Abstract
Background:
Human polypyrimidine tract binding protein 3 (PTBP3) was first discovered in 1999 and has been well characterised as a differentiation regulator. However, its role in human cancer has rarely been reported. Our previous study revealed increased PTBP3 protein level in gastric cancer tissues. Downregulation of PTBP3 suppressed the proliferation and differentiation of gastric cancer cells in vivo.
Methods:
PTBP3 mRNA levels in human gastric cancer and adjuvant non-tumour tissues were detected. Apoptosis and 5-FU effect were determined in PTBP3-silenced gastric cancer cells. Underlying molecular mechanisms were investigated.
Results:
MRNA expression of PTBP3 was upregulated in gastric cancer tissues, especially in those at an advanced stage. PTBP3 silencing led to apoptosis, under which modulation of PTB and thereby switch of Bcl-x pre-mRNA splicing pattern might be an important mechanism. Further research found that inhibition of PTBP3 expression enhanced the chemosensitivity of gastric cancer cells towards 5-FU treatment. This was mediated by reduced expression of histone deacetylase 6 (HDAC6), which further inhibited the phosphorylation of Akt and the expression of thymidylate synthase (TYMS), the critical determinant of 5-FU cytotoxicity.
Conclusions:
PTBP3 might serve as a biomarker of gastric cancer or potential target for anti-cancer therapy.
Similar content being viewed by others
Main
Alternative splicing (AS) is a post-transcriptional regulatory mechanism, which allows individual genes to produce multiple mRNA isoforms. Abnormalities in the expression and activity of splicing factors contribute to the pathogenesis of cancer and other human diseases (Chabot and Shkreta, 2016). Members of heterogeneous nuclear ribonucleoproteins (hnRNPs) family are important AS regulators, among which polypyrimidine tract binding protein (PTB, also named as PTBP1 or hnRNP I) is a well-investigated one (Wagner and Garcia-Blanco, 2001; Spellman et al, 2007). However, fewer reports have been published regarding the biofunction of its paralog PTBP3 (also known as the regulator of differentiation 1, ROD1).
Human PTBP3 was first discovered and isolated in 1999, and was found to regulate the differentiation and proliferation of hematopoietic cells and cardiac stem cells (Yamamoto et al, 1999; Hosoda et al, 2011). Further studies showed that PTBP3 could enhance the mitogenic activity of acidic amphipathic C-terminal peptide (C21) of thrombospondin-4 (Sadvakassova et al, 2009). Recently, PTBP3 has also been found to play important roles in lung adenocarcinoma (Tano et al, 2010), glioblastoma multiforme (Zhang et al, 2014) and squamous cell carcinoma (Ooi et al, 2014). However, the role of PTBP3 in gastric cancer remains further elucidation.
Our previous study has found higher level of PTBP3 protein in gastric cancer tissues. In addition, PTBP3-silenced MKN45 cells displayed poorer proliferation and differentiation in vivo (Chen et al, 2014). In the present study, we found that PTBP3 knockdown in gastric cancer cells induced apoptosis and cell cycle arrest at S-phase. Inhibition of PTBP3 expression also sensitised cancer cells towards 5-FU treatment. Further investigation on the underlying mechanism revealed that modulation of Bcl-x splicing event and HDAC6/Akt/TYMS pathway were involved in the anti-tumour effect of PTBP3 silencing. These results suggested that PTBP3 might be a promising diagnostic biomarker for gastric cancer and target for novel anti-tumour therapies.
Materials and Methods
Cell culture and transfection
Cell lines including AGS, BGC823, MGC803, MKN45, SGC7901 were purchased from Cell Bank of Type Culture Collection of Chinese Academy of Sciences. Cells were grown in RPMI-1640 medium (Life Technologies, Carlsbad, CA, USA) with 10% heat-inactivated calf serum, penicillin (100 U ml−1) and streptomycin (100 U ml−1), and incubated at 37 °C in a humidified atmosphere of 95% air and 5% CO2. Cell transfection was performed using Biotool DNA Transfection Reagent according to the manufacturer’s instruction.
Human gastric cancer samples
Human gastric cancer samples and adjacent tissues were collected at the time of surgical resection at Xinhua Hospital, Shanghai Jiaotong University. Samples were snap-frozen in liquid nitrogen immediately upon collection and then stored at −80 °C. Informed consents have been obtained from patients and use of human tissues was approved by the Ethical Committee of Xinhua Hospital based on the Declaration of Helsinki Principles.
MTT assays
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) was purchased from Sigma Chemical (St Louis, MO, USA). MTT assays were performed as described in the previous study (Lin et al, 2016). Briefly, the cells were transfected with indicated plasmids 24 h before the treatment of 5-FU for another 24 h. The efficiency of the plasmids at 48 h after transfection were tested beforehand. PTBP3-pSilencer plasmid was constructed in our previous study (Chen et al, 2014). PTBP3-expressing pcDNA 3.1(+) plasmid was purchased from Biogot Technology (Nanjing, China).
Flow cytometry analysis
Apoptosis of transfected cells was determined using Biotool Annexin V-FITC Detection Kit and a FACScalibur flow cytometer (Becton Dickinson, San Jose, CA, USA) as described in the previous study (Shen et al, 2013). As for the analysis of cellular DNA content, transfected cells were stained with propidium iodide (Vazyme, Nanjing, China) according to the manufacturer’s instructions, and analysed using the FACScalibur flow cytometer.
RNA extraction, RT–PCR, and qPCR
Total RNA extraction and reverse-transcription PCR (RT–PCR) were performed as described in the previous report (Mao et al, 2015). Quantitative PCR (qPCR) assays were conducted using SYBR Premix Ex Taq II (Tli RNaseH Plus; Takara, Duren, Germany) and a CFX96 real-time PCR detection system (Bio-Rad, Hercules, CA, USA).
Plasmid construction
PTB-targeting pSilencer plasmid was constructed as described in the previous report (Zheng et al, 2015). Briefly, pSilencer 2.1-U6 Neo vector and the non-targeting control plasmid were purchased from Invitrogen. The siRNA sequence used for PTB knockdown was 5′-GCGTGAAGATCCTGTTCAATA-3′, which was a validated sequence obtained from Sigma-Aldrich website. Hairpin shRNA strands were designed according to the manufacturer’s instruction and synthesised by Generay Biotech. Single-strand oligonucleotides of shPTB were then annealed into double strands and inserted into the pSilencer vector.
Western blot analysis
Extraction of protein were performed according to the previous report (Xu et al, 2013). Western blotting assays were performed as described in a previous study (Zheng et al, 2013). Primary antibodies used are listed as follows: anti-PTBP3 (sc-100845, Santa Cruz Biotechnology, Santa Cruz, CA, USA), anti-PARP-1 (GTX100573, GeneTex, Irvine, CA, USA), anti-cleaved caspase-3 (9664s, Cell Signaling Technology, Beverly, MA, USA), anti-cleaved caspase-9 (sc-22182, Santa Cruz Biotechnology), anti-PTB (12582-1-AP, Proteintech, Rosemont, IL, USA), anti-TYMS (15047-1-AP, Proteintech), anti-MRP1 (GTX116046, GeneTex), anti-MRP2 (7985-1, Epitomics, Burlingame, CA, USA), anti-p-gp (7719-1, Epitomics), anti-Akt (4691, Cell Signaling Technology), anti-p-Akt (4060, Cell Signaling Technology), and anti-HDAC6 (12834-1-AP, Proteintech). Propidium iodide (PI) and 5-bromo-4-chloro-3-indolyl-4phosphate/nitro blue tetrazolium (BCIP/NBT) were purchased from Sigma Chemical.
Statistical analysis
All Data were obtained from triplicate experiments and expressed as mean± s.d. As for the PTBP3 level in gastric cancer tissues, Mann Whitney tests were employed. For the data of apoptotic subpopulation obtained from flow cytometry assays, Chi-square tests were applied. To compare the effect of PTBP3 silencing on the viability of gastric cancer cells treated with 5-FU, two-way ANOVA tests were performed. Student’s t-tests were employed for analysing other data. The significance of difference was described as *P<0.05; **P<0.01; ***P<0.001.
Results
Expression level of PTBP3 in gastric cancer tissues and cells
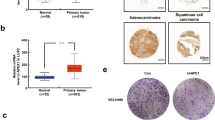
Our previous study has found that PTBP3 protein was overexpressed in gastric cancer tissues (Chen et al, 2014). To further confirm these results, we first tested the mRNA expression of PTBP3 in tissue samples from gastric cancer patients. Mann–Whitney test showed significantly higher PTBP3 level in tumour tissues than in adjuvant non-tumour (NT) tissues (Figure 1A). Comparison of PTBP3 level in paired cancer and NT tissues revealed that PTBP3 was upregulated in 23 cases (23/26, 88.5%) involved in this study (Figure 1B). Further, PTBP3 expression was found to be associated with lymph node metastasis and differentiation of the tumours. Consistently, higher PTBP3 was observed in tumour tissues from stage III patients. However, no significant difference of PTBP3 level was observed between subcategories divided by age (⩽60 or >60) or gender (Table 1).
Expression of PTBP3 in gastric cancer tissues, adjuvant non-tumour (NT) tissues and gastric cancer cell lines were detected. (A) Mann–Whitney test showed higher expression of PTBP3 in tumour tissues than in NT tissues. (B) PTBP3 was upregulated in 88.5% cases (23/26) involved in this study. (C) PTBP3 levels in gastric cancer cell lines.
Next, we tested PTBP3 expression in five gastric cancer cell lines. The results showed that PTBP3 was highly-expressed in BGC823 and MGC803 cells and relatively lowly-expressed in AGS, MKN45, and SGC7901 cells (Figure 1C). The expression of PTBP3 in gastric cancer cells was higher than that in NT tissues as well. Taken together, PTBP3 was upregulated in gastric cancer tissues and cell lines at the mRNA level, suggesting that it might play an important role in gastric cancer.
Inhibition of PTBP3 in gastric cancer cells induced apoptosis and cell cycle arrest
Further, we investigated the impact of PTBP3-silencing on gastric cancer cells. PTBP3 was knocked down in MKN45 cells using siRNAs or a pSilencer vector (PTBP3-pSilencer). Non-targeting siRNA (si-NC) and pSilencer 2.1-U6 neo negative control plasmid (NC-pSilencer) were employed as respective negative controls. Sequences of the siRNAs and that expressed by the pSilencer plasmid were distinct and non-overlapping. The results showed prompted cleavage of poly-ADP-ribose polymerase (PARP-1), caspase-3, and caspase-9 in PTBP3-silenced cells, suggesting that inhibition of PTBP3 expression led to activation of mitochondrial apoptotic pathway in MKN45 cells (Figure 2A). Similar results were observed in MGC803 and BGC823 cells with PTBP3 inhibited by PTBP3-pSilencer as well (Figure 2A).
PTBP3 inhibition induced apoptosis and cell cycle arrest in gastric cancer cells. (A) Cleavage of caspase-3, PARP-1, and caspase-9 were activated upon PTBP3-silencing. (B) Higher apoptotic subpopulations in gastric cancer cells upon PTBP3 silencing were detected via Annexin-FITC/PI staining and FACS analyses (***P<0.001, χ2-test). (C) PI staining and FACS analyses showed cell cycle arrest at S-phase in PTBP3-silenced gastric cancer cells (**P<0.01, ***P<0.001, Student’s t-test). KD, cells transfected with PTBP3-targeting pSilencer plasmid; NC, cells transfected with negative control pSilencer 2.1-U6 neo plasmid; si-NC, cells transfected with non-targeting siRNA; si-1 and si-2, cells transfected with different PTBP3-targeting siRNAs.
Additionally, PTBP3 silencing-induced apoptosis was confirmed by Annexin V-FITC/propidium iodide (PI) staining and flow cytometry analyses (Figure 2B). We also analysed the DNA content in cells transfected with PTBP3-pSilencer. Interestingly, PTBP3 inhibition led to S-phase arrest in all three cell lines detected (Figure 2C). Higher sub-G1 population was also observed in PTBP3-silenced cells as well, which confirmed the effect of PTBP3-silencing on cellular apoptosis (data not shown). Taken together, PTBP3 silencing induced apoptosis and arrested cell cycle progression in human gastric cancer cells, which may be responsible for the growth inhibition of PTBP3-silenced cells in vivo.
PTBP3 switched the AS of Bcl-x via modulating PTB expression
We further investigated the molecular mechanism underlying PTBP3’s regulatory effect on apoptosis. Activation of caspase-9 could trigger a caspase signalling cascade, such as the loss of mitochondrial membrane potential and the cleavage of anti-apoptotic Bcl-2, Bcl-xL, and Mcl-1 (Chen et al, 2007). Since caspase-9 was activated upon PTBP3 inhibition, we speculated that PTBP3 might be able to regulate the AS of Bcl-2 family members. Among these regulators, Bcl-x has been found to generate two mRNA isoforms via AS: anti-apoptotic Bcl-xL and pro-apoptotic Bcl-xS. We thus explored the effect of PTBP3 silencing on Bcl-x AS. Bcl-xL and Bcl-xS mRNAs were detected using specific primers and the results showed elevated Bcl-xS/Bcl-xL ratio in PTBP3-silenced gastric cancer cells (Figure 3A and B). Therefore, knockdown of PTBP3 promoted the generation of Bcl-xS, which was, at least partly, responsible for the apoptosis induced in PTBP3-silenced cells.
PTBP3 knockdown switched the splicing pattern of Bcl-x via modulating PTB expression. Inhibition of PTBP3 (A) in gastric cancer cells promoted the generation of Bcl-xS (B) and upregulated the expression of PTB (C, D) (*P<0.05, **P<0.01, ***P<0.001, Student’s t test). (E) PTB knockdown reversed the increase of Bcl-xS/Bcl-xL ratio resulting from PTBP3-silencing. * indicates the difference between the NC group and each of the rest groups; # indicates the difference between the PTBP3 KD and the PTB/PTBP3 KD group (*P<0.05, **P<0.01, ***P<0.001, ##P<0.01, Student’s t test).
However, it has been reported that PTBP3 protein could not bind to Bcl-x mRNA (Brazão et al, 2012), indicating that PTBP3 might modulate the AS of Bcl-x in an indirect way. Numerous splicing factors have been reported to regulate the splicing event of Bcl-x, among which PTB is a recently identified one (Bielli et al, 2014). Moreover, PTB has been found to regulate the expression of PTBP3 and itself via switch their splicing. We thus speculated that PTBP3 might also be able to modulate the expression level of PTB in a similar way and thereby affect the AS of Bcl-x. Consistent with our hypothesis, qPCR and Western Blot assays showed that in PTBP3-inhibited cells, both PTB mRNA and protein levels were increased (Figure 3C and D).
To further confirm the intermediate role of PTB, a PTB targeting pSilencer plasmid (PTB-pSilencer) was constructed, and MKN45 cells were transfected with PTBP3- and/or PTB-pSilencer. Consistent with previous reports (Spellman et al, 2007; Han et al, 2014), inhibition of PTB in MKN45 cells led to upregulation of PTBP3 mRNA and protein level (Figure 3E). Moreover, PTB silencing reversed the increase of Bcl-xS/Bcl-xL ratio resulting from PTBP3-silencing (Figure 3E). These results indicated that PTBP3 regulated the AS of Bcl-x in a way mediated by PTB.
PTBP3 modulated the chemosensitivity of gastric cancer cells towards 5-FU and regulated the expression of multi-drug resistance-related proteins
Resisting apoptosis is a critical mechanism underlying drug resistance (Wu et al, 2014). We thus further investigated whether PTBP3 knockdown could enhance the chemosensitivity of gastric cancer cells. MKN45, MGC803, and BGC823 cells were first transfected with NC- or PTBP3-pSilencer, and then treated with 5-FU. MTT assays showed that inhibition of PTBP3 could sensitise gastric cancer cells towards 5-FU and L-OHP treatment (Figure 4A and 5A). In addition, we employed a PTBP3-expressing plasmid (PTBP3-pcDNA 3.1(+)) to induce PTBP3 in SGC7901 cells. The efficiency of PTBP3-pcDNA 3.1(+) in SGC7901 cells was first validated by a western blot assay and pcDNA 3.1(+) empty vector was used as a control. MTT assays further showed that cells pre-transfected with PTBP3-pcDNA 3.1(+) were more resistant against 5-FU than those pre-transfected with pcDNA 3.1(+) vector (Figure 4B). These results indicated that PTBP3 contributed to 5-FU resistance in gastric cancer cells.
PTBP3 modulated the chemosensitivity of gastric cancer cells. (A) PTBP3 inhibition enhanced the cytotoxicity of 5-FU in gastric cancer cells (**P<0.01, ***P<0.001, #P<0.05, ##P<0.01, two-way ANOVA). (B) Ectopic expression of PTBP3 enhanced the resistance of SGC7901 cells against 5-FU (NC: cells transfected with pcDNA 3.1(+) empty vector; OE: cells transfected with PTBP3-pcDNA 3.1(+) plasmid, **P<0.01, ***P<0.001, #P<0.05, two-way ANOVA). (C) Lower expression of TYMS, MRP2, and p-gp was observed in PTBP3-silenced gastric cancer cells, while MRP1 level was not affected by PTBP3 modulation. Similar mRNA expression trends of TYMS (D), MRP2 (E), and p-gp (F) were detected (**P<0.01, ***P<0.001, Student’s t-test).
PTBP3 knockdown inhibited HDAC6/Akt pathway. (A) PTBP3 inhibition enhanced the cytotoxicity of L-OHP in gastric cancer cells (**P<0.01, ***P<0.001, #P<0.05, ##P<0.01, two-way ANOVA). (B) Inhibition of PTBP3 led to downregulation of HDAC6 protein and dephosphorylation of Akt in gastric cancer cells. (C) HDAC6 mRNA expression were downregulated in PTBP3-silenced cells as well (**P<0.01, ***P<0.001, Student’s t test). (D) TYMS and HDAC6 mRNA expression were upregulated in PTB-silenced MKN45 cells as well (**P<0.01, Student’s t-test). (E) Inhibition of PTB led to upregulation of HDAC6 protein and phosphorylation of Akt in MKN45 cells.
It has been reported that thymidylate synthase (TYMS) is a critical determinant of 5-FU cytotoxicity (Kawate et al, 2002). We thus explored the effect of PTBP3 silencing on the expression of TYMS. The results showed reduced TYMS mRNA and protein levels upon PTBP3 silencing (Figure 4C and D). This was consistent with our previous results that PTBP3 knockdown led to cell cycle arrest at S phase, since TYMS was found to play an important role in DNA biosynthesis (Carreras and Santi, 1995).
Moreover, we investigated whether PTBP3 may be involved in regulating the multi-drug resistance of gastric cancer cells. The protein levels of putative drug transporters MRP1, MRP2 and p-gp were detected. Lower protein levels of MRP2 and p-gp, but not MRP1, were found in PTBP3-silenced cells (Figure 4C). QPCR assays showed that mRNA expression of MRP2 and p-gp was also downregulated (Figure 4E and F).
PTBP3 inhibition-induced cell chemosensitisation was mediated by HDAC6/Akt pathway
We further wished to identify the mechanism by which PTBP3 modulated the expression of TYMS. It has been reported that Akt inhibitor (LY294002) decreased TYMS expression in chemoresistant colorectal cancer cells (Ahn et al, 2015). Consistently, PTBP3 knockdown in gastric cancer cells inhibited the phosphorylation of Akt as well (Figure 5B). Furthermore, PTB knockdown improved the phosphorylation of Akt (Figure 5E). Among all the proteins whose mRNA has been identified to physically interact with PTBP3 (Brazão et al, 2012), HDAC6 has been reported to be a regulator of p-Akt level (Balliu et al, 2015). We thus tested whether HDAC6 mediated PTBP3 inhibition-induced cell chemosensitisation. QPCR and Western Blot assays showed both HDAC6 mRNA and protein were downregulated in PTBP3-silenced cells, and upregulated in PTB-silenced cells (Figure 5B–E). Taken together, these results indicated that PTBP3 promoted the expression of HDAC6 in gastric cancer cells, which further enhanced the phosphorylation of Akt and thereby elevated the TYMS level.
Discussion
PTBP3 has been characterised as a regulator of cell differentiation since its first discovery in 1999 (Yamamoto et al, 1999). Recently, several studies have reported the potential role of PTBP3 in regulating other biological events. For example, PTBP3 was found to enhance hypoxia-induced cell death in HEK293 cells (Fasanaro et al, 2012). Moreover, physical interaction between PTBP3 and mitochondrial tRNA suggested that PTBP3 might be involved in regulating cellular apoptosis via a non-canonical way (Marnef et al, 2015).
The role of PTBP3 in human cancers appears to be tissue specific. PTBP3 was found to be significantly lower in glioblastoma multiforme tissues than in normal brain tissues (Zhang et al, 2014). However, higher expression of PTBP3 was observed in lung squamous cell carcinoma (Ooi et al, 2014). Moreover, downregulation of PTBP3 inhibited the migration of lung cancer cells (Tano et al, 2010). Our previous study showed elevated PTBP3 in gastric cancer tissues; knockdown of PTBP3 led to poorer growth of gastric cancer cells in vivo (Chen et al, 2014). In the present study, we confirmed that PTBP3 mRNA level was higher in gastric cancer tissues and cells. Further research found that inhibition of PTBP3 expression could induce apoptosis and inhibited drug resistance against 5-FU in gastric cancer cells.
Previous studies have reported that knockdown of PTB promoted PTBP3 exon2 skipping, which generated a isoform that cannot be translated efficiently (Han et al, 2014). Consistently, our results showed that the expression of PTBP3 was elevated upon PTB-silencing. Indeed, inhibition of PTBP3 exerted similar effect on the expression of PTB, suggesting a mutual regulation between these two paralogs. Notably, the inclusion of PTB exon 11 has been reported to be repressed by PTB protein itself, which generated a frameshifted mRNA degraded by nonsense-mediated decay (NMD; Wollerton et al, 2004). Since PTBP3 and PTB share more than 70% identical sequences and a common arrangement of RNA recognition motifs (RRMs; Spellman et al, 2007), they may function similarly at least in several splicing events. Therefore, PTBP3 may regulate the expression of PTB in the same way as PTB itself does.
A typical example of the splicing events within Bcl-2 family is the one of Bcl-x. Alternative usage of 5′ splice site in the exon 2 of Bcl-x pre-mRNA generates two variants with opposite functions. The longer isoform, Bcl-xL, has been reported to be anti-apoptotic whose overexpression was associated with drug resistance and poor prognosis (Zhou et al, 2015); on the contrary, the shorter one, Bcl-xS, was found to promote apoptosis (Plötz et al, 2012). The regulation of Bcl-x splicing has been proved to be complicated, which involved several splicing factors (Cloutier et al, 2008; Revil et al, 2009; Pedrotti et al, 2012; Bielli et al, 2014). Our exploration found that inhibition of PTBP3 expression indirectly promoted the generation of Bcl-xS via upregulating PTB.
It has been reported that HDACs are involved in the acquisition of 5-FU resistance (Lee et al, 2006; Iwahashi et al, 2011). Our exploration found that chemosensitisation induced by PTBP3 knockdown was mediated by HDAC6/Akt/TYMS pathway, which confirmed the role of HDAC6 in regulating the chemoresistance against 5-FU. Indeed, HDAC6 has been reported to form a complex with protein phosphatase 1 (PP1). Disruption of HDAC6/PP1 complex released active PP1 which could dephosphorylate Akt (Balliu et al, 2015). Inhibition of Akt was found to decrease the expression of TYMS (Ahn et al, 2015), critical determinant of 5-FU sensitivity in cancers. These reports were consistent with our findings that inhibition of PTBP3 in gastric cancer cells led to dephosphorylation of Akt and lower level of TYMS. The impact of PTBP3 knockdown on cell cycle progression may be mediated by TYMS as well since it catalyses the sole pathway of thymidylate biosynthesis (Chu et al, 2003). Inhibition of TYMS in PTBP3-silenced cells dysregulated the biosynthesis of DNA and thereby led to the S phase arrest.
Aside from inhibiting TYMS, PTBP3 silencing also reduced the expression of MRP2 and p-gp, but not that of MRP1. We hypothesised that this might also result from Akt inactivation, which has been reported to induce the expression of CHOP (Hyoda et al, 2006). Human MRP2 and MDR1 gene contains a C/EBPβ-binding site while MRP1 gene does not (Tanaka et al, 1999; Scotto, 2003; Baudis et al, 2006). Indeed, CHOP has been found to modulate the transcription of genes with C/EBPβ-binding site (Elizondo et al, 2009). Therefore, CHOP may mediate the downregulation of MRP2 and p-gp by PTBP3 silencing. This further confirmed the role of PTBP3 in regulating HDAC6/Akt pathway.
In summary, our present study identified the potential role of PTBP3 in regulating apoptosis and chemoresistance against 5-FU in gastric cancer cells. Further exploration revealed the involvement of PTB and HDAC6/Akt/TYMS pathway as underlying mechanisms. These results showed that PTBP3 may serve as a potent target for the clinical treatment of gastric cancer.
Change history
28 March 2017
This paper was modified 12 months after initial publication to switch to Creative Commons licence terms, as noted at publication
References
Ahn JY, Lee JS, Min HY, Lee HY (2015) Acquired resistance to 5-fluorouracil via HSP90/Src-mediated increase in thymidylate synthase expression in colon cancer. Oncotarget 6 (32): 32622–32633.
Balliu M, Guandalini L, Romanelli MN, D'Amico M, Paoletti F (2015) HDAC-inhibitor (S)-8 disrupts HDAC6-PP1 complex prompting A375 melanoma cell growth arrest and apoptosis. J Cell Mol Med 19 (1): 143–154.
Baudis M, Prima V, Tung YH, Hunger SP (2006) ABCB1 over-expression and drug-efflux in acute lymphoblastic leukemia cell lines with t(17;19) and E2A-HLF expression. Pediatr Blood Cancer 47 (6): 757–764.
Bielli P, Bordi M, Di Biasio V, Sette C (2014) Regulation of BCL-X splicing reveals a role for the polypyrimidine tract binding protein (PTBP1/hnRNP I) in alternative 5′ splice site selection. Nucleic Acids Res 42 (19): 12070–12081.
Brazão TF, Demmers J, van IJcken W, Strouboulis J, Fornerod M, Romão L, Grosveld F (2012) A new function of ROD1 in nonsense-mediated mRNA decay. FEBS Lett 586 (8): 1101–1110.
Carreras CW, Santi DV (1995) The catalytic mechanism and structure of thymidylate synthase. Annu Rev Biochem 64: 721–762.
Chabot B, Shkreta L (2016) Defective control of pre-messenger RNA splicing in human disease. J Cell Biol 212 (1): 13–27.
Chen B, Zhao AG, Shao J, Mu XY, Jiang L, Liu JW (2014) The effects of PTBP3 silencing on the proliferation and differentiation of MKN45 human gastric cancer cells. Life Sci 114 (1): 29–35.
Chen M, Guerrero AD, Huang L, Shabier Z, Pan M, Tan TH, Wang J (2007) Caspase-9-induced mitochondrial disruption through cleavage of anti-apoptotic BCL-2 family members. J Biol Chem 282 (46): 33888–33895.
Chu E, Callender MA, Farrell MP, Schmitz JC (2003) Thymidylate synthase inhibitors as anticancer agents: from bench to bedside. Cancer Chemother Pharmacol 52 (1): 80–89.
Cloutier P, Toutant J, Shkreta L, Goekjian S, Revil T, Chabot B (2008) Antagonistic effects of the SRp30c protein and cryptic 5′ splice sites on the alternative splicing of the apoptotic regulator Bcl-x. J Biol Chem 283 (31): 21315–21324.
Elizondo G, Medina-Díaz IM, Cruz R, Gonzalez FJ, Vega L (2009) Retinoic acid modulates retinaldehyde dehydrogenase 1 gene expression through the induction of GADD153-C/EBPβ interaction. Biochem Pharmacol 77 (2): 248–257.
Fasanaro P, Romani S, Voellenkle C, Maimone B, Capogrossi MC, Martelli F (2012) ROD1 is a seedless target gene of hypoxia-induced miR-210. PLoS One 7 (9): e44651.
Han A, Stoilov P, Linares AJ, Zhou Y, Fu X-D, Black DL (2014) De novo prediction of PTBP1 binding and splicing targets reveals unexpected features of its RNA recognition and function. PLoS Comput Biol 10 (1).
Hosoda T, Zheng H, Cabral-da-Silva M, Sanada F, Ide-Iwata N, Ogórek B, Ferreira-Martins J, Arranto C, D'Amario D, del Monte F (2011) Human cardiac stem cell differentiation is regulated by a mircrine mechanism. Circulation 123 (12): 1287–1296.
Hyoda K, Hosoi T, Horie N, Okuma Y, Ozawa K, Nomura Y (2006) PI3K-Akt inactivation induced CHOP expression in endoplasmic reticulum-stressed cells. Biochem Biophys Res Commun 340 (1): 286–290.
Iwahashi S, Ishibashi H, Utsunomiya T, Morine Y, Lkhaguva Ochir T, Hanaoka J, Mori H, Ikemoto T, Imura S, Shimada M (2011) Effect of histone deacetylase inhibitor in combination with 5-fluorouracil on pancreas cancer and cholangiocarcinoma cell lines. J Med Invest 58 (1, 2): 106–109.
Kawate H, Landis DM, Loeb LA (2002) Distribution of mutations in human thymidylate synthase yielding resistance to 5-fluorodeoxyuridine. J Biol Chem 277 (39): 36304–36311.
Lee JH, Park JH, Jung Y, Kim JH, Jong HS, Kim TY, Bang YJ (2006) Histone deacetylase inhibitor enhances 5-fluorouracil cytotoxicity by down-regulating thymidylate synthase in human cancer cells. Mol Cancer Ther 5 (12): 3085–3095.
Lin S, Lei K, Du W, Yang L, Shi H, Gao Y, Yin P, Liang X, Liu J (2016) Enhancement of oxaliplatin sensitivity in human colorectal cancer by hypericin mediated photodynamic therapy via ROS-related mechanism. Int J Biochem Cell B 71: 24–34.
Mao R, Zou F, Yang L, Lin S, Li Y, Ma M, Yin P, Liang X, Liu J (2015) The loss of MiR-139-5p promotes colitis-associated tumorigenesis by mediating PI3K/AKT/Wnt signaling. Int J Biochem Cell B 69: 153–161.
Marnef A, Jady BE, Kiss T (2015) Human polypyrimidine tract-binding protein interacts with mitochondrial tRNAThr in the cytosol. Nucleic Acids Res 44 (3): 1342–1353.
Ooi AT, Gower AC, Zhang KX, Vick JL, Hong L, Nagao B, Wallace WD, Elashoff DA, Walser TC, Dubinett SM (2014) Molecular profiling of premalignant lesions in lung squamous cell carcinomas identifies mechanisms involved in stepwise carcinogenesis. Cancer Prev Res 7 (5): 487–495.
Pedrotti S, Busa R, Compagnucci C, Sette C (2012) The RNA recognition motif protein RBM11 is a novel tissue-specific splicing regulator. Nucleic Acids Res 40 (3): 1021–1032.
Plötz M, Gillissen B, Hossini A, Daniel P, Eberle J (2012) Disruption of the VDAC2-Bak interaction by Bcl-xS mediates efficient induction of apoptosis in melanoma cells. Cell Death Differ 19 (12): 1928–1938.
Revil T, Pelletier J, Toutant J, Cloutier A, Chabot B (2009) Heterogeneous nuclear ribonucleoprotein K represses the production of pro-apoptotic Bcl-xS splice isoform. J Biol Chem 284 (32): 21458–21467.
Sadvakassova G, Dobocan MC, Difalco MR, Congote LF (2009) Regulator of differentiation 1 (ROD1) binds to the amphipathic C-terminal peptide of thrombospondin-4 and is involved in its mitogenic activity. J Cell Physiol 220 (3): 672–679.
Scotto KW (2003) Transcriptional regulation of ABC drug transporters. Oncogene 22 (47): 7496–7511.
Shen K, Zhang H, Sun L, Xu Y, Qian X, Liu J (2013) A ROS-mediated lysosomal-mitochondrial pathway is induced by a novel Amonafide analogue, 7c, in human Hela cervix carcinoma cells. Cancer Lett 333 (2): 229–238.
Spellman R, Llorian M, Smith CW (2007) Crossregulation and functional redundancy between the splicing regulator PTB and its paralogs nPTB and ROD1. Mol Cell 27 (3): 420–434.
Tanaka T, Uchiumi T, Hinoshita E, Inokuchi A, Toh S, Wada M, Takano H, Kohno K, Kuwano M (1999) The human multidrug resistance protein 2 gene: Functional characterization of the 5′-flanking region and expression in hepatic cells. Hepatology 30 (6): 1507–1512.
Tano K, Mizuno R, Okada T, Rakwal R, Shibato J, Masuo Y, Ijiri K, Akimitsu N (2010) MALAT-1 enhances cell motility of lung adenocarcinoma cells by influencing the expression of motility-related genes. FEBS Lett 584 (22): 4575–4580.
Wagner EJ, Garcia-Blanco MA (2001) Polypyrimidine tract binding protein antagonizes exon definition. Mol Cell Biol 21 (10): 3281–3288.
Wollerton MC, Gooding C, Wagner EJ, Garcia-Blanco MA, Smith CW (2004) Autoregulation of polypyrimidine tract binding protein by alternative splicing leading to nonsense-mediated decay. Mol Cell 13 (1): 91–100.
Wu Q, Yang Z, Nie Y, Shi Y, Fan D (2014) Multi-drug resistance in cancer chemotherapeutics: Mechanisms and lab approaches. Cancer Lett 347 (2): 159–166.
Xu K, Liang X, Cui D, Wu Y, Shi W, Liu J (2013) miR-1915 inhibits Bcl-2 to modulate multidrug resistance by increasing drug-sensitivity in human colorectal carcinoma cells. Mol Carcinog 52 (1): 70–78.
Yamamoto H, Tsukahara K, Kanaoka Y, Jinno S, Okayama H (1999) Isolation of a mammalian homologue of a fission yeast differentiation regulator. Mol Cell Biol 19 (5): 3829–3841.
Zhang S, Lai N, Liao K, Sun J, Lin Y (2014) MicroRNA-210 regulates cell proliferation and apoptosis by targeting regulator of differentiation 1 in glioblastoma cells. Folia Neuropathol 53 (3): 236–244.
Zheng Y, Le V, Cheng Z, Xie S, Li H, Tian J, Liu J (2013) Development of rapid and highly sensitive HSPA1A promoter-driven luciferase reporter system for assessing oxidative stress associated with low-dose photodynamic therapy. Cell Stress Chaperones 1–11.
Zheng Y, Zou F, Wang J, Yin G, Le V, Fei Z, Liu J (2015) Photodynamic therapy-mediated cancer vaccination enhances stem-like phenotype and immune escape, which can be blocked by thrombospondin-1 signaling through CD47 receptor protein. J Biol Chem 290 (14): 8975–8986.
Zhou W, Xu J, Gelston E, Wu X, Zou Z, Wang B, Zeng Y, Wang H, Liu A, Xu L, Liu Q (2015) Inhibition of Bcl-xL overcomes polyploidy resistance and leads to apoptotic cell death in acute myeloid leukemia cells. Oncotarget 6 (25): 21557–21571.
Acknowledgements
This work was supported by National Nature Science Funds of China (NO. 81173224, 81270555, 81373861). We appreciate the help of Professor Zhewei Fei and Mr Wen Xu at Shanghai Xinhua Hospital Affiliated to School of Medicine, Shanghai Jiaotong University in collecting and providing human gastric cancer samples.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License.
Rights and permissions
From twelve months after its original publication, this work is licensed under the Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/4.0/
About this article
Cite this article
Liang, X., Shi, H., Yang, L. et al. Inhibition of polypyrimidine tract-binding protein 3 induces apoptosis and cell cycle arrest, and enhances the cytotoxicity of 5- fluorouracil in gastric cancer cells. Br J Cancer 116, 903–911 (2017). https://doi.org/10.1038/bjc.2017.32
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/bjc.2017.32
Keywords
This article is cited by
-
Inhibition of histone deacetylase 6 destabilizes ERK phosphorylation and suppresses cancer proliferation via modulation of the tubulin acetylation-GRP78 interaction
Journal of Biomedical Science (2023)
-
MiR-297 inhibits tumour progression of liver cancer by targeting PTBP3
Cell Death & Disease (2023)
-
PTBP3 regulates proliferation of lung squamous cell carcinoma cells via CDC25A‐mediated cell cycle progression
Cancer Cell International (2022)
-
RETRACTED ARTICLE: LncRNA BCRT1 promotes breast cancer progression by targeting miR-1303/PTBP3 axis
Molecular Cancer (2020)
-
PTBP3 promotes migration of non-small cell lung cancer through regulating E-cadherin in EMT signaling pathway
Cancer Cell International (2020)