynthesis of NAD essential for metabolism and energy production. Tumour cells have high metabolic rate and NAD consumption and therefore depend on the production of NAD, and hypoxia has been shown to result in NAMPT induction (Bae et al, 2006). NAMPT small-molecule inhibitors are under investigation as a novel therapeutic, which reduce NAD levels resulting in ATP loss and inhibition of tumour cell proliferation (Xu et al, 2015). RUNX3 is downregulated in our samples with increasing hypoxic volume. RUNX3 has been shown to inhibit HIF-1α stability through enhancing the interaction between HIF-1α and PHD2, promoting HIF-1α degradation in gastric cancer cells (Lee et al, 2014) with resulting inhibition of angiogenesis. Its expression has been shown to be downregulated in response to hypoxia and is frequently inactivated in gastric cancer, resulting in stimulation of proliferation and suppression of apoptosis (Lee et al, 2009). Supplementary Table S5 summarises the genes and their function.
CA9 is a transmembrane glycoprotein that is induced by hypoxia and considered an endogenous marker of hypoxia. It is a downstream target of HIF-1 in the hypoxia response pathway and has been shown to be overexpressed in HNSCC due to hypoxia (Beasley et al, 2001). The association between the hypoxic volume and CA9, and between the 15-gene hypoxia classifier and CA9, further suggests that hypoxic volume is an important feature for understanding hypoxic response in this series of OPSCCs and that the gene expression from FFPE biopsy samples could reflect the hypoxia response of the tumour.
Among the miRNAs found to be associated with the hypoxic volume, miR-21 is a frequently dysregulated miRNAs in HNSCC (Chang et al, 2008). Upregulation of miR-21 promotes cell proliferation, migration and inhibition of apoptosis and is associated with poor prognosis (Lu et al, 2008). Mir-210, the hypoxia miRNA (Huang et al, 2010), had low base coverage in our analysis and was not associated with hypoxia expression signatures.
This study had several limitations. First, there was a small sample size that is typical of exploratory imaging-genomic studies with patients recruited from a single institution. However, a strong point of our study is that the PET studies were performed and analysed in a standardised way at a single PET centre. Second, the validation of our hypoxia-associated signature was limited to in silico functional investigations in the absence of a clinical validation set and warrants further study to elucidate the mechanisms underlying these associations. Third, Cu-ATSM as a radiotracer provides hypoxic-to-normoxic contrast of sufficient quality to define a hypoxic volume but the optimal method for hypoxic volume delineation requires further investigation. Owing to the existing clinical protocols for obtaining biopsy samples, it was not possible to attempt any registration (alignment) of biopsy sites and PET scans. Hypoxia responses detected in the diagnostic biopsy (CA9 hypoxia biomarker and gene expression profiling) were used to indicate the hypoxic status of the tumour and more directly reflects clinical practice. These factors may be considered a limitation, and although outside the scope of this study, further work is needed to understand the effect of tumour heterogeneity on hypoxia evaluation in OPSCCs.
In conclusion, by combining PET imaging and mRNA expression profiling, our study revealed that Cu-ATSM PET hypoxic volume is associated with hypoxia gene signatures in OPSCCs and suggests that PET could be a useful surrogate for hypoxia gene signatures in order to stratify patients for treatment. As with all exploratory biomarker studies, our findings now require prospective investigation in a larger number of patients.
Change history
11 April 2017
This paper was modified 12 months after initial publication to switch to Creative Commons licence terms, as noted at publication
References
Adams MC, Turkington TG, Wilson JM, Wong TZ (2010) A systematic review of the factors affecting accuracy of SUV measurements. AJR Am J Roentgenol 195: 310–320.
Aebersold DM, Burri P, Beer KT, Laissue J, Djonov V, Greiner RH, Semenza GL (2001) Expression of hypoxia-inducible factor-1alpha: a novel predictive and prognostic parameter in the radiotherapy of oropharyngeal cancer. Cancer Res 61: 2911–2916.
Anders S, Pyl PT, Huber W (2015) HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169.
Bae SK, Kim SR, Kim JG, Kim JY, Koo TH, Jang HO, Yun I, Yoo MA, Bae MK (2006) Hypoxic induction of human visfatin gene is directly mediated by hypoxia-inducible factor-1. FEBS Lett 580: 4105–4113.
Beasley NJ, Wykoff CC, Watson PH, Leek R, Turley H, Gatter K, Pastorek J, Cox GJ, Ratcliffe P, Harris AL (2001) Carbonic anhydrase IX, an endogenous hypoxia marker, expression in head and neck squamous cell carcinoma and its relationship to hypoxia, necrosis, and microvessel density. Cancer Res 61: 5262–5267.
Chang SS, Jiang WW, Smith I, Poeta LM, Begum S, Glazer C, Shan S, Westra W, Sidransky D, Califano JA (2008) MicroRNA alterations in head and neck squamous cell carcinoma. Int J Cancer 123: 2791–2797.
Chung CH, Parker JS, Karaca G, Wu J, Funkhouser WK, Moore D, Butterfoss D, Xiang D, Zanation A, Yin X, Shockley WW, Weissler MC, Dressler LG, Shores CG, Yarbrough WG, Perou CM (2004) Molecular classification of head and neck squamous cell carcinomas using patterns of gene expression. Cancer Cell 5: 489–500.
Connor KM, Hempel N, Nelson KK, Dabiri G, Gamarra A, Belarmino J, Van De Water L, Mian BM, Melendez JA (2007) Manganese superoxide dismutase enhances the invasive and migratory activity of tumor cells. Cancer Res 67: 10260–10267.
Donnelly PS, Liddell JR, Lim S, Paterson BM, Cater MA, Savva MS, Mot AI, James JL, Trounce IA, White AR, Crouch PJ (2012) An impaired mitochondrial electron transport chain increases retention of the hypoxia imaging agent diacetylbis(4-methylthiosemicarbazonato)copperII. Proc Natl Acad Sci USA 109: 47–52.
Grassi I, Nanni C, Cicoria G, Blasi C, Bunkheila F, Lopci E, Colletti PM, Rubello D, Fanti S (2014) Usefulness of 64Cu-ATSM in head and neck cancer: a preliminary prospective study. Clin Nucl Med 39: e59–e63.
Hendrickson K, Phillips M, Smith W, Peterson L, Krohn K, Rajendran J (2011) Hypoxia imaging with [F-18] FMISO-PET in head and neck cancer: potential for guiding intensity modulated radiation therapy in overcoming hypoxia-induced treatment resistance. Radiother Oncol 101: 369–375.
Horsman MR, Mortensen LS, Petersen JB, Busk M, Overgaard J (2012) Imaging hypoxia to improve radiotherapy outcome. Nat Rev Clin Oncol 9: 674–687.
Huang X, Le QT, Giaccia AJ (2010) MiR-210—micromanager of the hypoxia pathway. Trends Mol Med 16: 230–237.
Hueting R, Kersemans V, Cornelissen B, Tredwell M, Hussien K, Christlieb M, Gee AD, Passchier J, Smart SC, Dilworth JR, Gouverneur V, Muschel RJ (2014) A comparison of the behavior of (64)Cu-acetate and (64)Cu-ATSM in vitro and in vivo. J Nucl Med 55: 128–134.
Keck MK, Zuo Z, Khattri A, Stricker TP, Brown CD, Imanguli M, Rieke D, Endhardt K, Fang P, Bragelmann J, DeBoer R, El-Dinali M, Aktolga S, Lei Z, Tan P, Rozen SG, Salgia R, Weichselbaum RR, Lingen MW, Story MD, Ang KK, Cohen EE, White KP, Vokes EE, Seiwert TY (2015) Integrative analysis of head and neck cancer identifies two biologically distinct HPV and three non-HPV subtypes. Clin Cancer Res 21: 870–881.
Kozomara A, Griffiths-Jones S (2011) miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res 39: D152–D157.
Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359.
Lee SH, Bae SC, Kim KW, Lee YM (2014) RUNX3 inhibits hypoxia-inducible factor-1alpha protein stability by interacting with prolyl hydroxylases in gastric cancer cells. Oncogene 33: 1458–1467.
Lee SH, Kim J, Kim WH, Lee YM (2009) Hypoxic silencing of tumor suppressor RUNX3 by histone modification in gastric cancer cells. Oncogene 28: 184–194.
Lewis JS, McCarthy DW, McCarthy TJ, Fujibayashi Y, Welch MJ (1999) Evaluation of 64Cu-ATSM in vitro and in vivo in a hypoxic tumor model. J Nucl Med 40: 177–183.
Liu J, Hajibeigi A, Ren G, Lin M, Siyambalapitiyage W, Liu Z, Simpson E, Parkey RW, Sun X, Oz OK (2009) Retention of the radiotracers 64Cu-ATSM and 64Cu-PTSM in human and murine tumors is influenced by MDR1 protein expression. J Nucl Med 50: 1332–1339.
Lopci E, Grassi I, Rubello D, Colletti PM, Cambioli S, Gamboni A, Salvi F, Cicoria G, Lodi F, Dazzi C, Mattioli S, Fanti S (2015) Prognostic evaluation of disease outcome in solid tumors investigated with 64Cu-ATSM PET/CT. Clin Nucl Med 41 (2): e87–e92.
Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550.
Lu Z, Liu M, Stribinskis V, Klinge CM, Ramos KS, Colburn NH, Li Y (2008) MicroRNA-21 promotes cell transformation by targeting the programmed cell death 4 gene. Oncogene 27: 4373–4379.
Minagawa Y, Shizukuishi K, Koike I, Horiuchi C, Watanuki K, Hata M, Omura M, Odagiri K, Tohnai I, Inoue T, Tateishi U (2011) Assessment of tumor hypoxia by 62Cu-ATSM PET/CT as a predictor of response in head and neck cancer: a pilot study. Ann Nucl Med 25: 339–345.
Nordsmark M, Bentzen SM, Rudat V, Brizel D, Lartigau E, Stadler P, Becker A, Adam M, Molls M, Dunst J, Terris DJ, Overgaard J (2005) Prognostic value of tumor oxygenation in 397 head and neck tumors after primary radiation therapy. An international multi-center study. Radiother Oncol 77: 18–24.
Overgaard J, Eriksen JG, Nordsmark M, Alsner J, Horsman MR (2005) Plasma osteopontin, hypoxia, and response to the hypoxia sensitiser nimorazole in radiotherapy of head and neck cancer: results from the DAHANCA 5 randomised double-blind placebo-controlled trial. Lancet Oncol 6: 757–764.
Pyeon D, Newton MA, Lambert PF, den Boon JA, Sengupta S, Marsit CJ, Woodworth CD, Connor JP, Haugen TH, Smith EM, Kelsey KT, Turek LP, Ahlquist P (2007) Fundamental differences in cell cycle deregulation in human papillomavirus-positive and human papillomavirus-negative head/neck and cervical cancers. Cancer Res 67: 4605–4619.
R Core Team (2014) A language and environment for statistical computing http://www.R-project.org/.
Rischin D, Peters LJ, O’Sullivan B, Giralt J, Fisher R, Yuen K, Trotti A, Bernier J, Bourhis J, Ringash J, Henke M, Kenny L (2010) Tirapazamine, cisplatin, and radiation versus cisplatin and radiation for advanced squamous cell carcinoma of the head and neck (TROG 02.02, HeadSTART): a phase III trial of the Trans-Tasman Radiation Oncology Group. J Clin Oncol 28: 2989–2995.
Sato Y, Tsujikawa T, Oh M, Mori T, Kiyono Y, Fujieda S, Kimura H, Okazawa H (2014) Assessing tumor hypoxia in head and neck cancer by PET with (6)(2)Cu-diacetyl-bis(N(4)-methylthiosemicarbazone). Clin Nucl Med 39: 1027–1032.
Shackelford RE, Mayhall K, Maxwell NM, Kandil E, Coppola D (2013) Nicotinamide phosphoribosyltransferase in malignancy: a review. Genes Cancer 4: 447–456.
Sorensen BS, Knudsen A, Wittrup CF, Nielsen S, Aggerholm-Pedersen N, Busk M, Horsman M, Hoyer M, Bouchelouche PN, Overgaard J, Alsner J (2015) The usability of a 15-gene hypoxia classifier as a universal hypoxia profile in various cancer cell types. Radiother Oncol 116: 346–351.
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 102: 15545–15550.
Toustrup K, Sorensen BS, Nordsmark M, Busk M, Wiuf C, Alsner J, Overgaard J (2011) Development of a hypoxia gene expression classifier with predictive impact for hypoxic modification of radiotherapy in head and neck cancer. Cancer Res 71: 5923–5931.
Watson PH, Chia SK, Wykoff CC, Han C, Leek RD, Sly WS, Gatter KC, Ratcliffe P, Harris AL (2003) Carbonic anhydrase XII is a marker of good prognosis in invasive breast carcinoma. Br J Cancer 88: 1065–1070.
Wichmann G, Rosolowski M, Krohn K, Kreuz M, Boehm A, Reiche A, Scharrer U, Halama D, Bertolini J, Bauer U, Holzinger D, Pawlita M, Hess J, Engel C, Hasenclever D, Scholz M, Ahnert P, Kirsten H, Hemprich A, Wittekind C, Herbarth O, Horn F, Dietz A, Loeffler M, Leipzig H, Neck G (2015) The role of HPV RNA transcription, immune response-related gene expression and disruptive TP53 mutations in diagnostic and prognostic profiling of head and neck cancer. Int J Cancer 137: 2846–2857.
Winter SC, Buffa FM, Silva P, Miller C, Valentine HR, Turley H, Shah KA, Cox GJ, Corbridge RJ, Homer JJ, Musgrove B, Slevin N, Sloan P, Price P, West CM, Harris AL (2007) Relation of a hypoxia metagene derived from head and neck cancer to prognosis of multiple cancers. Cancer Res 67: 3441–3449.
Xu TY, Zhang SL, Dong GQ, Liu XZ, Wang X, Lv XQ, Qian QJ, Zhang RY, Sheng CQ, Miao CY (2015) Discovery and characterization of novel small-molecule inhibitors targeting nicotinamide phosphoribosyltransferase. Sci Rep 5: 10043.
Yoshii Y, Yoneda M, Ikawa M, Furukawa T, Kiyono Y, Mori T, Yoshii H, Oyama N, Okazawa H, Saga T, Fujibayashi Y (2012) Radiolabeled Cu-ATSM as a novel indicator of overreduced intracellular state due to mitochondrial dysfunction: studies with mitochondrial DNA-less rho0 cells and cybrids carrying MELAS mitochondrial DNA mutation. Nucl Med Biol 39: 177–185.
Acknowledgements
Patient tissue samples and data were provided by Guy’s and St Thomas’ Head and Neck Biobank–part of the KHP Cancer Biobank, which is supported by the Department of Health via the National Institute for Health Research (NIHR) comprehensive Biomedical Research Centre award and Guy’s and St Thomas’ NHS Foundation Trust. This work was supported by King’s Health Partner’s Research and Development Challenge Fund and The Rosetrees Trust (YS) and CRUK and EPSRC Comprehensive Cancer Imaging Centre at KCL and UCL (C1519/10331 and C1519/A16463; YS, KL).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License.
Supplementary Information accompanies this paper on British Journal of Cancer website
Supplementary information
Rights and permissions
From twelve months after its original publication, this work is licensed under the Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/4.0/
About this article
Cite this article
Suh, Ye., Lawler, K., Henley-Smith, R. et al. Association between hypoxic volume and underlying hypoxia-induced gene expression in oropharyngeal squamous cell carcinoma. Br J Cancer 116, 1057–1064 (2017). https://doi.org/10.1038/bjc.2017.66
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/bjc.2017.66
Keywords
This article is cited by
-
Inside the hypoxic tumour: reprogramming of the DDR and radioresistance
Cell Death Discovery (2020)
-
Positron Emission Tomography and Molecular Imaging of Head and Neck Malignancies
Current Radiology Reports (2020)
-
Clinical update on head and neck cancer: molecular biology and ongoing challenges
Cell Death & Disease (2019)
-
Hypoxia PET imaging techniques: data acquisition and analysis
Clinical and Translational Imaging (2017)