Abstract
Background:
Rhabdomyosarcoma (RMS) is the most common type of soft tissue sarcoma in children. The Hedgehog (HH) pathway is known to develop an oncogenic role in RMS. However, the molecular mechanism that drives activation of the pathway in RMS is not well understood.
Methods:
The expression of HH ligands was studied by qPCR, western blot and immunohistochemistry. Functional and animal model studies were carried out with cells transduced with shRNAs against HH ligands or treated with HH-specific inhibitors (Vismodegib and MEDI-5304). Finally, the molecular characterisation of an off-target effect of Vismodegib was also made.
Results:
The results showed a prominent expression of HH ligands supporting an autocrine ligand-dependent activation of the pathway. A comparison of pharmacologic Smoothened inhibition (Vismodegib) and HH ligand blocking (MEDI-5304) is also provided. Interestingly, a first description of pernicious off-target effect of Vismodegib is also reported.
Conclusions:
The clarification of the HH pathway activation mechanism in RMS opens a door for targeted therapies against HH ligands as a possible alternative in the future development of better treatment protocols. Moreover, the description of a pernicious off-target effect of Vismodegib, via unfolded protein response activation, may mechanistically explain its previously reported inefficiency in several ligand-dependent cancers.
Similar content being viewed by others
Main
Rhabdomyosarcoma (RMS) is the most common type of soft tissue sarcoma in children. Rhabdomyosarcoma is thought to derive from embryonic cells committed to developing into skeletal muscle and is divided into two major subtypes in childhood: embryonal and alveolar (eRMS and aRMS, respectively). The majority of aRMS (80–85%) contain one of the reciprocal chromosomal translocations: t(2;13)(q35;q14) or t(1;13)(p36;q14) (Barr et al, 1993; Davis et al, 1994). Conversely, no characteristic translocations have been described in eRMS, which is typically characterised by loss of heterozygosity on the short arm of chromosome 11 (11p15.5) (Loh et al, 1992) and gains in chromosomes 2, 7, 8, 11, 12, 13 and 17 (Bridge et al, 2000).
Hedgehog (HH) pathway-belonging proteins are considered to be key regulators of development. Likewise, HH signalling also plays important roles in adult organisms such as stem cell maintenance or tissue repair and regeneration. The three HH proteins present in mammals, Sonic (SHH), Indian (IHH) and Desert (DHH), are ligands of Patched receptors (PTCH1 and PTCH2). Ligand-free PTCH inhibits the activation of Smoothened (SMO) and Gli family zinc finger proteins (GLI1, GLI2 and GLI3) are proteosomically processed. Upon binding of an HH ligand, SMO becomes active and prevents GLI proteosomal processing. GLI is then translocated to the nucleus where regulates GLI-specific promoters to activate its specific targets (Hahn et al, 1998; Teglund and Toftgård, 2010).
HH signalling has been shown to be altered (either by mutation or deregulation) in many cancers including basal cell carcinoma (BCC), medulloblastoma, small-cell lung cancer and digestive tract tumours, among many others (Teglund and Toftgård, 2010). The relationship between HH signalling and RMS was first described in the Patched knockout mouse by Hahn et al in 1998 who reported that mice heterozygous for Ptch1 not only develop features consistent with Gorlin’s syndrome, but also have a high incidence of eRMS (Hahn et al, 1998). Consistent activation of the pathway is well established and generally accepted in RMS (Zibat et al, 2010; Pressey et al, 2011; Satheesha et al, 2015). However, controversy exists regarding the importance of mutation-driven constitutive activation of the pathway in RMS patients. Some works reported that neither PTCH mutations nor activating SMO mutations appear to be implicated in activation of the pathway (Calzada-Wack et al, 2002; Pressey et al, 2011); however, other authors reported losses in the PTCH region 9q22 in one third of eRMS and loss of SUFU has also been reported in 18% of eRMS (Bridge et al, 2000; 2002; Tostar et al, 2006). Additionally, genomic amplification of chromosomal region 12q13-15 containing GLI1 gene has been identified only in a subset of aRMS tumours (Bridge et al, 2002; Pressey et al, 2011). Consequently, the activator mutations reported to date can only account – in the best scenario – for the activation of the pathway in small subsets of patients. Moreover, a possible role of methylation of the PTCH1 promoter has been described and seems to be able to reduce WT allele expression in the Patched knockout mouse. However, this effect is only significant if concomitant with an oncogenic mutation of the first allele of this gene (Uhmann et al, 2005), thereby reducing its potential in tumours with no PTCH1 mutation; therefore, the mechanism of activation remains elusive for the majority of patients.
On the other hand, several publications with xenografted RMS have raised the possibility of effectively reducing tumour growth (Eichenmüller et al, 2010; Tostar et al, 2010; Yamanaka et al, 2011) or even an impaired tumour initiation (Zibat et al, 2010) after pharmacologic inhibition of the pathway. In fact, small molecular antagonists of SMO entered clinical phase I and II trials for HH-driven neoplasias (Von Hoff et al, 2009; Lin and Matsui, 2012). Recently, in 2012, the SMO inhibitor Vismodegib was the first HH signalling pathway-targeting agent approved by the U.S Food and Drug Administration for the treatment of metastatic or locally advanced BCC (Abidi, 2014).
Despite the knowledge gathered to date, the mechanism that drives activation of the pathway in RMS is not well understood and the possible role of DHH or IHH in the pathogenesis of RMS has not been studied. Although activation of the pathway is well known in RMS, herein we report for the first time a preponderant expression of DHH and IHH ligands in RMS, which supports the existence of an autocrine ligand-dependent activation of the HH pathway in this neoplasia. The possible benefits of pharmacologic SMO inhibition (Vismodegib) and HH ligand blocking (MEDI-5304) are also studied. Furthermore, the genetic silencing of the ligands led to a clear impairment of tumour growth of xenografted tumours. Finally, given that we observed a possible adverse effect of Vismodegib which promotes cell invasion in an HH-independent manner, the molecular characterisation of this putative undesired off-target effect is also provided.
Materials and methods
Human samples
Thirty-three frozen RMS tumours were obtained from our private collection (ISCIII C.0002311) and 20 from the CWS (‘Cooperative Weichteilsarkom Studiengruppe’) tissue bank (Stuttgart, Germany). Paraffin-embedded RMS tumour samples were obtained from our Pathology Department. Written informed consent according to the Declaration of Helsinki was obtained from all patients or their legal guardians and the study approved by the Vall d’Hebron Research Ethics Committee.
Cell culture and drug treatment
Cell lines RH30 and RH4 correspond to aRMS with PAX3/FOXO1 translocation, RH18 to a translocation-negative aRMS and RD and HTB82 to eRMS subtype. Some authors have attributed the HTB82 cell line to a rhabdoid tumour since it bears a mutation in the SMARCB1 gene (Khan et al, 2001; Hinson et al, 2013). All RMS cell lines were previously authenticated by STR-based DNA profiling, grown in MEM media (Biowest, Barcelona, Spain), supplemented with 10% foetal bovine serum (Sigma-Aldrich, Madrid, Spain), 2 mM L-glutamine, 1 mM sodium pyruvate and 1 × non-essential amino acids (all reagents from Biowest). All cell lines were obtained from American Type Culture Collection, except RH18 and RD cell lines which were a kind gift from Dr Beat Schäfer. The SMO inhibitor Vismodegib (GDC-0449) was purchased from Selleckchem (Madrid, Spain). The antibody MEDI-5304 was kindly provided by MedImmune (Cambridge, UK).
RNA extraction, reverse transcription and real-time PCR
Total RNA was extracted using the RNeasy Mini Kit (Qiagen, Hilden, Germany). Two micrograms of total RNA were reverse-transcribed using 200 U of MMLV (Promega, Madrid, Spain) and random primers (Invitrogen, Thermo Fisher Scientific, Madrid, Spain). PCRs based on the TaqMan assay (Applied Biosystems, Thermo Fisher Scientific, Madrid, Spain) was performed to detect SHH, IHH, DHH and GLI1 (Hs00179843_m1, Hs00745531_s1, Hs00368306_m1 and Hs00171790_m1). The housekeeping gene TBP (TATA-binding protein, Hs00172424_m1) was used as internal control. Relative levels of each mRNA were tested in triplicate and quantified by the method of Livak and Schmittgen (2001).
Western blot
Tumour samples were disrupted with a Pellet Pestle Motor (Sigma-Aldrich) in RIPA lysis buffer (Thermo Fisher Scientific, Madrid, Spain). Twenty micrograms of protein were separated on 8–15% SDS–PAGE gels and transferred to PVDF membranes (Life Sciences, Sigma-Aldrich, Madrid, Spain), blocked in 5% BSA and incubated with the appropriate antibodies: anti-SHH 06-1106 (1 : 2000, Millipore, Madrid, Spain), anti-IHH MABF23 (1 : 1000, Millipore), anti-DHH H00050846-B01P (1 : 500, Novus Biologicals, Abingdon, UK), anti-GLI1 clone EPR4523 (1 : 1000, Millipore), anti-GLI2 R770 (1 : 1000, Cell Signaling, Barcelona, Spain), anti-PTCH1 06-1102 (1 : 1000, Millipore), anti-Cyclin D2 sc-593 (1 : 200, Santa Cruz Biotechnology, Heildelberg, Germany), anti-TRIB3 antibody 07-2160 (1 : 1000, Millipore), anti-CHOP sc-7351 (1 : 200, Santa Cruz Biotechnology), anti-FAK #3285S (1 : 1000, Cell Signaling), anti-p-FAK #3283 (1 : 500, Cell Signaling), anti-ATF4 #11815 (1 : 1000, Cell Signaling) and anti-PARP1 #9542 (1 : 1000, Cell Signaling). Anti-αTubulin ab7291 (1 : 5000, Abcam, Cambridge, UK) or anti-Actin sc-1616 (1 : 10000, Santa Cruz Biotechnology) antibodies were used as loading control. After incubation with appropriate HRP-conjugated secondary antibodies, ECL Prime reagent (GE Healthcare, Barcelona, Spain) was used for chemiluminescence detection.
Immunohistochemistry
Paraffin-embedded tissues were sectioned, deparaffinised and rehydrated, followed by antigen retrieval in 10 mM citrate buffer (DAKO, Agilent Technologies, Barcelona, Spain). Samples were blocked and incubated with antibodies against SHH (06-1106, Millipore), IHH (MABF23, Millipore) or DHH (H00050846-B01P, Novus Biologicals), all diluted 1 : 200. The three HH antibodies yielded mainly membrane labelling, as expected. Samples were evaluated using a semiquantitative method (Rhodes et al, 2000) regarding the percentage of cells with membrane labelling, excluding blood vessels (0%, ⩽25%, 26–50%, 51–75% and ⩾75% corresponding to a score of 0, 1, 2, 3 and 4, respectively) plus intensity (from 0 to 3) of the labelling (total score range: 0 to 7).
Plasmids and lentiviral transduction
Genetic inhibition of the HH ligands, GLI1 and TRIB3 was performed by shRNAs cloned into the lentiviral vector pGIPZ (GE Dharmacon, Lafayette, CO, USA). Positively transduced cells were selected with puromycin (1 μg ml−1, Sigma-Aldrich). Knockdown efficiency of each shRNA was analysed by western blot. Six different shRNAs were tested by western blot with the aim of selecting the best option for each target. The GE Dharmacon IDs of selected clones were: V3LHS_82400 (shSHH), V3LHS_336297 (shIHH), V3LHS_401021 (shDHH), V2LHS_262249 (shGLI1). For TRIB3, four different shRNAs were tested and the selected option was V3LHS_644655 (shTRIB3). Functional assays were performed 7 days after infection.
WST-1 assay
Cell proliferation was measured by WST-1 colorimetric assay (Sigma-Aldrich), following the manufacturer’s instructions. Cells seeded at low density were allowed to proliferate for 4 days. WST-1 reagent was then added and, after 3 h, absorbance was measured at 440 nm. All conditions were assayed per triplicate in three independent experiments.
Transwell assay
Cell invasiveness was assessed by Transwell assay (Corning, Barcelona, Spain). Briefly, 105 cells were resuspended in serum-free MEM media supplemented with the appropriate drug and plated in the upper chamber previously coated with BD MatrigelTM (Corning) in an 8 μm pore size transwell (Corning). After 24 h, cells migrated to the lower surface of the membrane were stained in 5 ng ml−1 of Hoechst33342. Cells were counted under a light microscope in three independent wells for each experiment and three independent experiments were performed.
Animal models
Five-week-old female SCID and SCID-Beige mice were purchased from Charles River Laboratories (Barcelona, Spain). Animals were housed in specific-pathogen-free conditions with food and water available ad libitum. For the primary tumour model, 106 RD cells stably transduced with shRNA expressing lentiviral vectors were injected orthotopically in the right gastrocnemius muscle of SCID mice whereas the left muscle was injected with control cells (n=5 per condition). Limb volume and body weight were measured every 2 days. For the metastasis model, 106 RH30 cells (suspended by pipetting in PBS) were intravenously injected into tail veins of SCID-Beige mice. Animals were treated every other day with either DMSO (Control) or 40 mg kg−1 Vismodegib, as previously described (Chan et al, 2012; Moshai et al, 2014) and killed following three main criteria: (1) tumour size (when measurable, >2 cm), (2) weight loss (>10% of total body weight) or (3) poor general status of the animal. All animals were killed following these guidelines and none were ever allowed to suffer or reach a moribund state. Post-mortem analysis revealed that all animals killed during the study presented macroscopic metastasis. Conversely, all the animals that reached the end of the study as healthy were free of macroscopic metastasis. All procedures were previously approved by the Ethics Committee of Animal Experimentation of the Vall d’Hebron Research Institute (CEEA 30/14).
Microarray expression analysis
Microarray hybridisation was carried out with 200 ng of total mRNA in an Affymetrix (Thermo Fisher Scientific, Madrid, Spain) microarray 7000G platform and the Genechip Human Gene Array 2.0. Quality of isolated RNA was first assessed by Bioanalyzer Assay (Agilent, Barcelona, Spain). Briefly, sense ssDNA was generated from total RNA with the GeneChip WT Plus Reagent Kit from Affymetrix according to the manufacturer’s instructions. Results were submitted to the GEO microarray database (GSE77996).
Results
SHH is expressed in a small subset of RMS cell lines and tumours, whereas IHH and DHH are the predominant HH ligands in RMS
RMS cell lines
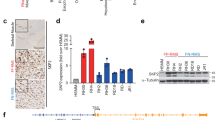
SHH mRNA levels were studied in five cell lines representing the main RMS subtypes. While the two eRMS cell lines expressed SHH, levels in the three aRMS cell lines were extremely low (Figure 1A). Conversely, IHH and DHH expression levels were noteworthy in all cell lines analysed without exception (Figure 1A). Additionally, the results shown by western blot (Figure 1B) concurred with the qPCR results. Thus, the SHH protein level was extremely low in all alveolar cell lines, and moderate in the embryonal cells. By contrast, IHH showed high levels in all cell lines. DHH showed high expression levels in the embryonal cell lines analysed and a moderate level of expression was shown for the alveolar cells (with the exception of RH30, with very low expression level).
IHH and DHH are widely distributed in RMS whereas SHH is only expressed in a small subset of patients. (A) mRNA levels of SHH, IHH and DHH in five cell lines (RH30, RH4, HTB82, RD and RH18). Values are expressed as −ΔCT value and the horizontal line depicts the mean of the values for the cell lines analysed. (B) Western blot showing the levels of the HH ligands in the same cell lines. The HEK cell line was used as a positive control. (C) Quantitative real-time PCR analysis of HH ligand expression in patients. Values are expressed as −ΔCT value and the horizontal line depicts the mean values (n=53). (D) Western blot showing HH ligand levels in eight RMS patients (aRMS, alveolar RMS; eRMS, embryonal RMS). Tubulin was used as loading control (TUB). The presence of SHH (E), IHH (F) and DHH (G) in archival human samples was examined by immunohistochemistry at low magnification for a general view of the section (left), and the area indicated by a square is shown magnified on the right for a more detailed view. Bars: 100 μm. (H) Semi-quantitative evaluation of the expression of the three ligands in patient samples (n=27). Horizontal lines depict the mean score for each marker.
RMS tumours
Concordantly with results in cell lines, SHH expression was negligible in the majority of RMS samples. However, approximately 30% of patients in a well-separated positive subpopulation showed remarkable mRNA SHH expression by real-time PCR (Figure 1C) and western blot (Figure 1D). In contrast, IHH and DHH levels were moderate, high or very high in all patients analysed by both qPCR (Figure 1C) and western blot (Figure 1D). No significant differences were observed between aRMS and eRMS (Supplementary Figure S1).
IHC in archival paraffin-embedded human RMS tumour sections also revealed predominant expression of DHH and IHH ligands in tumour cells
The majority of the tumours analysed showed very low or negligible SHH expression (15 out of 23, 65%), whereas the remaining 35% of samples showed low or moderate SHH levels (Figure 1H). Conversely, IHH and DHH were very highly expressed in the majority of samples, with a maximum expression of both proteins (semi-quantitative score 7) in 14 of 23 samples (61%), and semi-quantitative score values ⩾4 for IHH in 18 of 23 samples (78%) and 20 of 23 samples for DHH (87%). Representative photographs of SHH, IHH and DHH immunohistochemistries (E, F and G, respectively) are shown in Figure 1 with two magnifications.
The presence of HH ligands correlates with activation of the pathway, thereby suggesting an autocrine mechanism
The presence of HH ligands in human RMS tumours was correlated with the expression of one of the main downstream targets of the pathway, GLI1 (Figure 2A–D). The results showed significant correlation for SHH (P=0.0197), DHH (P=0.0268) and the sum of the three HH ligands (P=0.0189). No significant correlation was obtained for IHH, although a trend was also observed. Furthermore, the genetic downregulation by shRNAs of the HH ligands and GLI1 (decrease of ligands and GLI1 is shown in Supplementary Figure S2) revealed a significant downregulation of some components and downstream targets of the pathway in the cell lines RD and RH4. Thus, IHH, DHH and GLI1 downregulation resulted in a marked decrease in GLI1, GLI2 and PTCH1, whereas SHH downregulation did not affect GLI1 expression but clearly decreased GLI2 and PTCH1 (Figure 2E and F). Conversely, in RH30 cell line that bears a GLI1-amplification none of the ligands significantly changed GLI1 expression. However, despite GLI1 amplification, the deprivation of SHH, IHH and DHH produced a significant reduction in GLI2 and PTCH1 expression (Figure 2G).
qPCR and Western blot analysis reveal correlation between levels of the three HH ligands and activation status of the pathway. Degree of correlation between RNA expression (−ΔCT values) of GLI1 and SHH (A), IHH (B), DHH (C) and overall HH ligands expression (D). In (A) the plot shows only patients with significant SHH expression. The Pearson’s correlation r and P-values are included in each plot. (E–G) Western blots showing the effect of the genetic downregulation of SHH, IHH and DHH on the expression of components and target genes of the pathway (GLI1, GLI2 and PTCH1). Actin was used as an endogenous control. The same lysates were used to validate the efficacy of target knockdown for each shRNA (Supplementary Figure S2).
Deprivation of IHH and DHH rendered a reduction in cell proliferation
The effects of the shRNA-mediated IHH and DHH deprivation significantly reduced cellular proliferation of the cell lines RD and RH4 (Figure 3A and C). Unsurprisingly, no effects were observed for RH30 cells (Figure 3E). Interestingly, the pharmacologic effect of the HH ligand-blocking antibody MEDI-5304 produced comparable lessening in proliferation for RD and RH4. These cell lines were also sensitive to the SMO inhibitor Vismodegib and to the GLI1 shRNA. Once again, the RH30 cell line was completely insensitive to both ligand deprivation and MEDI-5304 antibody treatment, probably due to its GLI1-amplification. Only shRNA of GLI1 and Vismodegib treatment produced an antiproliferative effect on this cell line. Concordantly, Cyclin D2 levels were increased in cell lines RD and RH4 upon IHH, DHH and GLI1 deprivation (Figure 3B and D), thereby suggesting a cell cycle arrest at G1. No effects on this cell cycle effector were observed in the RH30 cell line (Figure 3F). Moreover, RD and RH4 RMS cells with downregulation of IHH and DHH showed a reduction in the percentage of cells in G2, thereby suggesting a cell cycle impairment (Figure 3G). Once again, the RH30 cell line showed no changes (not shown). Although a slight increase in early apoptotic cells and concordant PARP-1 cleavage was detected in the RD cell line, the results in the other two cell lines ruled out apoptosis as a general mechanism able to account for the decrease in the number of cells observed (Supplementary Figure S3). Moreover, no significant changes in total dead cells were observed for any of the cell line (not shown). The pharmacologic downregulation of the pathway was assessed by measuring Gli1 mRNA levels (Figure 3H–J).
Effects of the depletion of HH ligands on cell proliferation. (A, C and E) Relative proliferation, expressed as a reduction in WST uptake, compared to control cells (100%). The experiments were performed in the following samples: control (empty vector or vehicle) cells, cells transduced with shRNAs for SHH, IHH, DHH and GLI1 and, finally, cells treated with Vismodegib (50 μM) or the HH blocking antibody MEDI-5304 (30 μg/ml) after 4 days. The RMS cell lines analysed were RD, RH4 and RH30. (B, D and F) Western blots showing the accumulation of Cyclin D2 (CYC D2) in RD and RH4 cells. No changes were detected in RH30. (G) Cells transduced with shRNAs for IHH and DHH showed a slight reduction in the percentage of cells in G2, which indicates an impairment in the cell cycle. Percentages of cells in G1 and G2+S are indicated in the plots. Significance: *P<0.05, **P<0.01, ***P<0.001. (H–J) Relative GLI1 mRNA expression after Vismodegib or MEDI-5304 treatment.
Genetic impairment of HH ligand expression produced a severe tumour growth reduction in SCID mice
Six and a half weeks after intramuscular RD cell injection, excised tumours stably expressing shRNAs against SHH, IHH, DHH and GLI1 were clearly smaller – or even in some cases were unable to grow – compared to the control tumours (empty vector) (Figure 4A). The average weight of the tumours was also clearly lower (Figure 4B). The difference was particularly dramatic after SHH and GLI1 downregulation (<25%) but also noteworthy for IHH and DHH downregulation (<40%). The kinetics of growth were clearly reduced in all cases (Figure 4C–F), with statistically significant differences from 5 to 6 weeks onwards. All shRNA targets were found to be downregulated as expected in tumours at the end point (Figure 4G).
Decreased tumour volume of RMS cells with genetically impaired expression of SHH, DHH, IHH and GLI1 in SCID mice. (A) Photograph of all the tumours showing the reduction in tumour size after genetic depletion of SHH, DHH, IHH and GLI1 (NTG: no tumour growth). (B) Average tumour weight at the end point of the study in comparison to the control (empty vector pGIPZ, 100% value). (C–F) Kinetics of tumour growth expressed as mm3 of tumour volume compared to empty vector for SHH, DHH, IHH and GLI1 shRNAs. Significance: *P<0.05, **P<0.01. (G) Western blot to assess the downregulation of HH ligands in tumours at the experiment end-point. shRNA-positive tumours (+) showed downregulation of their specific targets (SHH, IHH or DHH) compared with controls (−).
SHH played a crucial role for the invasive phenotype in ligand-dependent RMS cells and paradoxical enhancement of invasiveness by Vismodegib
In contrast to the effects seen on cell proliferation, the genetic inhibition of IHH and DHH produced no effects on cell invasiveness. Surprisingly, the effect of the downregulation of SHH in terms of invasiveness was noteworthy in RD and RH4 cell lines (Figure 5A and B). Once again the cell line RH30 was refractory to ligand deprivation (Figure 5C). The effects of MEDI-5304 antibody on invasiveness were statistically not significant. Unexpectedly, Vismodegib enhanced the invasive properties of all three cell lines by two-fold. These surprising results, together with the fact that the invasiveness of the RH30 cell line was likewise increased despite its GLI1-amplification (downstream of SMO), suggested an off-target effect of Vismodegib on cell invasiveness. Similar results were seen in mice treated with Vismodegib that were more prone to developing metastasis after a tail vein injection of RH30 cells (Figure 5D). Thus, in the control group (vehicle), only 55% of mice developed metastasis, whereas in the group treated with Vismodegib, all mice metastasised.
Effects of HH ligand depletion or pharmacologic HH inhibition on RMS cells invasiveness. Cell invasiveness was studied after genetic (shRNAs indicated) and pharmacologic (Vismodegib 50 μM and MEDI-5304 30 μg/ml, 3 days of treatment) HH pathway inhibition. The cell lines were RD (A), RH4 (B) and RH30 (C). Values of invasiveness are expressed in percentages and referred to those of controls (empty vector or vehicle). (D) Overall survival of mice after Vismodegib treatment was reduced compared to mice treated with vehicle. All mice were intravenously injected with 106 RH30 cells. Significance: *P<0.05, **P<0.01, ***P<0.001.
The paradox effect of Vismodegib on the invasiveness of RMS cells is mediated by off-target activation of the unfolded protein response pathway and concomitant phosphorylation of FAK
A cDNA microarray was performed to identify pathways differentially regulated between untreated and Vismodegib-treated cells. The expression profile analysis was made in three cell lines (RD, RH4 and RH30). Results revealed an overexpression of several stem components of the unfolded protein response (UPR) pathway (PERK, CHOP, TRIB3) and some downstream components, particularly those involved in cell invasion such as ATG16/1, HMOX, STAT3, STAT5 or cell proliferation such as ASNS, FGFR2 and IL8 (Figure 6A–D). Western blots showed a very consistent increase in the expression of CHOP and TRIB3 in the three RMS cell lines analysed. Additionally, the stem component of the UPR pathway ATF4, which was not detected as overexpressed in the array, also showed an increase in both protein amount and phosphorylation (Figure 6D). Interestingly, the treatment with Vismodegib also produced a concomitant increase in phosphorylation of FAK (P-FAK), a protein often involved in invasion and metastasis. All cell lines analysed showed a very consistent increase in P-FAK, whereas total FAK levels remained stable in all cases (Figure 6E). Given that the cell lines studied covered all the main subtypes of RMS, the results strongly suggest that UPR activation may be a general mechanism of cell invasiveness in RMS under Vismodegib treatment and not a mere peculiarity of a single cell line. Furthermore, analysis of the Davicioni data set revealed the existence of interesting correlations between TRIB3 expression and RMS progression (Supplementary Figure S4). Moreover, the shRNA-mediated genetic inhibition of TRIB3 (shown in Figure 6F) together with Vismodegib treatment not only hindered the expected increase in invasion of cells treated with Vismodegib, but also produced a plunge in this cell invasion. TRIB3 genetic downregulation alone produced a moderate reduction in cell invasion (Figure 6G).
Vismodegib treatment resulted in activation of the UPR pathway by FAK phosphorylation. (A) cDNA microarray analysis revealed overexpression of several components of the UPR pathway under Vismodegib (50 μM) treatment for 3 days compared to the control in three RMS cell lines (RD, RH4 and RH30). (B) Schematic view of the main components of the pathway. The elements found overexpressed in the microarray are shown in red. (C) Western blots showing the increase in CHOP and TRIB3 levels in RMS cell lines under Vismodegib treatment. (D and E) Western blots showing that Vismodegib treatment increased total and phosphorylated ATF4 as well as phosphorylation of FAK. (F) Assessment of TRIB3 downregulation by western blot after shRNA transduction in RD cell line. The selected shRNA (sh4) is indicated by an arrow. (G) The shRNA-mediated genetic inhibition of TRIB3 produced a very clear impairment in cell invasion (Matrigel/Transwell assay), whereas Vismodegib treatment alone produced a notable increase. The TRIB3 genetic downregulation alone produced only a slight reduction in cell invasion. Significance: *P<0.05, ** or ##P<0.01, *** or ###P<0.001. (*) indicates differences to empty vector untreated control, whereas (#) indicates differences to treated empty vector.
Discussion
Mutation-driven cancers such as those associated with Gorlin’s syndrome were the first in which an oncogenic role of the HH pathway was described (Hahn et al, 1996; Johnson et al, 1996). The archetypical molecular basis for the development of the malignancies associated with Gorlin’s syndrome is the PTCH1 loss of function which leads to constitutively activated HH signalling even in the absence of a ligand. Given that the mutation of components of the HH pathway in RMS appears to be rare (Calzada-Wack et al, 2002) – except in Gorlin’s syndrome that accounts for a very low percentage of RMS patients – a plausible hypothesis is that, regarding HH pathway activation, the majority of RMS are ligand-dependent tumours. To prove this hypothesis, it is crucial to demonstrate the presence of functional HH ligands in the tumours. However, an earlier report on HH ligand expression showed very low or undetectable SHH levels in RMS tumours and cell lines (Calzada-Wack et al, 2002), thereby pointing to a ligand-independent activation of HH signalling in RMS. We here show for the first time that IHH and DHH – and, in a subset of cases, also SHH – are present in RMS tumour cells with concomitant variations in the expression of HH downstream targets, thus pointing to an autocrine mechanism of HH activation in which ligands may be both produced and responded to by the same tumour cells. Concordantly, the work of Satheesha et al (2015) ruled out paracrine activation of the pathway in RMS cells in in vivo mouse models. The reduction in proliferation upon IHH and DHH genetic deprivation in the cell lines RD and RH4 clearly indicates a crucial role of these ligands in the activation of the pathway and, therefore, is a further indication of ligand-dependent activation in these cell lines. The refractoriness of the cell line RH30, which bears a 10-fold GLI1 amplification (Calzada-Wack et al, 2002) is especially noteworthy. Presumably, GLI1 amplification makes the cell behave as ligand-independent, since the amplification bypasses the canonical ligand-dependent activation of the pathway. This cell line may be a paradigmatic example to illustrate the probable existence of a subset of patients with GLI1 amplification, as previously suggested (Calzada-Wack et al, 2002), which may be refractory to possible therapies based on ligand inhibition. According to that interesting previous work, GLI1 amplification seems to be alveolar-subtype specific (or at least is more frequently described in this subtype). However, the activation of the pathway in the majority of RMS tumours (including all eRMS and an undetermined proportion of aRMS) cannot be explained by this mechanism. On the other hand, cell lines RH4 and RD would represent tumours without mutations in HH pathway components, the oncogenicity of which strongly depends on ligand presence. The strong dependency on ligands, together with the fact that this phenotype presumably represents the majority of RMS patients, enables alternative therapeutic approaches based on ligand blocking in this neoplasia which may have wide translational potential.
The cell proliferation reduction drop observed in ligand-dependent cell lines after depletion of HH ligands strongly suggests that this is associated with a cell cycle alteration and the observed Cyclin D2 accumulation points to a G1-phase cell cycle arrest. Indeed, several of the genes activated by GLI proteins (i.e., Cyclin D) are key regulators of G1/S transition (Kasper et al, 2006). Interestingly, in the cell lines RD and RH4, the reduction in cell proliferation induced by pharmacologic inhibition is significant and comparable for both the ligand-blocking antibody MEDI-5304 and the SMO inhibitor Vismodegib. However, in RH30 cell line the inhibitory effect of Vismodegib on proliferation was probably an off-target effect, since GLI1 amplification is expected to result in insensitivity to SMO inhibitors. Conversely, the effect of MEDI-5304 appears to be pathway-specific since it did not impact on the RH30 cell line. Consequently, MEDI-5304 showed higher specificity and similar potency in the blocking of the HH pathway-induced proliferation. It is important to emphasise that the therapy of mutation-driven cancers by inhibition of the HH pathway constituents can only be successful if these are situated downstream of the acquired mutation or amplification (Heretsch et al, 2010). Conversely, ligand-dependent cancers are expected to be sensitive to therapeutic strategies based on ligand targeting. The results herein presented, as commented above, point to a reduction of RMS cell proliferation when treated with the antibody MEDI-5304. Interestingly, this antibody blocks SHH and IHH but fails to recognise DHH (Michaud et al, 2014). It is conceivable that the high level of expression of DHH in RMS may partially account for the moderate anti-proliferative effect of MEDI-5304. Thus, despite being the least active of the three HH ligands, DHH can significantly activate GLI1 expression (Pathi et al, 2001). Therefore, we cannot rule out that DHH in RMS plays a more important role than hitherto expected.
Although the shRNA-mediated downregulation of the ligands was not complete and performed separately for each ligand, the effects observed on tumour growth were remarkable. Thus, the downregulation of each single HH ligand clearly impaired tumour growth, suggesting that the participation of all ligands is crucial for RMS pathogenesis. The strong effect on tumour growth of SHH deprivation is particularly noteworthy. Because downregulation of SHH had no effect on cell proliferation in vitro, this ligand probably has to interact with the tumour microenvironment to execute its RMS-promoting functions. In this regard, the connection between an aberrant HH ligand secretion and the paracrine stimulation of stromal cells it is particularly interesting (Curran and Ng, 2008; Yauch et al, 2008). Indeed, also in RMS, SHH secretion may lead to a production of VEGF and IGF by the HH-stimulated stromal cells which, in turn, may affect angiogenesis of the growing tumour. Both factors, VEGF and IGF are clues to angiogenesis (Shigematsu et al, 1999; Bach, 2015).
The anti-oncogenic effect of the HH ligand deprivation is not restricted to cell proliferation. An essential role of HH pathway on epithelial to mesenchymal transition and/or invasiveness in a broad spectra of cancers (Lee et al, 2015; Lei et al, 2015; Liu et al, 2015) including RMS (Oue et al, 2013) has previously been well established. However, the possible influence of HH ligands on RMS cell invasion has not been elucidated to date. This work provides data to demonstrate for the first time a crucial role of SHH in the promotion of an invasive phenotype in RMS cells. Conversely, IHH and DHH appear not to influence invasion, pointing to a specific role of SHH in this aspect of tumour progression.
While SMO antagonists are active in BCC and in medulloblastoma, clinical results in other solid tumours, whereby mutational activation of the pathway is absent, have been less encouraging (McMillan and Matsui, 2012). For instance, in metastatic pancreatic cancer, a trial was prematurely halted after patients in the Saridegib plus Gemcitabine arm were found to have a higher rate of disease progression and lower median survival than those receiving Gemcitabine alone (McMillan and Matsui, 2012). Other attempts in advanced colorectal, ovarian and pancreatic cancers did not render significant positive results either (McMillan and Matsui, 2012; Catenacci et al, 2015). Indeed, in the study of Catenacci et al (2015), experiments in mice treated with Vismodegib revealed a worrying reduction in the survival median – albeit not statistically significant – compared to the control group. A plausible reason for these failures was that these cancers were ligand-dependent and not driven by mutations in specific components of the pathway. This raises the possibility of pharmacologic ligand inhibition, which may be considered an appealing therapeutic alternative in ligand-dependent cancers. Our findings in RMS show for the first time a relationship between an SMO inhibitor treatment and triggering of the intricate process of cell invasion via UPR pathway activation.
Activation of the UPR pathway has been suggested to play a role in cancer (Ma and Hendershot, 2004). UPR is thought to be initiated by three endoplasmic reticulum (ER) stress sensors: protein kinase RNA-like ER kinase (PERK), activating transcription factor 6 (ATF6) and inositol-requiring enzyme 1α (IRE1α) (Hetz et al, 2013). The fact that TRIB3, CHOP and ATF4 are essential downstream effectors of the pathway triggered by PERK and that they are activated (Figure 6) suggests that the activation of this response – after Vismodegib treatment – would be dependent on this UPR sensor, at least in RMS. To date, the potential UPR role in cancer is not completely understood and no relevant previous information on the implication of UPR in RMS progression was found; however, one of the most remarkable UPR functions in cancer progression is the promotion of invasiveness, including the promotion of some matrix metalloproteinases, extracellular matrix remodelling and angiogenesis (Dejeans et al, 2015). Interestingly, the concomitant increase in FAK phosphorylation (P-FAK) has been previously related to UPR activation as a mechanism able to induce cell survival, inhibit apoptosis and reduce chemosensitivity in cancer (Tang et al, 2012), thereby suggesting a pro-oncogenic potential associated with this mechanism. Moreover, the genetic inhibition of TRIB3 not only blocked the cell invasion increase but also promoted a clear sensitisation of cells to Vismodegib treatment, thereby pointing to a role for this protein in RMS chemoresistance. Taken together, the data reported and the discovery of this mechanism may help researchers and companies interested in the development and application of SMO inhibitors in cancer.
The clarification of the HH pathway activation mechanism in RMS, which has remained elusive to date, opens a door for targeted therapies against HH ligands as a possible alternative in the future development of better treatment protocols for RMS. Moreover, as a secondary point albeit not less important, the description of a pernicious off-target effect of Vismodegib may explain its previously reported inefficiency in several ligand-dependent cancers or even the premature halting of some of them owing to survival worsening. Therefore, the findings herein described may have significant translational implication not only for RMS but also for other HH ligand-dependent cancers.
Change history
26 October 2017
This paper was modified 12 months after initial publication to switch to Creative Commons licence terms, as noted at publication
References
Abidi A (2014) Hedgehog signaling pathway: a novel target for cancer therapy: vismodegib, a promising therapeutic option in treatment of basal cell carcinomas. Indian J Pharmacol 46: 3–12.
Bach LA (2015) Endothelial cells and the IGF system. J Mol Endocrinol 54: R1–13.
Barr FG, Gallili N, Holich J, Biegel JA, Rovera G, Emanuel BS (1993) Rearrangement of the PAX3 paired box gene in the paediatric solid tumor alveolar rhabdomyosarcomas. Nat Genet 3: 113–117.
Bridge JA, Liu J, Qualman SJ, Suijkerbuijk R, Wenger G, Zhang J, Wan X, Baker KS, Sorensen P, Barr FG (2002) Genomic gains and losses are similar in genetic and histologic subsets of rhabdomyosarcoma, whereas amplification predominates in embryonal with anaplasia and alveolar subtypes. Genes Chromosomes Cancer 33 (3): 310–321.
Bridge JA, Liu J, Weibolt V, Baker KS, Perry D, Kruger R, Qualman S, Barr F, Sorensen P, Triche T, Suijkerbuijk R (2000) Novel genomic imbalances in embryonal rhabdomyosarcoma revealed by comparative genomic hybridization: an Intergroup Rhabdomyosarcoma Study. Genes Chromosomes Cancer 27: 337–344.
Calzada-Wack J, Kappler R, Schnitzbauer U, Richter T, Nathrath M, Rosemann M, Wagner SN, Hein R, Hahn H (2002) Unbalanced overexpression of the mutant allele in murine Patched mutants. Carcinogenesis 23: 727–733.
Catenacci DV, Junttila MR, Karrison T, Bahary N, Horiba MN, Nattam SR, Marsh R, Wallace J, Kozloff M, Rajdev L, Cohen D, Wade J, Sleckman B, Lenz HJ, Stiff P, Kumar P, Xu P, Henderson L, Takebe N, Salgia R, Wang X, Stadler WM, de Sauvage FJ, Kindler HL (2015) Randomized phase Ib/II study of gemcitabine plus placebo or vismodegib, a hedgehog pathway inhibitor, in patients with metastatic pancreatic cancer. J Clin Oncol 33: 4284–4292.
Chan IS, Guy CD, Chen Y, Lu J, Swiderska-Syn M, Michelotti GA, Karaca G, Xie G, Krüger L, Syn WK, Anderson BR, Pereira TA, Choi SS, Baldwin AS, Diehl AM (2012) Paracrine Hedgehog signaling drives metabolic changes in hepatocellular carcinoma. Cancer Res 72 (24): 6344–6350.
Curran T, Ng JM (2008) Cancer: Hedgehog's other great trick. Nature 18: 293–294.
Davis RJ, D’Cruz CM, Lowell MA, Biegel JA, Barr FG (1994) Fusion of PAX7 to the FOXO1 by the variant t(1;13) (p36;q14) translocation in alveolar rhabdomyosarcomas. Cancer Res 54: 2869–2872.
Dejeans N, Barroso K, Fernandez-Zapico ME, Samali A, Chevet E (2015) Novel roles of the unfolded protein response in the control of tumor development and aggressiveness. Semin Cancer Biol 33: 67–73.
Eichenmüller M, Hemmerlein B, von Schweinitz D, Kappler R (2010) Betulinic acid induces apoptosis and inhibits hedgehog signaling in rhabdomyosarcoma. Br J Cancer 103: 43–51.
Hahn H, Wicking C, Zaphiropoulous PG, Gailani MR, Shanley S, Chidambaram A, Vorechovsky I, Holmberg E, Unden AB, Gillies S, Negus K, Smyth I, Pressman C, Leffell DJ, Gerrard B, Goldstein AM, Dean M, Toftgard R, Chenevix-Trench G, Wainwright B, Bale AE (1996) Mutations of the human homolog of Drosophila patched in the nevoid basal cell carcinoma syndrome. Cell 85: 841–851.
Hahn H, Wojnowski L, Zimmer AM, Hall J, Miller G, Zimmer A (1998) Rhabdomyosarcomas and radiation hypersensitivity in a mouse model of Gorlin syndrome. Nat Med 4: 619–622.
Heretsch P, Tzagkaroulaki L, Giannis A (2010) Modulators of the hedgehog signaling pathway. Bioorg Med Chem 18: 6613–6624.
Hetz C, Chevet E, Harding HP (2013) Targeting the unfolded protein response in disease. Nat Rev Drug Discov 12: 703–719.
Hinson AR, Jones R, Crose LE, Belyea BC, Barr FG, Linardic CM (2013) Human rhabdomyosarcoma cell lines for rhabdomyosarcoma research: utility and pitfalls. Front Oncol 3: 183.
Johnson RL, Rothman AL, Xie J, Goodrich LV, Bare JW, Bonifas JM, Quinn AG, Myers RM, Cox DR, Epstein EH Jr, Scott MP (1996) Human homolog of patched, a candidate gene for the basal cell nevus syndrome. Science 272: 1668–1671.
Kasper M, Regl G, Frischauf AM, Aberger F (2006) GLI transcription factors: mediators of oncogenic Hedgehog signalling. Eur J Cancer 42: 437–445.
Khan J, Wei JS, Ringnér M, Saal LH, Ladanyi M, Westermann F, Berthold F, Schwab M, Antonescu CR, Peterson C, Meltzer PS (2001) Classification and diagnostic prediction of cancers using gene expression profiling and artificial neural networks. Nat Med 7 (6): 673–679.
Lee J, Jeong S, Lee CR, Ku CR, Kang SW, Jeong JJ, Nam KH, Shin DY, Chung WY, Lee EJ, Jo YS (2015) GLI1 transcription factor affects tumor aggressiveness in patients with papillary thyroid cancers. Medicine 94: e998.
Lei J, Fan L, Wei G, Chen X, Duan W, Xu Q (2015) Gli-1 is crucial for hypoxia-induced epithelial-mesenchymal transition and invasion of breast cancer. Tumour Biol 36: 3119–3126.
Lin TL, Matsui W (2012) Hedgehog pathway as a drug target: Smoothened inhibitors in development. Onco Targets Ther 5: 47–58.
Liu Q, Sheng W, Dong M, Dong X, Dong Q, Li F (2015) Gli1 promotes transforming growth factor-beta1- and epidermal growth factor-induced epithelial to mesenchymal transition in pancreatic cancer cells. Surgery 158: 211–224.
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25: 402–408.
Loh WE Jr, Scrable HJ, Livanos E, Arboleda MJ, Cavenee WK, Oshimura M, Weissman BE (1992) Human chromosome 11 contains two different growth suppressor genes for embryonal rhabdomyosarcoma. Proc Natl Acad Sci USA 89: 1755–1759.
Ma Y, Hendershot LM (2004) The role of the unfolded protein response in tumour development: friend or foe? Nat Rev Cancer 4: 966–977.
McMillan R, Matsui W (2012) Molecular pathways: the hedgehog signaling pathway in cancer. Clin Cancer Res 18: 4883–4888.
Michaud NR, Wang Y, McEachern KA, Jordan JJ, Mazzola AM, Hernandez A, Jalla S, Chesebrough JW, Hynes MJ, Belmonte MA, Wang L, Kang JS, Jovanovic J, Laing N, Jenkins DW, Hurt E, Liang M, Frantz C, Hollingsworth RE, Simeone DM, Blakey DC, Bedian V (2014) Novel neutralizing hedgehog antibody MEDI-5304 exhibits antitumor activity by inhibiting paracrine hedgehog signaling. Mol Cancer Ther 13: 386–398.
Moshai EF, Wémeau-Stervinou L, Cigna N, Brayer S, Sommé JM, Crestani B, Mailleux AA (2014) Targeting the hedgehog-glioma-associated oncogene homolog pathway inhibits bleomycin-induced lung fibrosis in mice. Am J Respir Cell Mol Biol 51 (1): 11–25.
Oue T, Uehara S, Yamanaka H, Nomura M, Usui N (2013) Hedgehog signal inhibitors suppress the invasion of human rhabdomyosarcoma cells. Pediatr Surg Int 29: 1153–1158.
Pathi S, Pagan-Westphal S, Baker DP, Garber EA, Rayhorn P, Bumcrot D, Tabin CJ, Blake Pepinsky R, Williams KP (2001) Comparative biological responses to human Sonic, Indian, and Desert hedgehog. Mech Dev 106: 107–117.
Pressey JG, Anderson JR, Crossman DK, Lynch JC, Barr FG (2011) Hedgehog pathway activity in pediatric embryonal rhabdomyosarcoma and undifferentiated sarcoma: a report from the Children's Oncology Group. Pediatr Blood Cancer 57: 930–938.
Rhodes A, Jasani B, Balaton AJ, Miller KD (2000) Immunohistochemical demonstration of oestrogen and progesterone receptors: correlation of standards achieved on in house tumours with that achieved on external quality assessment material in over 150 laboratories from 26 countries. J Clin Pathol 53 (4): 292–301.
Satheesha S, Manzella G, Bovay A, Casanova EA, Bode PK, Belle R, Feuchtgruber S, Jaaks P, Dogan N, Koscielniak E, Schäfer BW (2015) Targeting hedgehog signaling reduces self-renewal in embryonal rhabdomyosarcoma. Oncogene 35 (16): 2020–2030.
Shigematsu S, Yamauchi K, Nakajima K, Iijima S, Aizawa T, Hashizume K (1999) IGF-1 regulates migration and angiogenesis of human endothelial cells. Endocr J 46: S59–S62.
Tang J, Guo YS, Zhang Y, Yu XL, Li L, Huang W, Li Y, Chen B, Jiang JL, Chen ZN (2012) CD147 induces UPR to inhibit apoptosis and chemosensitivity by increasing the transcription of Bip in hepatocellular carcinoma. Cell Death Differ 19 (11): 1779–1790.
Teglund S, Toftgård R (2010) Hedgehog beyond medulloblastoma and basal cell carcinoma. Biochim Biophys Acta 1805: 181–208.
Tostar U, Malm CJ, Meis-Kindblom JM, Kindblom LG, Toftgård R, Undén AB (2006) Deregulation of the hedgehog signalling pathway: a possible role for the PTCH and SUFU genes in human rhabdomyoma and rhabdomyosarcoma development. J Pathol 208: 17–25.
Tostar U, Toftgård R, Zaphiropoulos PG, Shimokawa T (2010) Reduction of human embryonal rhabdomyosarcoma tumor growth by inhibition of the hedgehog signaling pathway. Genes Cancer 1: 941–951.
Uhmann A, Ferch U, Bauer R, Tauber S, Arziman Z, Chen C, Hemmerlein B, Wojnowski L, Hahn H (2005) A model for PTCH1/Ptch1-associated tumors comprising mutational inactivation and gene silencing. Int J Oncol 27 (6): 1567–1575.
Von Hoff DD, LoRusso PM, Rudin CM, Reddy JC, Yauch RL, Tibes R, Weiss GJ, Borad MJ, Hann CL, Brahmer JR, Mackey HM, Lum BL, Darbonne WC, Marsters JC Jr, de Sauvage FJ, Low JA (2009) Inhibition of the hedgehog pathway in advanced basal-cell carcinoma. N Engl J Med 361: 1164–1172.
Yamanaka H, Oue T, Uehara S, Fukuzawa M (2011) Hedgehog signal inhibitor forskolin suppresses cell proliferation and tumor growth of human rhabdomyosarcoma xenograft. J Pediatr Surg 46: 320–325.
Yauch RL, Gould SE, Scales SJ, Tang T, Tian H, Ahn CP, Marshall D, Fu L, Januario T, Kallop D, Nannini-Pepe M, Kotkow K, Marsters JC, Rubin LL, de Sauvage FJ (2008) A paracrine requirement for hedgehog signalling in cancer. Nature 455: 406–410.
Zibat A, Missiaglia E, Rosenberger A, Pritchard-Jones K, Shipley J, Hahn H, Fulda S (2010) Activation of the hedgehog pathway confers a poor prognosis in embryonal and fusion gene-negative alveolar rhabdomyosarcoma. Oncogene 29: 6323–6330.
Acknowledgements
The authors wish to thank Dr Noel Monks (MedImmune) for the kind gift of the MEDI-5304 antibody and Ms Christine O’Hara for help with the English version of this manuscript. This work was supported by grants from Institut Català d’Oncologia (ICO), Instituto de Salud Carlos III (RTICC-RD12/0036/0016, /0020, /0035, /0057; and PI14/00647), Fundació A BOSCH, Fundació Amics Joan Petit, ajuts predoctorals del VHIR and RIS3CAT grants COMRDI15-1-0014 (ACCIÓ and FEDER).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License.
Supplementary Information accompanies this paper on British Journal of Cancer website
Rights and permissions
From twelve months after its original publication, this work is licensed under the Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/4.0/
About this article
Cite this article
Almazán-Moga, A., Zarzosa, P., Molist, C. et al. Ligand-dependent Hedgehog pathway activation in Rhabdomyosarcoma: the oncogenic role of the ligands. Br J Cancer 117, 1314–1325 (2017). https://doi.org/10.1038/bjc.2017.305
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/bjc.2017.305
Keywords
This article is cited by
-
TRIB3 silencing promotes the downregulation of Akt pathway and PAX3-FOXO1 in high-risk rhabdomyosarcoma
Experimental Hematology & Oncology (2024)
-
Hedgehog signaling in tissue homeostasis, cancers, and targeted therapies
Signal Transduction and Targeted Therapy (2023)
-
Context-dependent modulation of aggressiveness of pediatric tumors by individual oncogenic RAS isoforms
Oncogene (2021)
-
Up-regulation of GLI1 in vincristine-resistant rhabdomyosarcoma and Ewing sarcoma
BMC Cancer (2020)
-
Signaling pathways in Rhabdomyosarcoma invasion and metastasis
Cancer and Metastasis Reviews (2020)