Abstract
Background:
Dihydropyrimidine dehydrogenase (DPD) catabolises ∼85% of the administered dose of fluoropyrimidines. Functional DPYD gene variants cause reduced/abrogated DPD activity. DPYD variants analysis may help for defining individual patients’ risk of fluoropyrimidine-related severe toxicity.
Methods:
The TOSCA Italian randomised trial enrolled colon cancer patients for 3 or 6 months of either FOLFOX-4 or XELOX adjuvant chemotherapy. In an ancillary pharmacogenetic study, 10 DPYD variants (*2A rs3918290 G>A, *13 rs55886062 T>G, rs67376798 A>T, *4 rs1801158 G>A, *5 rs1801159 A>G, *6 rs1801160 G>A, *9A rs1801265 T>C, rs2297595 A>G, rs17376848 T>C, and rs75017182 C>G), were retrospectively tested for associations with ⩾grade 3 fluoropyrimidine-related adverse events (FAEs). An association analysis and a time-to-toxicity (TTT) analysis were planned. To adjust for multiple testing, the Benjamini and Hochberg’s False Discovery Rate (FDR) procedure was used.
Results:
FAEs occurred in 194 out of 508 assessable patients (38.2%). In the association analysis, FAEs occurred more frequently in *6 rs1801160 A allele carriers (FDR=0.0083). At multivariate TTT analysis, significant associations were found for *6 rs1801160 A allele carriers (FDR<0.0001), *2A rs3918290 A allele carriers (FDR<0.0001), and rs2297595 GG genotype carriers (FDR=0.0014). Neutropenia was the most common FAEs (28.5%). *6 rs1801160 (FDR<0.0001), and *2A rs3918290 (FDR=0.0004) variant alleles were significantly associated with time to neutropenia.
Conclusions:
This study adds evidence on the role of DPYD pharmacogenetics for safety of patients undergoing fluoropyrimidine-based chemotherapy.
Similar content being viewed by others
Main
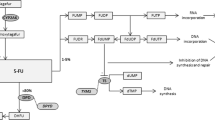
The pyrimidine analog 5-fluorouracil (5-FU) and its oral pro-drug capecitabine are among the most prescribed anti-cancer chemotherapeutic agents. Up to one-third of patients exposed to these drugs experience early-onset severe or life-threatening toxicity (Meulendijks et al, 2016). The narrow therapeutic index may be even more unfavorable when 5-FU and capecitabine are used in the adjuvant setting, where potentially cured patients undergo a prophylactic treatment strategy.
Dihydropyrimidine dehydrogenase (DPD) catabolises ∼85% of the administered dose of fluoropyrimidines and its activity is highly variable (∼8–21-fold) in the population (van Kuilenburg et al, 1999). Functional dihydropyrimidine dehydrogenase (DPYD) gene variants have been found to be associated with reduced/abrogated DPD activity (Meulendijks et al, 2016). Retrospective and prospective pharmacogenetic studies have emphasised the possible predictive role of DPYD variants for 5-FU and capecitabine toxicity. This information and the prediction of an individual patients’ risk of severe toxicity could allow for an adequate monitoring and improve overall management and quality of care (Meulendijks et al, 2016).
To date, three DPYD genetic variants have been consistently associated with fluoropyrimidine risk of toxicity (Caudle et al, 2013): *2A rs3918290 G>A, which causes the skipping of the entire exon 14; *13 rs55886062 T>G, which causes an Ile56Ser aminoacid change in a flavine binding domain of DPD; and the rs67376798 A>T, which results in a Asp949Val aminoacid change near an iron-sulfur motif. In a recent review with clinical practice guidelines, fluoropyrimidine dose omission or reductions were recommended in carriers of homozygous and heterozygous carriers of these three ‘core’ variants (Caudle et al, 2013). Because of the very low frequency of these risk alleles there is still debate on their relevance and cost-effectiveness in a ‘real world’ pre-treatment screening strategy (Deenen et al, 2016). Also, the frequencies of the risk ‘core’ variants in the general population are ∼0.1–1%, but these figures cannot explain the estimated 10–15% of DPD-linked fluoropyrimidine-related adverse events (FAEs; Caudle et al, 2013; Meulendijks et al, 2016). Therefore, additional DPYD risk variants need to be investigated for broadening the spectrum of DPYD genotyping in the clinical practice. The analyses from randomised clinical trial represent a unique opportunity for evaluating association between genetic variants and clinical outcomes and they are necessary for confirming the predictive role for toxicity of candidate polymorphisms. Three or six colon adjuvant (TOSCA) is a large randomised trial addressing the role of a shorter duration of an adjuvant oxaliplatin/fluoropyrimidines regimen in surgically resected stage III and high-risk stage II colorectal cancer (Lonardi et al, 2016).
In 2006, we planned an ancillary pharmacogenetic study to the TOSCA clinical trial for investigating genetic variants with possible predictive role for chemotherapy-related toxicity. The early study plan did not include the analysis of DPYD genetic variants. Patients from the main clinical trial were accrued in the ancillary pharmacogenetic study, which evaluated 17 polymorphisms in 11 genes (Ruzzo et al, 2014). In 2014, we planned an additional retrospective analysis in the original study population and devoted to DPYD genetic variants for fluoropyrimidine-related toxicity.
Materials and methods
The TOSCA trial
Patients included in this study represent a subgroup of the 3.759 patients with surgically resected, stage III and high-risk stage II colorectal cancer recruited in TOSCA trial between 2007 and 2011 (Lonardi et al, 2016). This is an Italian intergroup, multicentre, randomised, non-inferiority phase III study in high-risk stage II and stage III colon cancer patients treated with 3 or 6 months of either FOLFOX-4 (intravenous oxaliplatin 85 mg/m2 on day 1 and a 2-hour infusion of L-folinic acid 100 mg/m2 followed by bolus 5-FU 400 mg/m2 and a 22-hour continuous infusion of 5-FU 600 mg/m2 for two consecutive days with treatment repeated every two weeks) or XELOX (intravenous oxaliplatin 130 mg/m2 on day 1, followed by capecitabine 1000 mg/m2 per os twice daily on days 1–14 with cycles were repeated every 21 days) adjuvant chemotherapy, sponsored by GISCAD (Italian Group For The Study Of Gastrointestinal Cancer) and supported by Italian Medicines Agency (AIFA; Lonardi et al, 2016). Selected haematologic and non-haematologic toxicities (anaemia, leukopenia, neutropenia, thrombocytopenia, asthenia, diarrhoea, mucositis stomatitis, vomiting, nausea, hepatic toxicity, skin toxicity, and neurotoxicity) were assessed at the start of each cycle using Common Toxicity Criteria for Adverse Events (CTCAE) version 3.0.
All adverse events at any time were monitored and reported. Toxicity was managed as follows; in case of grade ⩾3 haematologic toxicity or persistent grade 2 the dose of all drugs was reduced by 25%. In case of grade ⩾3 non-haematologic toxicity the dose of the related drugs was reduced by 50%. In case of grade ⩾3 or persistent grade 2 neurotoxicity, oxaliplatin dose was reduced by 20%. Oxaliplatin was definitely stopped if grade ⩾2 neurosensory symptoms persisted between cycles.
Patients eligible for the TOSCA study were asked to give further and specific written informed consent to be enrolled in the pharmacogenetic studies. All experiments were performed in accordance with relevant guidelines and regulations and the Local Ethics Committee of each Institution approved the Study.
DPYD assessments
The retrospective DPYD analysis in the ancillary pharmacogenetic study to the TOSCA clinical trial was planned in 2014 after the publication of the pharmacogenetic analysis in the QUASAR2 study (Rosmarin et al, 2014). Genetic markers of toxicity for capecitabine monotherapy were selected after systemic review and then investigated in the QUASAR2 patients population of Caucasian individuals. We aimed at re-evaluating the DPYD panel of the QUASAR2 study (*4 rs1801158, *5 rs1801159, *6 rs1801160, *9A rs1801265, rs2297595, *2A rs3918290, *13 rs55886062, and rs67376798) in the homogeneous population of patients of the TOSCA trial who underwent adjuvant fluoropyrimidine/oxaliplatin combination chemotherapy. At the time of the planning of our DPYD analysis, two additional variants, rs17376848 and rs75017182, showed promising predictive role for fluoropyrimidine-related toxicity (van Kuilenburg et al, 2010; Teh et al, 2013; Froehlich et al, 2015). These genetic variants were included in our panel (Table 1) considering that: (A) the polymorphisms had some degree of likelihood to alter the structure or the expression of the gene in a biologically relevant manner; (B) the ‘q’ allele frequency was expected to be >1%; and (C) the polymorphisms were established and well-documented.
Genomic DNA was extracted from 2 ml whole blood by using the QiaAmp kit (Qiagen, Valencia, CA, USA). rs75017182 was analysed by Real-Time PCR assay using the Easy DPYD kit (Diatech Pharmacogenetics, Jesi, Italy), while the other nine DPYD variants were all included in the MYRIAPOD ADMET kit (Diatech Pharmacogenetics), and analysed on the MassARRAY System (Agena Bioscience). The MassARRAY protocol is characterised by three main steps: polymerase chain reaction (PCR), single-base primer extension (SBE), and separation of the products on a matrix-loaded silicon chip by matrix-assisted laser desorption ionization time of life mass spectrometry (MALDI-TOF MS). After the amplification of the region of interest, a primer extension reaction with oligos that bind adjacent to the targeted polymorphic site and all four nucleotide terminators (iPLEX) was carried out. The extension reaction generated different products for different alleles: primers extended with the terminator dNTP complementary to the targeted polymorphic site. All iPLEX products, each with its unique mass, were then identified using mass spectrometry. PCR and SBE reactions were performed in a thermal cycler (Labcycler, SensoQuest), whereas the extension products were analysed using the MALDI-TOF MassARRAY Analyzer 4 (Agena Bioscience), according to the MYRIAPOD ADMET kit’s instructions for use and using all reagents and consumables contained in the SQ TYPING 960 Kit (Diatech Pharmacogenetics). The genotype call was performed with the iGENETICS MYRIAPOD software (Diatech Pharmacogenetics).
All laboratory analyses were performed blind to the patients' treatment and clinical outcomes. Genetic data were then transferred to and independently analysed at IRCCS Istituto di Ricerche Farmacologiche ‘Mario Negri’.
Statistics
Conforming to previously FAEs definition (Lee et al, 2014; Boige et al, 2016) and to the planned management of toxicity in the TOSCA trial, grade ⩾3 neutropenia, diarrhoea, asthenia, nausea, vomiting, leukopenia, thrombocytopenia, mucositis, stomatitis, and skin toxicity were deemed as severe FAEs. The treatment compliance was described in terms of treatment interruption and dose intensity, defined as the dose given in mg per m2 per week.
According to the results of DPYD analysis, patients were categorised in three genotype groups: carriers of the homozygous wild type (p2); heterozygous (pq); and homozygous variant (q2). The possible association of DPYD variant with FAEs was analysed in the codominant model (p2, pq, and q2 genotypes considered separately) and in a dominant model with merged heterozygous (pq) and homozygous (q2) risk variant genotype carriers.
To test the effect of DPYD genotypes on toxicity, two analyses were planned: an association analysis and a time-to-toxicity (TTT) analysis. This choice was made because a conventional analysis with a binary outcome describing only the occurrence of severe toxicity may be inaccurate in the case of few observations (due to the rarity of some genotypes), and it may not capture potential clinically meaningful differences also in terms of time of toxicity onset (Thanarajasingam et al, 2016). The association analysis compared the rate of FAEs across DPYD genotypes by means of a Fisher’s test in contingency tables. The TTT was defined as the time from date of randomisation in TOSCA trial to the date of severe FAEs occurrence. Subjects without severe FAEs at the time of analysis were censored at the date they were last known to be event-free while on treatment. TTT curves were estimated using the Kaplan–Meier method. Cox proportional hazard models stratified for treatment duration (6 or 3 months) were used to assess the effects of DPYD genotypes on TTT. Multivariate analysis stratified for treatment duration was performed to adjust the identified effect for age, gender, stage and treatment (FOLFOX-4 or XELOX). Results were provided as the hazard ratio (HR) with 95% confidence interval (95% CI).
All reported P-values were two-sided with P<0.05 value considered statistically significant. However, to adjust the analyses for multiple testing, the Benjamini and Hochberg’s False Discovery Rate (FDR) procedure was used, considering both the dominant and codominant model.
Assuming the prevalence of a high-risk allele of at least 10% and FAEs in about one-third of the study population, 188 events would allow the detection of a HR of at least 2 associated to the group with unfavorable genotypes (90% power and 5% type I error in a bilateral test). Detection of significant association for the three ‘core’ variants (*2A rs3918290, *13 rs55886062, and rs67376798) would require higher HR values given the expected frequencies of their risk alleles below 10%.
A χ2 test was used for checking the Hardy–Weinberg equilibrium. Linkage disequilibrium (LD), defined as a non-random association of alleles adjacent loci, was assessed and both D′ and r2 measures were provided. D′ can take any value from 0 (random co-inheritance of alleles) to 1 (complete LD); r2 also ranges from 0 (random co-inheritance of alleles) to 1 (perfect LD). Values of r2<0.33 suggest absence of strong LD (Ardlie et al, 2002). Analyses were performed with SAS 9.4 (SAS Institute, Cary, NC, USA) and the SNPStats package (Solè et al, 2006).
Results
Patient characteristics and toxicity
From July 2007 to October 2011, 534 patients from 26 experimental centers entered the study. This figure represents 81% of patients randomised in the same period and by the same centers in the main study. Twenty-six patients were not assessable for the following reason: 5 patients were never treated, for 2 patients the treatment data were unavailable, and for 19 patients the blood sampling was not assessable due to technical problems. Therefore, the analysis was conducted in 508 patients.
Characteristics of the 508 patients are shown in Table 2. Patients’ baseline characteristics were consistent with those of the whole trial population (Lonardi et al, 2016). Most patients were randomised to FOLFOX-4 because option for XELOX regimen was introduced in TOSCA trial only during the late phase of accrual of this ancillary study. Toxicity related to adjuvant chemotherapy is reported in Table 3. Again, the spectrum and the frequency of toxicities did not differ from those observed in whole trial population (Lonardi et al, 2016). One hundred ninety-four (38.2%) patients experienced at least one FAE. Neutropenia was the commonest among FAEs occurring in 145 patients (28.5%). As shown in Supplementary Table S1, analysis of dose intensity did not show differences across treatment arms.
Genetic assessments
Table 1 lists the studied genetic variants and the distribution of genotypes of patients successfully assessed for each polymorphism. Consistent with previous observations, genotype frequency did not differ from those observed in Caucasian population. The *13 rs55886062 G allele was not found in the studied population and therefore, this variant was excluded from subsequent analyses. Allele frequencies of the remaining polymorphisms were consistent with the Hardy–Weinberg equilibrium (P>0.05). Results of LD analyses are shown in Supplementary Table S2.
DPYD variants and FAEs
The prevalence of DPYD high-risk alleles was heterogeneous, ranging from 0% of the *13 rs55886062 G allele to 37.5% of the *9A rs1801265 C allele. Therefore, 194 events would allow detection of an HR of at least 8.3 and an HR of at least 1.5 for a prevalence of a high-risk allele equal to 1% and to 35%, respectively (power of 90% and a I type error of 5%, for a bilateral test). A statistically significant association was found between *6 rs1801160 genotypes and FAEs (FDR=0.0083 in both the dominant and codominant models). No additional significant associations were detected (data not shown).
Results about the effect of DPYD variants on TTT are shown in Table 4. At univariate analysis, *6 rs1801160 (codominant model: FDR=0.0022), rs2297595 (codominant model: FDR=0.0413), *2A rs3918290 (codominant model: FDR=0.0001) correlated with TTT. Specifically, *6 rs1801160 GA genotype carriers and A allele carriers were at risk for shorter TTT (HR 1.99, 95% CI 1.38–2.86, FDR=0.0002 and HR 2.01, 95% CI 1.42–2.86, FDR=0.0006, respectively). Median TTT for *6 rs1801160 GG, GA and AA genotype carriers were 7.0, 3.0 and 2.1 months, respectively. Also, the rs2297595 GG genotype (HR 4.28, 95% CI 1.35–13.55, FDR=0.0136) and the *2A rs3918290 GA genotype (HR 15.34, 95% CI 4.72–49.89, FDR=0.0001) showed a shorter TTT. Median TTT for rs2297595 AA, AG and GG genotype carriers were 7.0, 6.6 and 1.2 months, respectively. Median TTT for *2A rs3918290 GG and GA genotype carriers were 7.0 and 0.9 months, respectively. Figure 1 depicts Kaplan–Meier curves of ‘q’ allele carriers vs ‘p2’ genotype carriers of rs2297595 and *6 rs1801160. At multivariate analyses the associations with DPYD variants identified in the univariate analyses were confirmed.
Kaplan-Meier curves. (A) TTT curves of the *6 rs1801160 minor A allele carriers (merged heterozygous plus homozygous minor allele carriers) and homozygous GG genotype carriers. (B) TTT curves of the rs2297595 minor G allele carriers (merged heterozygous plus homozygous minor allele carriers) and homozygous AA genotype carrier.
Neutropenia was the commonest FAEs, occurring in 145 patients (28.5%). The second one was diarrhoea, which occurred in 33 patients (6.5%). Therefore, univariate and multivariate Cox analyses to address the effect of DPYD variants on TTT for specific FAEs were performed only for neutropenia (Table 5). At univariate analysis, associations with time to neutropenia were found for *6 rs1801160 and *2A rs3918290. In detail, *6 rs1801160 GA genotype carriers in the codominant model and A allele carriers in the dominant model were at risk for shorter time to neutropenia (HR 2.19, 95% CI 1.46–3.28, FDR=0.0002 and HR 2.18, 95% CI 1.47–3.24, FDR=0.0024, respectively). The codominant model analysis for *2A rs3918290 showed significant association with short time to neutropenia for GA variant genotype carriers (HR 10.74, 95% CI 2.59–44.61, FDR=0.0054). The impact of all this DPYD variants was confirmed at multivariate analysis.
Discussion
As shown in Table 6, this study is added to previous pharmacogenetic analyses for DPYD, which were incorporated in randomised clinical trials of fluoropyrimidine-based chemotherapy in colorectal cancer (Deenen et al, 2011; Lee et al, 2014; Rosmarin et al, 2014; Del Re et al, 2015; Boige et al, 2016; Lee et al, 2016). These studies offer a unique opportunity for performing pharmacogenetics in an optimal setting, where the genotyped patient population is well characterised and uniformly assessed for clinical/pathologic characteristics and the monitoring of toxicity. Unfortunately, these studies cannot be uniformly evaluated because of the substantial differences in disease stage (adjuvant vs metastatic), chemotherapy protocols (often with biologics), panels of DPYD variants, and methodology for assessing putative pharmacogenetics associations. To this regard, we introduced the TTT analysis in addition to a standard genotypes/FAEs distribution analysis, which was commonly adopted in studies listed in Table 6. The TTT analysis for detecting pharmacogenetic associations with FAEs may help to disclose potential clinical impact of DPYD variants, which could be lost in a common binary analysis of genotype frequencies in contingency tables. The TTT analysis adds the dimension of time, and therefore, it allows for detection of ‘more and early’ toxicity events (Thanarajasingam et al, 2016). In fact, if severe toxicity occurs after multiple cycles of chemotherapy, it may also represent a cumulative effect and the stress of the system after several doses of the drugs. On the contrary, if severe toxicity events occur early, they are more likely related to innate defects, often linked with catabolic pathways (Sahota et al, 2016). Notably, some clinical analyses on DPYD variants and fluoropyrimidine-related toxicity were based on FAEs occurring within the first 3 cycles of therapy (Gross et al, 2008; Deenen et al, 2011, Froehlich et al, 2015). The TTT approach avoids the need of defining such a cut-point and it may better characterise a gene-linked toxicity profile. Also, it should be considered that some functional DPYD variants may not induce a dramatic loss of enzyme function like the *2A rs3918290, and therefore, in these cases, TTT analysis may be more sensitive for detecting the risk of toxicity determined by DPYD variants with moderate functional effects.
In the present study population, potential baseline confounders for early toxicity could be excluded since the administration of adjuvant combination chemotherapy was per-protocol proposed to high-risk colon cancer patients without evidence of metastatic disease, no major comorbidity, long life expectancy, and good performance status. Furthermore, only 2 patients interrupted treatment due to disease progression and in these patients no fluoropyrimidine-related toxicity was observed.
In our population of patients, the observed frequencies of the rare deleterious DPYD variant alleles *2A rs3918290, *13 rs55886062, and rs67376798 were 0.6%, 0%, and 1.2%, respectively. Only *2A rs3918290 showed significant association with FAEs in the TTT analysis achieving an HR equal to 14.98, and a significant impact on time to neutropenia (Tables 4 and 5, respectively). However, even if they all had shown significant HRs for FAEs, they cannot explain the overall estimated contribution of functional DPYD variants in causing severe fluoropyrimidine toxicity. DPD deficiency has been described in ∼40–60% of patients with ⩾3 grade fluoropyrimidine-induced toxicity (Meulendijks et al, 2015). However, DPD deficiency cannot always be traced back to a currently known DPYD variant associated with reduced enzyme activity (Meulendijks et al, 2015). Therefore, other detrimental variants should be identified to improve sensitivity of DPYD genotyping (Gentile et al, 2016). Indeed, among the seven additional DPYD studied variants, two (*6 rs1801160 and rs2297595) showed associations with FAEs.
The DPYD *6 rs1801160 was analysed within the DPYD panel of three studies listed in Table 6 (Deenen et al, 2011; Rosmarin et al, 2014; Boige et al, 2016). Notably, in the large PETACC-8 study, *6 rs1801160 showed statistically significant association with grade 3 or greater FAEs and neutropenia in particular (Boige et al, 2016). In the QUASAR2 (Rosmarin et al, 2014) and the CAIRO-2 (Deenen et al, 2011) studies, *6 rs1801160 did not show predictive role for FAEs. However, it should be considered that the QUASAR2 analysis (Rosmarin et al, 2014) was performed in patients treated with capecitabine mono-chemotherapy only. As far as the CAIRO-2 is concerned, the high probability of developing FAEs (85%) was considered as a major reason for not detecting significant associations between FAEs and all tested DPYD variants in this study (Deenen et al, 2011). If we look at risk associations between *6 rs1801160 and FAEs in the present study and the PETACC-8 study (Boige et al, 2016), it should be noted a significant but moderate effect size attributed to the *6 rs1801160 A risk allele. Results from the pharmacogenetics analysis by Kleibl et al suggested an impact of the *6 rs1801160 A allele in determining fluoropyrimidine toxicity especially in the context of specific DPYD haplotypes (Kleibl et al, 2009). Notably, in the whole DPYD panel, the *6 rs1801160 locus did not show strong LD, thus excluding that the association of the variant with toxicity may be only the results of LD with a neighboring etiologic variant. These aspects would suggest direct but mild impact on phenotype of the *6 rs1801160, which cumulates with other variants and/or emerges in specific chemotherapy regimen because of toxicity synergy between fluoropyrimidine and other drugs (i.e., oxaliplatin; Offer and Diasio, 2016). In the sub-type analysis of FAEs, the *6 rs1801160 variant showed detrimental effect on time to neutropenia. We observed grade ⩾3 neutropenia in the 28.5% of patients and this figure is slightly lower than the toxicity rates previously reported in patients treated with XELOX and FOLFOX regimens (up to 40%; Eng (2009)). These figures would exceed the expected frequency of ⩾3 grade neutropenia if the sum of neutropenia rates in single-agent studies of oxaliplatin, capecitabine and bolus/infusional 5-FU (<10% of patients) would be applied for prediction. The array of interactions and synergisms between fluoropyrimidines and oxaliplatin in humans may explain this discrepancy. In this context, a DPYD variant, which depresses, but does not abrogate the enzyme function may significantly increase the risk of severe toxicity (neutropenia) when the fluorpyrimidine is combined with other drugs.
DPYD pharmacogenetics in the PETACC-8 study (Boige et al, 2016) included the rs2297595, but without detecting significant associations with FAEs. In the present study, the homozygous rs2297595 GG genotype was associated with a significant relatively large effect (HR 6.77) in the TTT analysis, whereas the heterozygous genotype did not. This behavior would suggest an ‘allele-dosage’ effect and a clinically meaningful DPD deficient phenotype in carriers of the ‘q2’ genotype. This hypothesis parallels previous findings in a retrospective pharmacogenetic study by Gross et al (2008), and it is compatible with the putative functional effect of the rs2297595 variant. The methionine-valine exchange, as consequence of the non-synonymous sequence variation occurs in a highly conserved site during evolution, which may be critical to enzyme structure and function (Mattison et al, 2002). Moreover, LD analyses showed that the *6 rs1801160 and rs2297595 loci are not co-inherited, and therefore they may act independently.
The analysis of the median TTT values contributes to the understanding of the clinical impact of the *6 rs1801160, *2A rs3918290, and rs2297595 variants. Median TTT was 7 months among common homozygous genotypes carriers, whereas it was significantly shortened (between 0.9 and 2.1 months) in carriers of the homozygous variant *6 rs1801160 and rs2297595 and the *2A rs3918290 heterozygous genotypes. Notably, shortened TTT was detectable in *6 rs1801160, but not rs2297595 heterozygous genotype carriers, thus corroborating the hypothesis of a different effect of the two variants in depressing/altering the DPD function. The early onset of toxicity corroborates the hypothesis of an underlying enzymatic defect and the opportunity of verifying DPYD variants/DPD status in patients with early severe FAEs after fluoropyrimidine exposure.
As far as ethnicity is concerned, the frequency of the *6 rs1801160 A risk allele seems comparable in Caucasian, Middle-Eastern, and African-American, whereas it seems less frequent in Asian populations (Caudle et al, 2013). The clinical impact of the rs2297595 variant may be more relevant to populations of African ancestry, where its frequency seems to double in comparison with Caucasian populations (Aminkeng et al, 2014).
It is still matter of debate whether DPYD genotyping should be incorporated in the routine pre-treatment screening of patients undergoing fluoropyrimidine-based chemotherapy. To this regard, the recent guidelines of the European Society for Medical Oncology (ESMO) consider the testing as an option, which is indicated in the case of patients who experience severe toxicity and before the fluoropyrimidine is re-introduced (van Cutsem et al, 2016). We disagree with statement, especially when possible cautions could be adopted in treatment settings with narrow therapeutic window. Since the DPYD assessment was not incorporated in our original study plan, we could not perform a reliable cost-effectiveness analysis. However, available analyses suggest that DPYD-genotype guided dosing according to *2A rs3918290 (Deenen et al, 2016), or *2A rs3918290, *13 rs55886062, and rs67376798 (Cortejoso et al, 2016) may significantly improve safety of fluoropyrimidine therapy and being cost saving.
It should be considered that additional tests have been developed for assessing the activity of the DPD enzyme (DPD activity in peripheral blood monuclear cells, Uracil breath test, endogenous plasma/urine Uracil/Dihydrouracil, sampling PK model after 5-fluoruracil test dose; van Staveren et al, 2013, 2016; Del Re et al, 2017). These phenotyping tests seem to possess better predictivity then genotyping for fluoropyrimidine toxicity (van Staveren et al, 2013, 2016). In a recent analysis in 550 patients, Meulendijks et al found that high pre-treatment uracil concentrations were strongly associated with severe fluoropyrimidine-related toxicity, whereas DPYD genotypes did not (Meulendijks et al, 2017). However, in this study, DPYD genotyping was limited to rs67376798, *13 rs55886062, rs75017182, and *4 rs1801158. In general, as with the genotyping strategy, the phenotyping tests suffer from suboptimal sensitivity and specificity. Notably, a test for detecting DPD deficiency and preventing fluoropyrimidine toxicity requires high sensitivity. On the other side, low specificity may cause unnecessary dose reduction and suboptimal exposure to effective chemotherapy. To this end, as pointed out by Boisdron-Celle et al (2007), DPYD genotyping and DPD phenotyping tests could be integrated in a two-step strategy for screening selected patients.
In conclusion, this study remarks the role of DPYD *2A rs3918290 for fluoropyrimidine-related toxicity. It also indicates that *6 rs1801160 and rs2297595 produce additional DPYD genotypes, which may be predictive of toxicity in the same setting. TTT analysis in pharmacogenetic studies may help to characterise the clinical impact of risk alleles causing reduced DPD function.
References
Aminkeng F, Ross CJ, Rassekh SR, Brunham LR, Sistonen J, Dube MP, Ibrahim M, Nyambo TB, Omar SA, Froment A, Bodo JM, Tishkoff S, Carleton BC, Hayden MR ( 2014 ) Canadian Pharmacogenomics Network for Drug Safety Consortium. Higher frequency of genetic variants conferring increased risk for ADRs for commonly used drugs treating cancer, AIDS and tuberculosis in persons of African descent . Pharmacogenomics J 14 (2) : 160 – 170 .
Ardlie KG, Kruglyak L, Seielstad M ( 2002 ) Patterns of linkage disequilibrium in the human genome . Nat Rev Genet 3 : 299 – 309 .
Boige V, Vincent M, Alexandre P, Tejpar S, Landolfi S, Le Malicot K, Greil R, Cuyle PJ, Yilmaz M, Faroux R, Matzdorff A, Salazar R, Lepage C, Taieb J, Laurent-Puig P ( 2016 ) DPYD genotyping to predict adverse events following treatment with flourouracil-based adjuvant chemotherapy in patients with stage III colon cancer: a secondary analysis of the PETACC-8 randomized clinical trial . JAMA Oncol 2 : 655 – 662 .
Boisdron-Celle M, Remaud G, Traore S, Poirier AL, Gamelin L, Morel A, Gamelin E ( 2007 ) 5-Fluorouracil-related severe toxicity: a comparison of different methods for the pretherapeutic detection of dihydropyrimidine dehydrogenase deficiency . Cancer Lett 249 (2) : 271 – 282 .
Caudle KE, Thorn CF, Klein TE, Swen JJ, McLeod HL, Diasio RB, Schwab M ( 2013 ) Clinical pharmacogenetics implementation consortium guidelines for dihydropyrimidine dehydrogenase genotype and fluoropyrimidine dosing . Clin Pharmacol Ther 94 : 640 – 645 .
Cortejoso L, García-González X, García MI, García-Alfonso P, Sanjurjo M, López-Fernández LA ( 2016 ) Cost-effectiveness of screening for DPYD polymorphisms to prevent neutropenia in cancer patients treated with fluoropyrimidines . Pharmacogenomics 17 (9) : 979 – 984 .
Deenen MJ, Tol J, Burylo AM, Doodeman VD, de Boer A, Vincent A, Guchelaar HJ, Smits PH, Beijnen JH, Punt CJ, Schellens JH, Cats A ( 2011 ) Relationship between single nucleotide polymorphisms and haplotypes in DPYD and toxicity and efficacy of capecitabine in advanced colorectal cancer . Clin Cancer Res 17 : 3455 – 3468 .
Deenen MJ, Meulendijks D, Cats A, Sechterberger MK, Severens JL, Boot H, Smits PH, Rosing H, Mandigers CM, Soesan M, Beijnen JH, Schellens JH ( 2016 ) Upfront genotyping of DPYD*2A to individualize fluoropyrimidine therapy: a safety and cost analysis . J Clin Oncol 34 : 227 – 234 .
Del Re M, Cremolini C, Loupakis F, Marmorino F, Citi V, Palombi M, Bergamo F, Schirripa M, Rossini D, Cortesi E, Tomasello G, Spadi R, Buonadonna A, Amoroso D, Vitello S, Di Donato S, Granetto C, D'Amico M, Danesi R, Falcone A ( 2015 ) DPYD c.1905+1G>A and c.2846A>T and UGT1A1*28 allelic variants as predictors of toxicity: Pharmacogenetic translational analysis from the phase III TRIBE study in metastatic colorectal cancer . J Clin Oncol 33, (suppl): abstr 3532 . http://meetinglibrary.asco.org/record/112386/abstract .
Del Re M, Restante G, Di Paolo A, Crucitta S, Rofi E, Danesi R ( 2017 ) Pharmacogenetics and metabolism from science to implementation in clinical practice: the example of dihydropyrimidine dehydrogenase . Curr Pharm Des 23 (14) : 2028 – 2034 .
Eng C ( 2009 ) Toxic effects and their management: daily clinical challenges in the treatment of colorectal cancer . Nat Rev Clin Oncol 6 (4) : 207 – 218 .
Froehlich TK, Amstutz U, Aebi S, Joerger M, Largiadèr CR ( 2015 ) Clinical importance of risk variants in the dihydropyrimidine dehydrogenase gene for the prediction of early-onset fluoropyrimidine toxicity . Int J Cancer 136 (3) : 730 – 739 .
Gentile G, Botticelli A, Lionetto L, Mazzuca F, Simmaco M, Marchetti P, Borro M ( 2016 ) Genotype-phenotype correlations in 5-fluorouracil metabolism: a candidate DPYD haplotype to improve toxicity prediction . Pharmacogenomics J 16 : 320 – 325 .
Gross E, Busse B, Riemenschneider M, Neubauer S, Seck K, Klein HG, Kiechle M, Lordick F, Meindl A ( 2008 ) Strong association of a common dihydropyrimidine dehydrogenase gene polymorphism with fluoropyrimidine-related toxicity in cancer patients . PLoS One 3 (12) : e4003 .
Kleibl Z, Fidlerova J, Kleiblova P, Kormunda S, Bilek M, Bouskova K, Sevcik J, Novotny J ( 2009 ) Influence of dihydropyrimidine dehydrogenase gene (DPYD) coding sequence variants on the development of fluoropyrimidine-related toxicity in patients with high-grade toxicity and patients with excellent tolerance of fluoropyrimidine-based chemotherapy . Neoplasma 56 : 303 – 316 .
Lee AM, Shi Q, Pavey E, Alberts SR, Sargent DJ, Sinicrope FA, Berenberg JL, Goldberg RM, Diasio RB ( 2014 ) DPYD variants as predictors of 5-fluorouracil toxicity in adjuvant colon cancer treatment (NCCTG N0147) . J Natl Cancer Inst 106 : dju298 .
Lee AM, Shi Q, Alberts SR, Sargent DJ, Sinicrope FA, Berenberg JL, Grothey A, Polite B, Chan E, Gill S, Kahlenberg MS, Nair SG, Shields AF, Goldberg RM, Diasio RB ( 2016 ) Association between DPYD c.1129-5923 C>G/hapB3 and severe toxicity to 5-fluorouracil-based chemotherapy in stage III colon cancer patients: NCCTG N0147(Alliance) . Pharmacogenet Genomics 26 : 133 – 137 .
Lonardi S, Sobrero A, Rosati G, Di Bartolomeo M, Ronzoni M, Aprile G, Scartozzi M, Banzi M, Zampino MG, Pasini F, Marchetti P, Cantore M, Zaniboni A, Rimassa L, Ciuffreda L, Ferrari D, Barni S, Zagonel V, Maiello E, Rulli E, Labianca R ( 2016 ) TOSCA (Three or Six Colon Adjuvant) Investigators. Phase III trial comparing 3-6 months of adjuvant FOLFOX4/XELOX in stage II-III colon cancer: safety and compliance in the TOSCA trial . Ann Oncol 27 : 2074 – 2081 .
Mattison LK, Johnson MR, Diasio RB ( 2002 ) A comparative analysis of translated dihydropyrimidine dehydrogenase cDNA; conservation of functional domains and relevance to genetic polymorphisms . Pharmacogenetics 12 (2) : 133 – 144 .
Meulendijks D, Cats A, Beijnen JH, Schellens JH ( 2016 ) Improving safety of fluoropyrimidine chemotherapy by individualizing treatment based on dihydropyrimidine dehydrogenase activity—ready for clinical practice? Cancer Treat Rev 50 : 23 – 34 .
Meulendijks D, Henricks LM, Sonke GS, Deenen MJ, Froehlich TK, Amstutz U, Largiadèr CR, Jennings BA, Marinaki AM, Sanderson JD, Kleibl Z, Kleiblova P, Schwab M, Zanger UM, Palles C, Tomlinson I, Gross E, van Kuilenburg AB, Punt CJ, Koopman M, Beijnen JH, Cats A, Schellens JH ( 2015 ) Clinical relevance of DPYD variants c.1679T>G, c.1236G>A/HapB3, and c.1601G>A as predictors of severe fluoropyrimidine-associated toxicity: a systematic review and meta-analysis of individual patient data . Lancet Oncol 16 : 1639 – 1650 .
Meulendijks D, Henricks LM, Jacobs BAW, Aliev A, Deenen MJ, de Vries N, Rosing H, van Werkhoven E, de Boer A, Beijnen JH, Mandigers CMPW, Soesan M, Cats A, Schellens JHM ( 2017 ) Pretreatment serum uracil concentration as a predictor of severe and fatal fluoropyrimidine-associated toxicity . Br J Cancer 116 (11) : 1415 – 1424 .
Offer SM, Diasio RB ( 2016 ) Biomarkers of fluorouracil toxicity: insight from the PETACC-8 trial . JAMA Oncol 2 : 662 – 663 .
Rosmarin D, Palles C, Church D, Domingo E, Jones A, Johnstone E, Wang H, Love S, Julier P, Scudder C, Nicholson G, Gonzalez-Neira A, Martin M, Sargent D, Green E, McLeod H, Zanger UM, Schwab M, Braun M, Seymour M, Thompson L, Lacas B, Boige V, Ribelles N, Afzal S, Enghusen H, Jensen SA, Etienne-Grimaldi MC, Milano G, Wadelius M, Glimelius B, Garmo H, Gusella M, Lecomte T, Laurent-Puig P, Martinez-Balibrea E, Sharma R, Garcia-Foncillas J, Kleibl Z, Morel A, Pignon JP, Midgley R, Kerr D, Tomlinson I ( 2014 ) Genetic markers of toxicity from capecitabine and other fluorouracil-based regimens: investigation in the QUASAR2 study, systematic review, and meta-analysis . J Clin Oncol 32 : 1031 – 1039 .
Ruzzo A, Graziano F, Galli F, Giacomini E, Floriani I, Galli F, Rulli E, Lonardi S, Ronzoni M, Massidda B, Zagonel V, Pella N, Mucciarini C, Labianca R, Ionta MT, Veltri E, Sozzi P, Barni S, Ricci V, Foltran L, Nicolini M, Biondi E, Bramati A, Turci D, Lazzarelli S, Verusio C, Bergamo F, Sobrero A, Frontini L, Magnani M ( 2014 ) Genetic markers for toxicity of adjuvant oxaliplatin and fluoropyrimidines in the phase III TOSCA trial in high-risk colon cancer patients . Sci Rep 5 : 6828 .
Sahota T, Danhof M, Della Pasqua O ( 2016 ) Pharmacology-based toxicity assessment: towards quantitative risk prediction in humans . Mutagenesis 31 : 359 – 374 .
Solè X, Guinò E, Valls J, Iniesta R, Moreno V ( 2006 ) SNPStats: a web tool for the analysis of association studies . Bioinformatics 22 : 1928 – 1929 .
Teh LK, Hamzah S, Hashim H, Bannur Z, Zakaria ZA, Hasbullani Z, Shia JK, Fijeraid H, Md Nor A, Zailani M, Ramasamy P, Ngow H, Sood S, Salleh MZ ( 2013 ) Potential of dihydropyrimidine dehydrogenase genotypes in personalizing 5-fluorouracil therapy among colorectal cancer patients . Ther Drug Monit 35 (5) : 624 – 630 .
Thanarajasingam G, Atherton PJ, Novotny PJ, Loprinzi CL, Sloan JA, Grothey A ( 2016 ) Longitudinal adverse event assessment in oncology clinical trials: the Toxicity over Time (ToxT) analysis of Alliance trials NCCTG N9741 and 979254 . Lancet Oncol 17 : 663 – 670 .
van Cutsem E, Cervantes A, Adam R, Sobrero A, Van Krieken JH, Aderka D, Aranda Aguilar E, Bardelli A, Benson A, Bodoky G, Ciardiello F, D'Hoore A, Diaz-Rubio E, Douillard JY, Ducreux M, Falcone A, Grothey A, Gruenberger T, Haustermans K, Heinemann V, Hoff P, Köhne CH, Labianca R, Laurent-Puig P, Ma B, Maughan T, Muro K, Normanno N, Österlund P, Oyen WJ, Papamichael D, Pentheroudakis G, Pfeiffer P, Price TJ, Punt C, Ricke J, Roth A, Salazar R, Scheithauer W, Schmoll HJ, Tabernero J, Taïeb J, Tejpar S, Wasan H, Yoshino T, Zaanan A, Arnold D ( 2016 ) ESMO consensus guidelines for the management of patients with metastatic colorectal cancer . Ann Oncol 27 (8) : 1386 – 1422 .
van Kuilenburg AB, van Lenthe H, Blom MJ, Mul EP, Van Gennip AH ( 1999 ) Profound variation in dihydropyrimidine dehydrogenase activity in human blood cells: major implications for the detection of partly deficient patients . Br J Cancer 79 (3–4) : 620 – 626 .
van Kuilenburg AB, Meijer J, Gökcay G, Baykal T, Rubio-Gozalbo ME, Mul AN, de Die-Smulders CE, Weber P, Mori AC, Bierau J, Fowler B, Macke K, Sass JO, Meinsma R, Hennermann JB, Miny P, Zoetekouw L, Roelofsen J, Vijzelaar R, Nicolai J, Hennekam RC (2010) Dihydropyrimidine dehydrogenase deficiency caused by a novel genomic deletion c.505_513del of DPYD. Nucleosides Nucleotides Nucleic Acids 29: 509–514 .
van Staveren MC, Guchelaar HJ, van Kuilenburg AB, Gelderblom H, Maring JG ( 2013 ) Evaluation of predictive tests for screening for dihydropyrimidine dehydrogenase deficiency . Pharmacogenomics J 13 (5) : 389 – 395, Review. Erratum in: (2014) Pharmacogenomics J 14(4): 400 .
van Staveren MC, van Kuilenburg AB, Guchelaar HJ, Meijer J, Punt CJ, de Jong RS, Gelderblom H, Maring JG ( 2016 ) Evaluation of an oral uracil loading test to identify DPD-deficient patients using a limited sampling strategy . Br J Clin Pharmacol 81 (3) : 553 – 561 .
Acknowledgements
This work was partially supported by the TERPAGE Project ‘POR MARCHE FESR 2007–2013’. AR, Francesco G, and MM conceived and performed the study design, performed the manuscript preparation and data interpretation. Fabio G performed coordination study. Francesca G, Fabio G, and ER performed statistical analysis, data interpretation, and manuscript preparation. SL, MR, BM, VZ, NP, CM, RL, MTI, EV, PS, SB, V R, IB, LF, MN, EB, AB, DT, SL, CV, FB,AS, and LF collected samples and patients’ data, and commented the manuscript. RL, LF, and AS participated in the study design and data interpretation, and helped to draft the manuscript. All authors reviewed the manuscript.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
This paper is dedicated to the memory of our friend and colleague, Irene Floriani.
Supplementary Information accompanies this paper on British Journal of Cancer website
Supplementary information
Rights and permissions
This work is licensed under the Creative Commons Attribution-Non-Commercial-Share Alike 4.0 International License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/4.0/
About this article
Cite this article
Ruzzo, A., Graziano, F., Galli, F. et al. Dihydropyrimidine dehydrogenase pharmacogenetics for predicting fluoropyrimidine-related toxicity in the randomised, phase III adjuvant TOSCA trial in high-risk colon cancer patients. Br J Cancer 117, 1269–1277 (2017). https://doi.org/10.1038/bjc.2017.289
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/bjc.2017.289
Keywords
This article is cited by
-
Precision fluoropyrimidines dosing in a compound heterozygous variant carrier of the DPYD gene: a case report
Cancer Chemotherapy and Pharmacology (2023)
-
Challenges in prevention, early detection, and management of febrile neutropenia in adult patients with solid tumors
memo - Magazine of European Medical Oncology (2023)
-
Introducing a simple and cost-effective RT-PCR protocol for detection of DPYD*2A polymorphism: the first study in Kurdish population
Cancer Chemotherapy and Pharmacology (2022)
-
Comprehensive pharmacogenetic analysis of DPYD, UGT, CDA, and ABCB1 polymorphisms in pancreatic cancer patients receiving mFOLFIRINOX or gemcitabine plus nab-paclitaxel
The Pharmacogenomics Journal (2021)
-
Targeted exon sequencing in deceased schizophrenia patients in Denmark
International Journal of Legal Medicine (2020)