Abstract
Background:
The current study aimed to establish a novel nomogram to predict the overall survival of individual Chinese patients with gastroenteropancreatic neuroendocrine neoplasms (GEP-NENs). Furthermore, this study sought to externally validate this nomogram using the Surveillance, Epidemiology, and End Results (SEER) database.
Methods:
The records of 1183 patients with GEP-NENs treated at five high-capacity institutions in China between 2005 and 2015 were retrospectively analysed. In addition, 10 236 GEP-NEN cases from the SEER database were included as an external validation set.
Results:
A multivariate analysis using Cox proportional hazards (PHs) regression was performed, and a nomogram was constructed. Discrimination, calibration, and external validation were performed using the SEER data set. The multivariate Cox model indicated that age, tumour size, differentiation, lymph node metastases, and distant metastases were independent covariates associated with survival. With respect to the training set, the nomogram exhibited better discrimination power than TNM classification (Harrell’s concordance index (C-index): 0.837 vs 0.784, P=0.006). Discrimination was also excellent and superior to that of TNM classification for the SEER-based validation set (C-index: 0.808 vs 0.717, P<0.001). The calibrated nomogram predicted a survival rate that closely corresponded to the actual survival rate.
Conclusions:
We developed a nomogram that predicted the 3- and 5-year overall survival rates of patients with GEP-NENs. Validation revealed excellent discrimination and calibration for this nomogram, suggesting that it exhibits satisfactory clinical utility that might improve individualised predictions of survival risks and lead to the creation of additional clinical therapies.
Similar content being viewed by others
Main
Neuroendocrine neoplasms (NENs), a heterogeneous group of rare tumours, consist of a spectrum of malignancies that can arise from neuroendocrine cells throughout the body. Neuroendocrine neoplasms are primarily derived from the gastroenteropancreatic system (i.e., GEP-NENs) and lungs (Modlin et al, 2008). From 1973 to 2004, the reported annual age-adjusted incidence of NENs significantly increased from 1.09/100 000 to 5.25/100 000 in the United States (Yao et al, 2008). Because of their complex and heterogeneous biological behaviour and associated treatments, prognoses for patients with GEP-NENs remain unclear. To date, the most common predictive systems for NENs are the American Joint Committee on Cancer (AJCC) and European Neuroendocrine Tumour Society (ENETS) TNM staging system, which is based on the depth of tumour invasion (T), the number of metastatic lymph nodes (N), and the distant metastases (M). Using the World Health Organisation (WHO) classification proposed in 2010, mitotic count and the Ki-67 proliferative index have also provided potential prognoses of this special type of malignancy (Bosman et al, 2010). Furthermore, other clinicopathological features such as gender, age at diagnosis, tumour location, and primary tumour size can also influence patient outcomes (Greene and Sobin, 2008). To assist with clinical practice, decision making, and clinical trial design, accurate prognoses must identify homogeneous, high-risk patient groups.
Of the available decision-making tools, nomograms are currently the most accurate and discriminatory for predicting outcomes among patients with cancer (Shariat et al, 2008). According to a statistical definition, a nomogram is a graphical calculation or algorithm that incorporates several continuous variables to predict a particular end point using traditional statistical methods (e.g., logistic or Cox proportional hazards (PHs) regression models) (Kattan, 2002). Nomograms use continuous scales to calculate the probability of a particular outcome. The effect of these continuous variables on specific outcomes is represented on the axes, and risk points are attributed based on the prognostic importance of the variable of interest.
Previous studies have successfully quantified risk by combining the prognostic factors for certain malignancies (Wierda et al, 2007, 2008; Rudloff et al, 2010; Wang et al, 2011; Han et al, 2012; Hirabayashi et al, 2014). To date, however, few studies have used a nomogram to predict outcomes for patients with GEP-NENs; these studies have included relatively small samples of patients with small intestinal NENs (Modlin et al, 2010), pancreatic NENs (Ellison et al, 2014; Ruzzenente et al, 2016; Ye et al, 2016), or gastric NENs (Cao et al, 2016).
The present study sought to develop a more elaborative nomogram to predict 3- and 5-year overall survival rates based on a relative large cohort of patients with GEP-NENs from multiple centres in China. Furthermore, this study aimed to externally validate this nomogram using the Surveillance, Epidemiology, and End Result (SEER) database.
Materials and methods
Patients
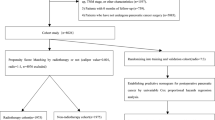
We retrieved all data collected from patients with GEP-NENs who were treated between January 2005 and December 2015 at five high-capacity medical centres in Southern China (henceforth referred to as the GD-NEN data set), including Sun Yat-sen University Cancer Center (SYSUCC, n=482), the First Affiliated Hospital of Sun Yat-sen University (SYSUFH, n=309), Guangdong General Hospital (GGH, n=143), Nanfang Hospital of Southern Medical University (SMUNH, n=131), and Sun Yet-san Memorial Hospital of Sun Yat-sen University (SYSUMH, n=118), using the medical management systems. The above five hospitals belong to Guangdong Neuroendocrine Neoplasms Cooperative Group and members of Chinese Study Group for Neuroendocrine Tumors (CSNET). Wei Wang and Cheng Fang reviewed and crosschecked all deidentified data for inconsistencies. The above centres are large-capacity hospitals in southern China that have shown a long-term and productive regional cooperation that led to the development of specific, multidisciplinary diagnosis and treatment teams focusing on GEP-NENs. Gastroenteropancreatic NENs (GEP-NENs) were defined and classified histopathologically according to the 2010 WHO classification criteria. These criteria include complete demographic data, clinical parameters, pathological findings, staging information, therapeutic procedure records, and full follow-up results. In total, the data of 1183 patients of Chinese cohort were enrolled and analysed in the present study.
Regarding the SEER data sets, we screened the cases of GEP-NENs using the newest database, ‘Incidence – SEER 18 Regs Research Data+Hurricane Katrina Impacted Louisiana Cases, Nov 2015 Sub (1973–2013 varying)’. The International Classification of Diseases for Oncology (ICD-O-3) was used to identify cases of NENs. The following ICD-O-3 codes for histological type were used: large cell carcinoma (8012–8013), small cell carcinoma (8041–8044), islet cell carcinoma (8150), malignant insulinoma (8151), malignant glucagonoma (8152), malignant gastrinoma (8153), vipoma (8155), malignant somatostatinoma (8156), malignant enteroglucagonoma (8157), carcinoid tumour (8240), argentaffin carcinoid tumour (8241), enterochromaffin cell tumour (8242), mucocarcinoid tumour (8243), mixed adenoneuroendocrine carcinoma (8244), adenocarcinoid tumour (8245), neuroendocrine carcinoid (8246), and atypical carcinoid tumour (8249). The ‘Site recode ICD-O-3/WHO 2008’ data were used to filter by tumour location, including stomach, pancreas, small intestine, cecum, appendix, ascending colon, hepatic flexure, transverse colon, splenic flexure, descending colon, sigmoid colon, large intestine, rectum/rectosigmoid junction, rectum, colon, and rectum. Year of diagnosis was set to range from 2005 to 2013. The TNM staging data were retrieved based on the following codes: derived AJCC TNM stage group 7th ed. (2010+), derived AJCC TNM stage group 6th ed. (2004+), CS tumour size (2004+), CS lymph nodes (2004+), and CS mets at dx (2004+). Survival information was retrieved using the phrases the ‘vital status recode’ and ‘survival months’. Totally the data of 10 236 cases of SEER data set were enrolled and analysed in the present study.
The institutional review boards of SYSUCC, SYSUFH, GGH, SMUNH, and SYSUMH approved the study protocol.
Nomogram construction
For nomogram construction and external validation, the Chinese patient data set was used as the training set, and the SEER database was used as the external validation set. A multivariate analysis was conducted using a Cox PHs model without violating the PH assumption.
The clinicopathological characteristics of the training and validation sets were evaluated. The PH and linearity assumptions for the continuous variables (i.e., age, tumour size) were examined using restricted cubic splines (Hess, 1994). To maximise predictive ability, continuous variables were transformed to adequately fit the PH and linearity assumptions, when possible. The categorical variables (i.e., tumour location, tumour invasion, and distant metastases) were grouped for clinical reasons, and the decisions regarding grouping were made before modelling. A log–log survival plot of the categorical variables was used to determine whether the PH assumption was appropriate, and all variables were fit to the PH assumption. Variables were selected using the forward stepwise selection method in the Cox PH regression model. Based on the predictive model using the identified prognostic factors, a nomogram was constructed to predict the 3- and 5-year overall survival rates.
Nomogram validation
The performance of the nomogram included its discrimination and calibration using the SEER external validation set. Discrimination was evaluated using a concordance index (C-index), which quantifies the probability that of two random patients, the patient who relapses first had a higher probability of the event of interest. Harrell’s C-index, which is appropriate for censored data, was used to evaluate discrimination (Harrell et al, 1996). Calibration was performed by comparing the mean predicted survival rate with the mean actual survival rate determined using a Kaplan–Meier analysis after grouping the nomogram-predicted survival by decile. In addition, we applied a bootstrapped resample with 1000 iterations to verify the accuracy of the nomogram. Furthermore, the precision of the 3- and 5-year survival predictions was evaluated using the area under receiver operating characteristic (ROC) curve (AUC). Significance was set as P<0.05 in a two-tailed test. All analyses were performed using SPSS version 20 (IBM, Armonk, NY, USA) and R version 2.13.2 (http://www.r-project.org) via the design and survival packages.
Results
The demographic features and clinicopathological characteristics of the training and validation sets are presented in Table 1. Then, univariate analysis were performed to filter prognostic factors (Table 2). According to the Cox PH regression model, which included those filtered factors based on univariate analysis, five clinicopathological factors were independently correlated with prognosis in both the GD-NEN and SEER data sets, including age, tumour size, tumour differentiation, lymph node metastases, and distant metastases. Table 3 presents the selected variables with their associated hazard ratios (HRs). However, prognosis was not independently correlated with gender, tumour location, or tumour invasion.
Figure 1 illustrates the predictive nomogram established for the 3- and 5-year overall survival rates based on the selected parameters in the training set. Considering clinical utility and tumour biology, we retained tumour location as a parameter in the nomogram model even though this parameter did not exhibit independent prognostic significance. A patient’s probability of individual survival can easily be calculated by adding the scores for each selected variable.
We compared the discrimination of the nomogram with that of the seventh AJCC TNM classification in the training set. The nomogram discrimination was 0.837 (95% CIs=0.813–0.860), which was superior to that of the seventh AJCC TNM classification (0.784, 95% CIs=0.758–0.810, P=0.006). The bootstrapped resample (1000 iterations) used to verify the accuracy of the nomogram via the training set was 0.833. Discrimination was also enhanced compared with the seventh AJCC TNM staging with regard to the SEER validation set (C-index=0.808, 95% CIs=0.798–0.818 vs 0.717, 95% CIs=0.706–0.728, P<0.001).
Furthermore, the two AUC models of the 3- and 5-year overall survival rate regarding the prediction ability of the two data sets were compared (Figure 2). For the GD-NENs data sets, the AUCs of the nomogram predicting the 3- and 5-year overall survival rates were 0.876 and 0.887, respectively, whereas the AUCs of the 7th AJCC TNM staging for predicting the 3- and 5-year overall survival rates were 0.813 and 0.816, respectively. Regarding the SEER data set, the AUCs of the nomogram for predicting the 3- and 5-year overall survival rates were 0.794 and 0.747, respectively, whereas the AUCs of the 7th AJCC TNM staging for predicting the 3- and 5-year overall survival rates were 0.713 and 0.684, respectively. As Figure 2 shows, the nomogram exhibited superior survival predictive ability to that of the 7th AJCC TNM staging system.
The calibration of the nomograms using the GD-NEN training set and SEER validation set. The x axis represents the nomogram-predicted survival rate, whereas the y axis represents the actual survival rate. The 95% CIs were measured via a Kaplan–Meier analysis. All predictions lie within a 10% margin of error (within the dashed lines). (A) Three-year survival rate according to the GD-NENs data set. (B) Five-year survival rate according to the GD-NENs data set. (C) Three-year survival rate according to the SEER data set. (D) Five-year survival rate according to the SEER data set.
A calibration plot was generated to validate the similarities between the survival rates predicted by the nomogram and the actual survival rate (Figure 3). The x axis represents the survival rate predicted by the nomogram, whereas the y axis presents the actual survival rate obtained using the Kaplan–Meier method. The results demonstrate that the predicted 3- and 5-year overall survival rates closely correspond with the actual survival rates within a 10% margin of error represented by the dotted lines according to both the training and validation sets.
Comparison of the AUCs of the nomogram and AJCC TNM staging system. Area under the curves of the two models to predict overall survival at 3 years (A) and 5 years (B) using the GD-NENs training set as well as at 3 years (C) and 5 years (D) using the SEER validation set. The red lines represent nomogram-predicted overall survival rates, whereas the black lines represent AJCC TNM stage-predicted overall survival rates.
Discussion
Previous studies have shown the predictive ability of nomograms regarding NENs with liver metastases (Ruzzenente et al, 2016) and those of the small intestine (Modlin et al, 2010), pancreas (Ellison et al, 2014; Ye et al, 2016), and stomach (Cao et al, 2016). These results demonstrate that a specific, consistent, and clinically applicable nomogram can accurately estimate the prognosis of patients with NENs. However, the aforementioned studies contained limited patient cohorts (70 to 326 patients) or lacked external validation. The current study established a novel nomogram model to predict the overall survival rate associated with GEP-NENs based on the largest Chinese cohort to date. This nomogram was more significantly predictive than the seventh AJCC stage grouping, with a C-index of 0.837 (95% CIs=0.813–0.860). For the first time, external validation using the SEER database also verified that the nomogram exhibited excellent predictive ability compared with the classic TNM staging system. In addition, the ROC curve demonstrated that the nomogram showed better predictive ability than the seventh AJCC staging method. Furthermore, the calibration plots demonstrated that the predicted 3- and 5-year overall survival rates closely corresponded to the actual survival rates with regard to both the training and validation sets.
Currently, the criteria for assessing prognosis and predicting progression for GEP-NENs differ for distinct clinical systems (e.g., the TNM staging system) and pathological differentiation statuses. An algorithm-based assessment of the clinical, pathological, and biochemical parameters is not available to provide a multivariate assessment of the weighting of the numerous variables associated with GEP-NENs. For other types of solid tumours, the TNM staging system provides the most important prognostic values; in contrast, for GEP-NENs, tumour differentiation is the most significant prognostic metric for course and progression (Gatta et al, 2006). Using both the training and validation sets, the multivariate Cox regression model revealed that the tumour differentiation showed a higher HR than the other variables in the model (HR=5.037 and 4.447, respectively).
According to our nomogram, patients older than 50 years would likely have a higher disease-specific death prediction than younger patients. In addition, our nomogram illustrates the magnitude of poor prognosis as tumour size increases. The nomogram also clearly showed that patients with lymph node metastases and distant metastases are more likely to die than those without metastases. Our nomogram revealed an interesting phenomenon: The classic ‘T’ stage failed to show independent prognostic significance. In contrast, in the nomogram model, points for tumour differentiation extended across the full range of the point axis, and tumour differentiation exhibited more points than any other variable. These results clearly identify differences between prognoses predicted using the traditional TNM staging system and those predicted based on the tumour differentiation statuses of GEP-NENs. For example, patients with GEP-NENs with histologically high or moderate differentiation and minimal additional adverse indices might have low-to-intermediate death prediction scores even if their tumours have metastasised; however, patients with poorly differentiated disease might face substantially increased risks of death even in the absence of lymph node or distant metastases. For instance, if there are two patients with pancreatic NEN both with the same age (60 years; 25 points), tumour size (4 cm; 48 points), N1 stage (35 points), and M1 stage (68 points), but having different histological differentiation, well differentiation (Patient A; 0 point), and poor differentiation (Patient B; 100 points), their individual total points would be 176 and 276 points for Patients A and B, respectively according, to our nomogram. Their resulting expected 3-year overall survival rate would be 60% and 10%, while their expected 5-year overall survival rate 50% and <10%, respectively. Comparatively, according to the TNM classification, both patients would be categorised as stage IV with similar 3- and 5-year overall survival rate of 46.8% and 34.6%, respectively (based on our data analysis, data not shown).
Furthermore, based on the total points calculated by the nomogram, clinicians might recommend certain instructions. For example, the ENETS guidelines suggest that patients with poor-differentiating histology would undergo palliative chemotherapy because of their low life expectance. Those with well-differentiating histology on the other hand have better prognosis and therefore can opt for debulking surgery. However, solely depending on the TNM classification for patient selection may be ambiguous and doctors would have to rely on their clinical experience. Using the proposed nomogram of this study, oncologists would be in a better position of selecting patients with better survival rate for they would bear a higher probability of benefiting from palliative resection as it consists of a larger combination of clinicopathological parameters. Moreover, for patients with similar clinical characteristics but different tumour location, for instance, rectum and colon, where guidelines may have not proposed specific treatment recommendations, the treatment options may differ based on their different life expectancy according to the nomogram.
Thus, using nomogram to identify subgroups of patients with a more homogeneous prognosis, physicians can assess a diverse range of parameters with more objectiveness and precision for GEP-NENs so that the interpretation of clinical trial outcomes becomes clearer. The nomogram can also be used to assess individual clinical outcomes and the potential for specific GEP-NEN treatments.
This study has limitations. The major limitation of the present study is that the parameter of tumour location, which does not exhibit independent prognostic significance, was nonetheless included in the training and validation sets for the nomogram. We hypothesise that prognostic differences across specific tumour locations are subtle among patients with GEP-NENs, particularly relative to differences related to other clinicopathological parameters such as differentiation and distant metastases. However, for clinical utility and with the knowledge of tumour biology of NENs, the univariate analysis indicated that patients with colon NENs had the worst prognoses, but that patients with rectal NENs experienced the best outcomes. Theoretically, different tumour locations might represent distinct biological origins; however, our results failed to illuminate this possibility. On the other hand, our nomogram provided a general predictive tool for patients with GEP-NENs. Another limitation is that although the Ki-67 index was categorised as ‘well differentiated’, ‘moderately differentiated’, or ‘poorly differentiated/undifferentiated’ in the SEER database, we did not regard this parameter as categorical rather than continuous in the predictive model. Once an external validation set is undertaken, it might be necessary to treat the parameters consistently between the training and validation sets. We hypothesise that the nomogram will improve if the cutoff value of the Ki-67 index is changed in the future. Thus, the use of a continuous Ki-67 index variable might be better for establishing the nomogram. The third limitation is that our nomogram was established using only six clinicopathological factors. We acknowledge that certain additional variables (e.g., biological markers or genes) might also provide potential prognostic information; however, these variables were not included in the nomogram because these data are not currently available at all clinical centres. The purpose of creating this nomogram was to provide a potentially objective clinical tool that physicians can use to predict the prognosis of patients with GEP-NENs around the world.
In conclusion, the present study established and externally validated a novel nomogram to predict the prognoses of patients with GEP-NENs. Because our nomogram included only five common clinicopathological variables, it can be used to accurately provide useful information to both physicians and patients, allowing tailored treatments for GEP-NENs.
Change history
07 November 2017
This paper was modified 12 months after initial publication to switch to Creative Commons licence terms, as noted at publication
References
Bosman F, Carneiro F, Hruban R, Theise N (2010) WHO Classification of Tumours of the Digestive System. International Agency for Research on Cancer: Lyon, France.
Cao LL, Lu J, Lin JX, Zheng CH, Li P, Xie JW, Wang JB, Chen QY, Lin M, Tu RH, Huang CM (2016) A novel predictive model based on preoperative blood neutrophil-to-lymphocyte ratio for survival prognosis in patients with gastric neuroendocrine neoplasms. Oncotarget 7: 42045–42058.
Ellison TA, Wolfgang CL, Shi C, Cameron JL, Murakami P, Mun LJ, Singhi AD, Cornish TC, Olino K, Meriden Z, Choti M, Diaz LA, Pawlik TM, Schulick RD, Hruban RH, Edil BH (2014) A single institution's 26-year experience with nonfunctional pancreatic neuroendocrine tumors: a validation of current staging systems and a new prognostic nomogram. Ann Surg 259: 204–212.
Gatta G, Ciccolallo L, Kunkler I, Capocaccia R, Berrino F, Coleman MP, De Angelis R, Faivre J, Lutz JM, Martinez C, Möller T, Sankila R EUROCARE Working Group (2006) Survival from rare cancer in adults: a population-based study. Lancet Oncol 7: 132–140.
Greene FL, Sobin LH (2008) The staging of cancer: a retrospective and prospective appraisal. Cancer J Clin 58: 180–190.
Han DS, Suh YS, Kong SH, Lee HJ, Choi Y, Aikou S, Sano T, Park BJ, Kim WH, Yang HK (2012) Nomogram predicting long-term survival after d2 gastrectomy for gastric cancer. J Clin Oncol 30: 3834–3840.
Harrell FEJ, Lee KL, Mark DB (1996) Multivariable prognostic models: issues in developing models, evaluating assumptions and adequacy, and measuring and reducing errors. Stat Med 15: 361–387.
Hess KR (1994) Assessing time-by-covariate interactions in proportional hazards regression models using cubic spline functions. Stat Med 13: 1045–1062.
Hirabayashi S, Kosugi S, Isobe Y, Nashimoto A, Oda I, Hayashi K, Miyashiro I, Tsujitani S, Kodera Y, Seto Y, Furukawa H, Ono H, Tanabe S, Kaminishi M, Nunobe S, Fukagawa T, Matsuo R, Nagai T, Katai H, Wakai T, Akazawa K (2014) Development and external validation of a nomogram for overall survival after curative resection in serosa-negative, locally advanced gastric cancer. Ann Oncol 25: 1179–1184.
Kattan MW (2002) Nomograms. Introduction. Semin Urol Oncol 20: 79–81.
Modlin IM, Gustafsson BI, Pavel M, Svejda B, Lawrence B, Kidd M (2010) A nomogram to assess small-intestinal neuroendocrine tumor ('carcinoid') survival. Neuroendocrinology 92: 143–157.
Modlin IM, Oberg K, Chung DC, Jensen RT, de Herder WW, Thakker RV, Caplin M, Delle Fave G, Kaltsas GA, Krenning EP, Moss SF, Nilsson O, Rindi G, Salazar R, Ruszniewski P, Sundin A (2008) Gastroenteropancreatic neuroendocrine tumours. Lancet Oncol 9: 61–72.
Rudloff U, Jacks LM, Goldberg JI, Wynveen CA, Brogi E, Patil S, Van Zee KJ (2010) Nomogram for predicting the risk of local recurrence after breast-conserving surgery for ductal carcinoma in situ. J Clin Oncol 28: 3762–3769.
Ruzzenente A, Bagante F, Bertuzzo F, Aldrighetti L, Ercolani G, Giuliante F, Ferrero A, Torzilli G, Grazi GL, Ratti F, Cucchetti A, De Rose AM, Russolillo N, Cimino M, Perri P, Cataldo I, Scarpa A, Guglielmi A, Iacono C (2016) A novel nomogram to predict the prognosis of patients undergoing liver resection for neuroendocrine liver metastasis: an analysis of the Italian Neuroendocrine Liver Metastasis Database. J Gastrointest Surg 21: 41–48.
Shariat SF, Karakiewicz PI, Suardi N, Kattan MW (2008) Comparison of nomograms with other methods for predicting outcomes in prostate cancer: a critical analysis of the literature. Clin Cancer Res 14: 4400–4407.
Wang SJ, Lemieux A, Kalpathy-Cramer J, Ord CB, Walker GV, Fuller CD, Kim JS, Thomas Jr CR (2011) Nomogram for predicting the benefit of adjuvant chemoradiotherapy for resected gallbladder cancer. J Clin Oncol 29: 4627–4632.
Weiser MR, Landmann RG, Kattan MW, Gonen M, Shia J, Chou J, Paty PB, Guillem JG, Temple LK, Schrag D, Saltz LB, Wong WD (2008) Individualized prediction of colon cancer recurrence using a nomogram. J Clin Oncol 26: 380–385.
Wierda WG, O'Brien S, Wang X, Faderl S, Ferrajoli A, Do KA, Cortes J, Thomas D, Garcia-Manero G, Koller C, Beran M, Giles F, Ravandi F, Lerner S, Kantarjian H, Keating M (2007) Prognostic nomogram and index for overall survival in previously untreated patients with chronic lymphocytic leukemia. Blood 109: 4679–4685.
Yao JC, Hassan M, Phan A, Dagohoy C, Leary C, Mares JE, Abdalla EK, Fleming JB, Vauthey JN, Rashid A, Evans DB (2008) One hundred years after ‘carcinoid’: epidemiology of and prognostic factors for neuroendocrine tumors in 35,825 cases in the United States. J Clin Oncol 26: 3063–3072.
Ye L, Ye H, Zhou Q, Li Z, Lin Q, Tan L, Gao W, Fu Z, Zheng S, Chen R (2016) A retrospective cohort study of pancreatic neuroendocrine tumors at single institution over 15 years: new proposal for low- and high-grade groups, validation of a nomogram for prognosis, and novel follow-up strategy for liver metastases. Int J Surg 29: 108–117.
Acknowledgements
The Natural Science Foundation of Guangdong Province (2015A030313089), the Major Program of Collaborative Innovation of Guangzhou (201508030042), and the Medical Research Foundation of Guangdong Province (B2014160) supported this work.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License.
Rights and permissions
From twelve months after its original publication, this work is licensed under the Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/4.0/
About this article
Cite this article
Fang, C., Wang, W., Feng, X. et al. Nomogram individually predicts the overall survival of patients with gastroenteropancreatic neuroendocrine neoplasms. Br J Cancer 117, 1544–1550 (2017). https://doi.org/10.1038/bjc.2017.315
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/bjc.2017.315
Keywords
This article is cited by
-
Lymph node ratio and hematological parameters predict relapse-free survival in patients with high grade rectal neuroendocrine neoplasms after radical resection: a multicenter prognostic study
World Journal of Surgical Oncology (2023)
-
Potent molecular-targeted therapies for gastro-entero-pancreatic neuroendocrine carcinoma
Cancer and Metastasis Reviews (2023)
-
Construction and validation of the prognostic model for patients with neuroendocrine cervical carcinoma: a competing risk nomogram analysis
BMC Cancer (2022)
-
Postoperative locoregional recurrence pattern and treatment management of stage pT4 sigmoid colon cancer: a retrospective cohort study
Radiation Oncology (2022)
-
The Number of Lymph Nodes Examined is Associated with Survival Outcomes of Neuroendocrine Tumors of the Jejunum and Ileum (siNET): Development and Validation of a Prognostic Model Based on SEER Database
Journal of Gastrointestinal Surgery (2022)