Abstract
Background:
Diaphanous-related formins (DRFs), actin necleator, have been known to participate in the progression of cancer cells. We previously reported that FMNL2 (Formin-like2), a member of DRFs, was a positive regulator in colorectal cancer (CRC) metastasis, yet proteins and pathways required for the function of this pro-invasive DRFs remain to be identified.
Methods:
The relationship between FMNL2 and COMMD10 was examined using Co-IP, GST pull-down, immunofluorescence and in vitro ubiquitination assay. The in vitro and in vivo function of COMMD10 in CRC was evaluated using CCK-8 proliferation assay, plate colony formation, cell cycle, apoptosis and animal models. The inhibition of NF-κB signalling by COMMD10 was detected using dual-luciferase reporter assay and western blotting. Co-IP, GST pull-down and nuclear protein extraction assay were performed to evaluate the effect on p65 by COMMD10. Real-time PCR and western blotting were performed to detect expressions of FMNL2, COMMD10 and p65 in paired tissues.
Results:
FMNL2 targets COMMD10 for ubiquitin-mediated proteasome degradation in CRC cells. COMMD10 targets p65 NF-κB (nuclear factor-κB) subunit and reduces its nuclear translocation, thereby leading to the inactivation of NF-κB pathway and suppression of CRC invasion and metastasis. Inhibition of NF-κB signalling by COMMD10 is necessary for FMNL2-mediated CRC cell behaviours. Downregulation of COMMD10 predicts poor prognosis of CRC patients. The expressions of FMNL2, COMMD10 and p65 are highly linked in CRC tissues.
Conclusions:
These data demonstrate that the FMNL2/COMMD10/p65 axis acts as a critical regulator in the maintenance of metastatic phenotypes and is strongly associated with negative clinical outcomes.
Similar content being viewed by others
Main
Colorectal cancer (CRC) is one of the most common types of malignant tumours worldwide with about 1.2 million new cases diagnosed every year, of which around 600 000 ended in death (Siegel et al, 2016). Although there are many diagnostic strategies for CRC, tumour metastasis is the main reason for the high morbidity and poor survival of CRC patients (Brenner et al, 2014). So it is urged to better understand the mechanism of CRC metastasis for reducing the morbidity of clinical patients.
Formins are an actin nucleating protein family with diverse actin-regulating and potentially pro-invasive functions (Faix and Grosse, 2006). Formin-like2 (FMNL2), a member of formins, is dysregulated in several cancers such as CRC, melanoma and involved in invasive behaviours and progression of cancer cells (Zhu et al, 2008, 2011; Wang et al, 2015; Peladeau et al, 2016). FMNL2 drives amoeboid invasive cell motility downstream of RhoC (Kitzing et al, 2010). Our previous study showed that FMNL2 promotes motility, invasion and metastasis of CRC cells (Li et al, 2010; Zhu et al, 2011; Liang et al, 2013), but the underlying molecular mechanisms and interacting proteins are not well understood.
Copper metabolism MURR1 domain (COMMD) is a newly discovered protein family. They are characterised by the possession of a conserved and unique motif named the COMM domain, which functions as an interface for interactions between proteins (de Bie et al, 2006). COMMD family has been reported to regulate numerous biology processes such as the transportation of copper and sodium (van De Sluis et al, 2002), the transcription activity of NF-κB, cell proliferation and adaption to hypoxia (de Bie et al, 2006; Maine and Burstein, 2007a, 2007b). There are 10 members of the family, among which COMMD1 has been most extensively studied. COMMD1 inhibits human tumour cell invasion (van de Sluis et al, 2010). COMMD1 regulates inflammation and colitis-associated cancer progression (Li and Burstein, 2014). However, the role of COMMD10 in tumours remains unknown.
In this study, we identify that FMNL2 targets COMMD10 for ubiquitin-mediated proteasome degradation, which is required for FMNL2-induced invasion of metastasis of CRC. COMMD10 triggers less nuclear accumulation of p65, resulting in the termination of NF-kB pathway in CRC cells. In addition, we show high expression correlations of FMNL2 with COMMD10 and p65 in CRC tissues and the significance of COMMD10 as a valuable prognostic maker for CRC patients.
Materials and methods
Construction of plasmids and transfection
The N-terminal (NT) domain, the FH1 and FH2 regions and the mutant of FMNL2 were amplified respectively by PCR utilising the primers (Supplementary Table S1) and inserted into pCDNA3.1-FLAG vector (Invitrogen, Foster city, CA, USA). The complete sequence in open reading frame of COMMD10 was amplified by PCR using the primers (Supplementary Table S1) and subcloned into the pEGFP-C1 vector at the KpnI-XhoI site. The C-terminal (CT) and NT domains were amplified by PCR using the primers (Supplementary Table S1) and the fragments of COMMD10 digested with BamHI and XhoI were subcloned into the pEGX-6p-1 vector at the BamHI-XhoI site. The PCR conditions were as follows: 95 °C for 3 min, followed by 30 cycles of amplification (94 °C for 30 s, 55 °C for 40 s with outer primers or 68 °C for 40 s with inner primers, and 72 °C for 2 min). The correct coding regions of all plasmids were confirmed by sequencing. Transient transfection was performed using Lipofectamine2000 as a vehicle according to the manufacturer’s instructions (Invitrogen, Beijing, China). Lentiviral construct expressing or repressing COMMD10 was purchased from GeneCopoeia (Guangzhou, China) and was used to infect CRC cell lines SW480 and HT29. The eukaryotic expression vectors of pENTER-FLAG-RELA and pENTER-FLAG-RELB were purchased from Vigene Biosciences (Shandong, China).
Immunoblotting, immunofluorescence
Immunoblotting and immunofluorescence were performed as previously described (Moody et al, 2005). The minimum amount of cells used for analysis was 20 in immunofluorescence experiments. The primary antibodies used were as follows: 1 : 100 mouse anti-FMNL2 (Santa cruz, Guangzhou, China), 1 : 1 000 rabbit anti-COMMD10 and 1 : 200 rabbit anti-NF-κB p65 (Abcam, London, UK), 1 : 1000 rabbit anti-GST and 1 : 1000 rabbit anti-GAPDH and 1 : 1000 anti-mouse β-actin and 1 : 200 rabbit anti-Flag (Origene, Rockville, MD, USA), 1 : 200 goat anti-rabbit or anti-mouse Dylight 488 and 594 (Earthox, Beijing, China), 1 : 1000 DAPI (Sigma, Shanghai, China).
Co-immunoprecipitation (Co-IP)
Cell extracts were incubated 2 h at 4 °C with IgG and protein A/G Agarose to get rid of unspecific binding. FMNL2 (2 μg) was then added at 4 °C overnight. The protein A/G-agarose was collected by centrifugation. The beads were suspended in 2 × SDS sample buffer and heated to 100 °C for 10 min. Immunoprecipitated proteins were analysed by SDS–PAGE (12%, Minigel) followed by transfer to a polyvinylidene difluoride membrane at 100 V for 1.5 h. Membranes were blocked overnight. COMMD10 antibody (Abcam) was diluted respectively and incubated with membranes at 4 °C overnight. The secondary antibodies were then incubated for 1 h at room temperature. Finally, the blots were detected using the Odyssey Infrared Imaging System (LiCorBioSciences, Lincoln, NE, USA).
GST pull-down assay
GST-pEGX-6p-1-COMMD10 (1–132a) and (133–202a) were transformed into colibacillus BL21 (DE3), and induced for expression by IPTG. GST-COMMD10 (1–132a), GST-COMMD10 (133–202a) and GST proteins were purified by Glutathione Sepharose4B (GE Healthcare, Little Chalfont, UK) according to the manufacturer’s instructions. After purification, the GST fusion proteins were then incubated with the cell lysates (SW480 transfected with FLAG-FMNL2-CT) for 4 h at 4 °C with constant shaking. The beads were washed three times with cold PBS (pH 7.4) and then added with elution buffer (50 mM Tris-Cl, 10 mM reduced glutathione, pH 8.0) shaking at 4 °C for 30 min. The eluted proteins were subjected to western blot identification.
Luciferase activity assays
Cells were co-transfected with COMMD10 overexpressing vector (GeneCopoeia, Guangzhou, China) or COMMD10 shRNA (RIBOBIO, Guangzhou, China) vector and 100 ng per well REPONF-κB reporter plasmid (Genomeditech, Shanghai, China) in a six-well plate by using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol. Firefly and Renilla luciferase activities were measured consecutively by using Dual-Luciferase Reporter Assay System (Promega, Shanghai, China) 48 h post transfection. All assays were performed in triplicate, and all data shown are representative of at least three independent experiments.
Proliferation, plate colony formation, cell cycle, apoptosis, cell migration, cell invasion assays in vitro
The CCK8 (Cell Counting Kit-8) proliferation assay, plate colony formation, cell cycle, apoptosis, migration and invasion of transfected CRC cells were determined as previously described (Liang et al, 2013; Ren et al, 2016).
Subcutaneous tumour implantation and metastasis assay
All animal care and experiments were approved by the Institutional Animal Care and Use Committee of Nanfang hospital, Southern Medical University, Guangzhou. A number of 5 × 106 stable transfected cells were injected into the left and right flanks of nude mice for subcutaneous tumour implantation and allowed to grow for 28 days before mice were killed. Primary tumour size was measured every 7 days. For metastasis assay, a total of 1 × 106 stable transfected cells were injected into vein and mice were killed after 40 days. All tissues were embedded in paraffin, sectioned, stained with haematoxylin and eosin. Images were taken by Olympus DP72 upright microscope and were outputted by DP2-BSW software.
Statistical analysis
All statistical analyses were performed using SPSS 20.0 statistical software. Quantitative analyses of the experiments are expressed as the mean±s.d. Statistical significance was determined using independent samples T-test or one-way ANOVA. The degree of the linear relationship of gene expression levels was determined by Spearman’s correlation coefficient. P<0.05 was considered as statistically significant.
Results
FMNL2 targets COMMD10 for ubiquitin-mediated proteasome degradation in CRC cells
Our previous study revealed that FMNL2 was a positive regulator in CRC metastasis (Zhu et al, 2011). To elucidate the molecular mechanisms of FMNL2 involved in tumour metastasis, we performed yeast two-hybrid assay to obtain potential FMNL2-interacting proteins (Supplementary Figure S1A). Among seven FMNL2-interacting proteins initially identified, we focused on COMMD10, which is a member of COMMD protein family and its association with tumour has not been reported. We examined whether COMMD10 interacts with FMNL2 physically in CRC cells. Results of Co-IP and immunofluorescence analyses showed that FMNL2 bound to COMMD10 (Figure 1A) and co-localisation of FMNL2 and COMMD10 was observed in the cytoplasm of SW480 and HCT116 cells (Figure 1B). After that, we generated a series of constructs of FMNL2 and COMMD10. Results of GST pull-down showed that FH1 domain of FMNL2 interacted with NT domain of COMMD10 (Figure 1C and D). We also constructed FMNL2 mutant lacking FH1 domain and found that deletion of FH1 fragment of FMNL2 (FH1-mut) failed to interact with COMMD10 in CRC cells (Figure 1E and F). In addition, we detected whether the interaction between FMNL2 and COMMD10 affects protein stability in CRC cells. Results of western blot analyses showed that ectopic FMNL2 in SW480 and HT29 cells reduced the level of COMM10, while knockdown of FMNL2 in SW480 and SW620 cells led to increased expression of COMMD10 (Figure 1G). However, either overexpression or knockdown of COMMD10 did not affect the expression of FMNL2 in CRC cells (Figure 1H). Since FMNL2 decreased the protein level of COMMD10 in CRC cells and some peptides at COMMD10 protein which might be targeted for ubiquitination were predicted by the CPLM (Compendium of Protein Lysine Modifications) database (Supplementary Figure S1B), we raised the possibility that FMNL2 targets COMMD10 for ubiquitin-mediated proteasome degradation. We examined whether MG132, an inhibitor of proteasome, could prevent COMMD10 from degradation induced by FMNL2. Results of western blot showed that COMMD10 was downregulated by FMNL2, while MG132 could reverse the effect (Figure 1I), which was most obvious at 48 h after stimulation (Supplementary Figure S1C). Results of in vitro ubiquitination assay showed that overexpression of FMNL2 led to increased recovery of ubiquitinated COMMD10 (Figure 1J). Utilising a complementary approach, we further corroborated that the ubiquitinated material recovered was COMMD10 itself (Supplementary Figure S1D). Increased expression of FMNL2 accelerated COMMD10 ubiquitination, whereas depletion of FMNL2 reduced the amount of ubiquitinated COMMD10 (Supplementary Figure S1D and F). In addition, immunofluorescence analyses showed the co-localisation effect between FMNL2 and COMMD10 was greatly enhanced after MG132 stimulation (Figure 1K). These above data suggest that ubiquitination of COMMD10 is largely, if not exclusively, mediated by binding to FMNL2.
FMNL2 targets COMMD10 for ubiquitin degradation in CRC cells. (A) Lysates from SW480 and HCT116 cells were immunoprecipitated with anti-FMNL2 or anti COMMD10 and immunoblotted with the indicated antibodies. (B) Immunofluorescence microscopy analyses of co-localisation of FMNL2 and COMMD10 in SW480 and HCT116 cells. Scale bars represented 50 μm. (C) Lysates from SW480 cells expressing ectopic FMNL2 truncate (525–616a), FMNL2 (617–1092a) or FMNL2 (1–524a) were immunoprecipitated with anti-FLAG and immunoblotted with the indicated antibody. (D) Pull-down of FMNL2 by GST-tagged COMMD10 fragments with indicated boundaries. (E) Pull-down of FH1-wt or FH1-mut of FMNL2- by GST-tagged COMMD10 fragment (1–132a). (F) Schematics outlines of the binding sites of FMNL2 on COMMD10 and COMMD10 on FMNL2 as well. (G) Expressions of FMNL2 and COMMD10 in FMNL2 overexpression or depleting cells by western blotting and were normalised by β-actin expression. (H) Expressions of FMNL2 and COMMD10 in COMMD10 overexpression or depleting cells by western blotting and were normalised byβ-actin expression. (I) Expression of COMMD10 was detected in FMNL2 overexpression SW480 cells before and after MG132 stimulation by western blotting. (J) COMMMD10 was immunoprecipitated from SW480 cell lysates (cells were transfected with FMNL2 or vector control or shFMNL2 or scrambled control) and the recovered material was immunoblotted for ubiquitin (Ubi). (K) Immunofluorescence microscopy analyses of co-localisation of FMNL2 and COMMD10 after MG132 stimulation in HCT116 cells. Scale bars represented 50 μm. A full colour version of this figure is available at the British Journal of Cancer journal online.
COMMD10 inhibits the proliferation, invasion and metastasis of CRC cells
To determine the function of COMMD10 in the progression of CRC, we performed a series of gain-of-function and loss-of-function assays. According to endogenous COMMD10 expression in six CRC cells (Supplementary Figure S2A and B), we overexpressed COMMD10 in SW620 and SW480 cells and silenced COMMD10 in HT29 and SW480 cells. Results of real-time RT–PCR and western blot validated high transfection efficiency (Supplementary Figure S2C–F). CCK-8 assay showed that overexpression of COMMD10 caused a significant decrease of the growth rate in SW480 and SW620 cells, while COMMD10 depletion enhanced the proliferative abilities of SW480 and HT29 cells (Figure 2A, P<0.001). Similarly, overexpression of COMMD10 reduced the number of colony formation in SW480 cells (P<0.001), while the inhibition of COMMD10 had the adverse effects (Figure 2B, P<0.001). We also assessed the effect of COMMD10 on CRC cell cycle and apoptosis. Forced expression of COMMD10 in SW480 cells induced an evident increase in the percentage of cells in the G1 peak (P<0.001) and a decrease in the percentage of cells in the S and G2/M peaks (Supplementary Figure S3A, P<0.001; P<0.01). Contrarily, depletion of COMMD10 in SW480 cells led to the reverse effect (Supplementary Figure S3B, P=0.002; P=0.016; P=0.035). Ectopic COMMD10 in SW480 cells increased the rate of early apoptosis (Supplementary Figure S3C, P=0.009), while depletion of COMMD10 had the opposite effect (Supplementary Figure S3D, P=0.009). These results indicate that COMMD10 negatively regulates cell proliferation by arresting the tumour cells at the G1 phase and accelerating apoptosis of tumour cells. In addition, results of Boyden chamber assay showed that invasive abilities were decreased in COMMD10 expressing SW480 and SW620 cells, and contrarily, sharply increased invasive abilities were observed in COMMD10 depleting cells (Figure 2C, P<0.001).
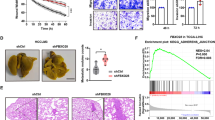
COMMD10 inhibits the proliferation, invasion and metastasis of CRC cells. (A) Effects of COMMD10 overexpression and knockdown on CRC cell proliferation in vitro. (B) Effect of COMMD10 overexpression and knockdownon on plate colony formation of CRC cells. *P<0.001. (C) Effects of COMMD10 overexpression and knockdown on CRC cell invasion in vitro by Boyden chamber. Scale bars represented 50 μm. *P<0.001. (D) Effect of COMMD10 knockdown on subcutaneous tumour growth in vivo. **P<0.001. (E) Effect of COMMD10 knockdown on lung metastasis in nude mice. Error bars represent mean±s.d. from three independent experiments. Scale bars represent 50 μm. *P=0.033. A full colour version of this figure is available at the British Journal of Cancer journal online.
After that, we carried out in vivo tumour growth and metastasis experiments and found that decreased expression of COMMD10 enhanced tumour growth in vivo (Figure 2D, P<0.001). Moreover, COMMD10 depleting cells developed more lung metastases than mock cells (Figure 2E, P=0.033). Collectively, these data demonstrate that COMMD10 profoundly suppresses proliferation, invasion and metastasis in CRC cells.
COMMD10 inhibits the proliferation and invasion of CRC cells by inactivating NF-κB pathway
NF-κB signalling pathway plays an important role in a variety of cancer and participates in tumour proliferation, survival and metastasis (Xia et al, 2014). COMMD1 was recently reported to be capable of strongly inhibiting NF-κB-mediated transcription (Zoubeidi et al, 2010; Bartuzi et al, 2013). Thus we tested whether COMMD10 is involved in the regulation of NF-κB signalling pathway in CRC cells. Results showed that a consistent reduction of luciferase activity of NF-κB was observed in COMMD10 expressing SW480 and SW620 cells (P=0.005; P=0.028), while TNF-α stimulation increased the luciferase activity in COMMD10 expressing cells (Figure 3A, P<0.001; P=0.030). On the contrary, depletion of COMMD10 induced an increase of luciferase activity in SW480 and HT29 cell lines (P<0.001; P<0.001), while transfection of IκBα-mut, an inhibitor of the NF-κB signalling pathway, eliminated the changes induced by COMMD10 knockdown (Figure 3B, P<0.001; P<0.001). Consistent with the results obtained from the luciferase activity assays, the levels of phosphorylated IKKα/β and IκBα as well as p65 in nucleus were obviously decreased, while the levels of IKKα/β and IκBα in total were greatly increased in COMMD10 expressing SW480 cells (Figure 3C). However, knockdown of COMMD10 showed the opposite results and it could be reversed by IκBα-mut (Figure 3D). We then investigated whether COMMD10 affects CRC cell behaviours through inhibiting NF-κB signalling pathway. Results of CCK8 assay showed that transfection of IκBα-mut obviously abolished the promotion of COMMD10 knockdown in cell proliferation (Figure 3E, P<0.001). IκBα-mut also decreased invasive abilities of CRC cells induced by COMMD10 knockdown (Figure 3F, P<0.01). The above results make it clear that COMMD10 inactivates NF-κB signalling to suppress CRC cell proliferation and invasion.
COMMD10 inhibits the proliferation and invasion of CRC cells by inactivating NF- κ B pathway. (A) Effects of COMMD10, TNF-α stimulation and the combinations of COMMD10 and TNF-α stimulation on the luciferase activity of NF-κB in SW480 and SW620 cells by luciferase reporter assay. (B) Effects of COMMD10 knockdown and co-transfection of shCOMMD10 and IκBα-mut on the luciferase activity of NF-κB in SW480 and HT29 cells by luciferase reporter assay. Error bars represent mean±s.d. from three independent experiments. **P<0.001; *P<0.05. (C) Western blot analyses of the levels of p-IKKα/β, p-IκBα and total IKKα/β, IκBα, as well as p65 in total, in the nucleus or cytoplasm in SW480 cells treated with TNF-α or the combination of TNF-α and COMMD10. (D) Western blot analyses of the levels of p-IKKα/β, p-IκBα and total IKKα/β, IκBα, as well as p65 in total, in the nucleus or cytoplasm in SW480 cells treated with shCOMMD10 or co-transfection of shCOMMD10 and IκBα-mut. (E) Effects of shCOMMD10 and co-transfection of shCOMMD10 and IκBα-mut on cell proliferation by CCK8 assay. (F) Effects of shCOMMD10 and co-transfection of shCOMMD10 and IκBα-mut on cell invasion by Boyden chamber assay. The level of COMMD10 downregulation was measured by western blot. Error bars represented mean±s.d. from three independent experiments. Scale bars represented 50 μm. *P<0.01. A full colour version of this figure is available at the British Journal of Cancer journal online.
COMMD10 reduces the nuclear translocation of NF-κB subunit p65 in CRC cells
Recent studies reveal that the ability of COMMD1 to inhibit NF-κB-mediated transcription depends on its association with the NF-κB complex (Xia et al, 2014). COMMD1 is able to interact with all five subunits of NF-κB, whereas other COMMD members interact with NF-κB subunits selectively (Maine and Burstein, 2007a; Zoubeidi et al, 2010). To understand how COMMD10 inhibits NF-κB signalling, we observed the interactions between COMMD10 with the NF-κB complex. Results of Co-IP showed that COMMD10 was capable of binding NF-κB complex, particularly NF-κB subunits p105, RelB and p65 (Figure 4A and B). We focused on p65 and RelB for the reason that among the three subunits detected in the co-immunoprecipitation assay, both of them had the transcriptional activation domain (TAD) which is important for the transcription of NF-κB target genes (Xia et al, 2014). Results of GST pull-down assays further validated that only p65 presented the direct interaction with COMMD10 (Figure 4C). After that, we examined whether COMMD10 regulates nuclear translocation of p65 in CRC cells. Results showed that ectopic COMMD10 decreased the level of p65 in total, nuclear and cytoplasm, and contrarily, depletion of COMMD10 resulted in greater accumulation of p65 in nuclear (Figure 4D). Similarly, an increase of nuclear p65 was observed in COMMD10 depleting SW620 cells by immunofluorescence (Figure 4E). Moreover, depletion of COMMD10 in SW480 cells resulted in prolonged nuclear p65 levels upon TNF-α stimulated NF-κB activation (Figure 4F), which coincided with increased and sustained expression of a specific group of NF-κB target genes such as CCL2, IL1A, CXCL2 and CXCL8 in COMMD10 depleting SW480 cells (Figure 4G). All the results demonstrate that COMMD10 suppresses NF-κB activity by reducing the nuclear translocation of p65 in CRC.
COMMD10 reduces the nuclear translocation of NF- κ B subunit p65 in CRC cells. (A) Endogenous COMMD10 was immunoprecipitated from SW480 cell lysates and immunoblotted for detection of NF-κB subunits NF-κB1/p105, NF-κB2/p100, RelB and p65. (B) Endogenous NF-κB subunits were immunoprecipitated from SW480 cell lysates and immunoblotted for detection of COMMD10. (C) Pull-down of COMMD10 by GST-tagged p65 full-length. (D) Effects of COMMD10 overexpression or knockdown on the nuclear translocation of p65 by western blotting. (E) Effects of COMMD10 knockdown on the nuclear translocation of p65 by immunofluorescence microscopy. Scale bars represented 50 μm. (F) Effect of COMMD10 knockdown on the accumulation of nuclear p65 after TNF treatment. Representative results of three independent experiments are shown. (G) Effect of COMMD10 knockdown on the mRNA level of a specific group of NF-κB-dependent genes by real-time PCR. Error bars represent mean±s.d. from three independent experiments. A full colour version of this figure is available at the British Journal of Cancer journal online.
Inhibition of NF-κB signalling by COMMD10 is necessary for FMNL2-mediated CRC cell behaviours
To ascertain whether FMNL2 could functionally target COMMD10 thereby regulating CRC invasion and metastasis, we performed the rescue experiments to evaluate the role of COMMD10 in FMNL2-mediated NF-κB activity and CRC cell behaviours. Results of western blot showed that overexpression of FMNL2 in SW480 cells increased the level of p-IKKα/β, p-IκBα and p65 in nucleus. However, these effects were compromised when COMMD10 was introduced in FMNL2-expressing cells (Figure 5A). On the contrary, knockdown of FMNL2 showed the opposite effects and it could be reversed by COMMD10 depletion (Figure 5B), indicating that FMNL2 activates NF-κB signalling through COMMD10. In vitro functional rescue results showed that constitutive COMMD10 expression obviously reversed the promotive effects of FMNL2 on cell proliferation and invasion (Figure 5C and D, P<0.001). Similarly, knockdown of COMMD10 rescued the proliferative and invasive phenotypes in FMNL2-depleting cells (Figure 5C and D, P<0.001). In vivo tumour growth and metastasis assays further revealed that depletion of FMNL2 significantly inhibited tumour growth and lung metastases(P=0.001, P=0.007), while these effects were significantly abolished when COMMD10 was knocked down in FMNL2-depleting cells (Figure 5E and F, P=0.001, P=0.033). Thus, these results indicate that COMMD10 is required for FMNL2-mediated CRC cell proliferation, invasion and metastasis.
Inhibition of NF- κ B signalling by COMMD10 is necessary for FMNL2-mediated CRC cell behaviours. (A) Effects of FMNL2 and FMNL2/COMMD10 on the levels of key proteins of NF-κB signalling pathway. (B) Effects of shFMNL2 and shFMNL2/shCOMMD10 on the levels of key proteins of NF-κB signalling pathway. (C) Effects of FMNL2, FMNL2/COMMD10, shFMNL2 and shFMNL2/shCOMMD10 on the proliferation of SW480 or SW620 cells in vitro by CCK-8 arrays. *P<0.001. (D) Effect of FMNL2, FMNL2/COMMD10, shFMNL2 and shFMNL2/shCOMMD10 on the invasion of SW480 or SW620 cells in vitro by Boyden chamber. Morphological comparison of cells penetrating the artificial basement membrane was also shown. Scale bars represented 50 μm. (E) Images of subcutaneous tumours of mice injected with SW620/NC or SW620/shFMNL2 or SW620/shFMNL2/shCOMMD10 stable transfected cells. The expressions of FMNL2, COMMD10 and p65 in the shFMNL2 tumours were measured by western blot. (F) Effects of shFMNL2 and shFMNL2/shCOMMD10 on lung metastasis in vivo. SW620/NC or SW620/shFMNL2 or SW620/shFMNL2/shCOMMD10 stable transfected cells were injected through tail vein. The number of lung metastasis nodules was counted. The tumour sections were stained with H&E. Error bars represent mean±s.d. Scale bars represented 50 μm. **P<0.01; *P<0.05. A full colour version of this figure is available at the British Journal of Cancer journal online.
Downregulation of COMMD10 predicts poor prognosis of CRC patients
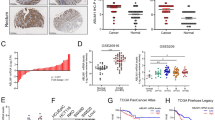
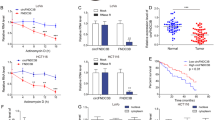
To assess the expression pattern and clinicopathological value of COMMD10 in CRC tissues, we detected the expression of COMMD10 in 120 cases of clinical paraffin-embedded CRC tissues by IHC. Positive signal of COMMD10 was located in the cytoplasm. Strong staining of COMMD10 was observed mainly in adjacent normal tissues, while negative or weak COMMD10 staining was observed in CRC tissues (Figure 6A). The expression of COMMD10 was significantly lower in CRC tissues or lymphatic metastatic tissues than that in paired normal tissues, respectively (Supplementary Table S2, P<0.001, χ2=−4.921; P<0.001, χ2=−6.335). There were no significant differences of COMMD10 expression between CRC tissues with lymph metastasis and lymphatic metastatic tissues or those without lymph metastasis (Supplementary Table S2, P>0.05; P=0.064, χ2=−1.853). Clinicopathologic analyses showed that COMMD10 expression was relevant with lymphatic metastasis and distant metastasis (P=0.013, P=0.014, Supplementary Table S3). By Kaplan–Meier curve assessment, CRC patients with low COMMD10 expression had a lower 5-year survival rate than those with high COMMD10 protein level (Figure 6B, P<0.01). From univariate analyses, the significant prognostic factors were COMMD10 expression (P=0.026), serosal invasion (P=0.024), lymph metastasis (P<0.001), Dukes’ stage (P<0.001), remote metastasis (Supplementary Table S4, P<0.001). Multivariate analyses results showed that remote metastasis might play an important role in predicting the overall survival in CRC patients (Supplementary Table S5, P<0.001). The above data clearly show that low COMMD10 expression level was accompanied by a worse clinical outcome, and that it could be an independent prognostic marker for survival of CRC patients.
FMNL2, COMMD10 and p65 expressions are highly linked in CRC tissues. (A) Expression of COMMD10 in normal mucosa, CRC tissues and metastatic carcinoma of lymph nodes. Also the representative pictures of CRC tissues with low and high COMMD10 levels. Scale bars represented 250 μm in (a, c, e, g, i, k) and 50 μm in (b, d, f, h, j, l). (B) Kaplan–Meier survival analysis of COMMD10 expression in CRC patients after surgical resection. P<0.01. (C) Expression of COMMD10 in the 31 paired human CRC tissues by real-time PCR. **P<0.01 (D) Western blotting analyses of FMNL2, COMMD10 and p65 expressions in the same 31 paired CRC tissues. N=normal mucosa and T=tumour. (E) Expressions of FMNL2, COMMD10 and p65 in the 31 paired human CRC tissues by real-time PCR. **P<0.01. (F) Spearman’s correlation analyses among relative mRNA expression levels of FMNL2, p65 and COMMD10 in CRC tissues. P<0.01. A full colour version of this figure is available at the British Journal of Cancer journal online.
The expressions of FMNL2, COMMD10 and p65 are highly linked in CRC tissues
To evaluate the expression correlation of FMNL2, COMMD10 and p65 in CRC tissues, we detected the expressions of FMNL2, COMMD10 and p65 in 31 paired samples of CRC patients. Results of real-time RT–PCR and western blotting showed that FMNL2 and p65 was upregulated in CRC tissues, while COMMD10 was downregulated in CRC tissues (Figure 6C and D, t=5.226, P<0.001). Moreover, the expressions of FMNL2 and p65 were negatively correlated with COMMD10, and FMNL2 expression was positively correlated with p65 (Figure 6E and F, P=0.0001, r=−0.6306; P=0.002, r=−0.5335; P<0.0001, r=0.7689). Together, these data provide further support for direct and functional associations among FMNL2, COMMD10 and NF-κB pathway in CRC tissues.
Discussion
Diaphanous-related formins (DRFs) are a highly conserved family of cytoskeletal remodelling proteins. A growing body of evidence suggests that DRFs play key roles in the progression and spread of a variety of cancers (Schwartzberg, 2007). FMNL2 is a novel member of DRFs and has been strongly implicated in driving tumourigenesis and invasion of specific tumours (Kitzing et al, 2010; Wang et al, 2015; Gardberg et al, 2016; Peladeau et al, 2016). We previously found that FMNL2 was upregulated in metastatic CRC tissue and a positive regulator in CRC cell motility and metastasis (Zhu et al, 2008, 2011; Li et al, 2010; Liang et al, 2013). However, the underlying mechanisms and interacting proteins of FMNL2 in the regulation of cancer metastasis need to be further investigated.
In our study, COMMD10 was identified as one of FMNL2 interacting proteins by yeast two-hybrid assay. Our presented data perfectly validated that N-terminal domain of COMMD10 specifically bound to FH1 domain of FMNL2. Also, the results displayed that the protein expression of COMMD10 was decreased by FMNL2 by the ubiquitin-mediated proteasome pathway. Since FMNL2 is not an ubiquitin ligase and cause accelerated ubiquitinaion of COMMD10 directly. There might be some other ubiquitin ligases involved in the ubiquitination of COMMD10 induced by FMNL2, which is worth further exploration. The COMMD family has been reported to be involved in several biological processes including the regulation of copper and sodium transport, NF-κB activity and cell cycle progression (Maine and Burstein, 2007b). Regarding the involvement of the COMMD family in tumour development, only COMMD1 and COMMD5 were described (Solban et al, 2000; van de Sluis et al, 2010; Zoubeidi et al, 2010; Gkiafi and Panayotou, 2011; Li and Burstein, 2014; Taskinen et al, 2014; Yeh et al, 2016), while little is known about COMMD10. Recent studies of the COMMD family have mostly focused on the prototype of this family, COMMD1, demonstrating its dysregulation and molecular mechanism in several human cancer types (van de Sluis et al, 2010; Li and Burstein, 2014; Taskinen et al, 2014). Here, our accumulating data including in vitro and in vivo functional experiments showed that COMMD10 inhibited CRC cell proliferation, invasion and metastasis.
Having established COMMD10 as a suppressor in the progression of CRC, we investigated possible pathways by which COMMD10 could be involved in CRC. The COMMD family has been identified as a new group of proteins involved in NF-κB termination (Maine and Burstein, 2007a). All 10 COMMD members seem to play distinct and non-redundant roles in various physiological processes, including NF-κB signalling (Maine and Burstein, 2007a). NF-κB signalling plays a critical role in many biological processes including cell survival, inflammation and oncogenesis (Karin and Lin, 2002). NF-κB-mediated transcription is mainly regulated by the cytoplasmic sequestration of NF-κB dimeric complexs bound to the inhibitory IκB proteins (Baldwin, 1996). Thus, we examined whether COMMD10 regulates NF-κB signalling and found that COMMD10 inactivated NF-κB signalling pathway in CRC cells. Moreover, COMMD10 inhibited CRC cell proliferation and invasion by terminating NF-κB signalling.
In recent years more insight into the mechanisms of COMMD family inhibiting NF-κB activity has been obtained. COMMD1 controls the ubiquitination of NF-κB subunits, an event linked to transcriptional termination (Maine and Burstein, 2007a). In contrast with COMMD1, COMMD6 does not bind to IκBα (de Bie et al, 2006), indicating that different COMMD proteins inhibit NF-κB in an overlapping, but not completely similar, manner. Then we investigated that how COMMD10 mediates NF-κB activity in CRC cells. We found that COMMD10 inhibited NF-κB by targeting NF-κB subunit p65 and reducing its nuclear translocation in CRC. However, the total p65 was decreased after overexpressing COMMD10, suggesting that COMMD10 might accelerate the ubiquitination of p65 as COMMD1 does. It needs to be further investigated in our research. Moreover, we chose some NF-κB target genes that involved in immunoregulatory, inflammatory, vascular cell adhesion and cell proliferation processes (Song et al, 2012) and found that CXCL2, CXCL8, IL1A and CCL2 were increased upon COMMD10 silencing, while VCAM and MAPK1 were decreased in COMMD10 depleting cells. CXCL2 and CXCL8 are involved in the outcomes of metastatic CRC patients due to the oxaliplatin (OXA)-acquired resistance through NF-κB signalling pathway (Ruiz et al, 2016). IL1A and CCL2 participate in immune responses and inflammatory processes. CCL2 is a pro-inflammatory chemokine with known roles in metastasis (Volpato et al, 2016). IL1A has pro-inflammatory effect through NF-κB signalling pathway (Papacleovoulou et al, 2011). Upregulations of CXCL2, CXCL8, IL1A and CCL2 after depletion of COMM10 further demonstrate that COMMD10 can suppress the NF-κB signalling pathway. Interesting, VCAM and MAPK1 were decreased in COMMD10 depleting cells in CRC cells. VCAM-1 is stimulated by TNF-α and accelerated monocyte adhesion in a NF-κB-dependent manner (Li et al, 2017). MAPK1 is involved in a wide variety of cellular processes, such as proliferation, differentiation, transcription regulation and development. Some studies also suggest that MAPK1 acts as a transcriptional repressor independent of its kinase activity (Hu et al, 2009). These above data provide new mechanistic insight of COMMD10 into the regulation of NF-κB activity. After that, we examined the involvement of COMMD10 in FMNL2-mediated cell behaviours in CRC. Our data proved that FMNL2 activated NF-κB signalling through COMMD10 and enhanced NF-κB-mediated cellular responses in CRC.
Finally, we detected the expression correlations of FMNL2, COMMD10 and p65 in CRC cells. High FMNL2 level is linked with invasive behaviours of CRC and melanoma cells and predicts worse outcome of patients (Zhu et al, 2008; Peladeau et al, 2016). However, FMNL2 is downregulated in hepatocellular cancer (Liang et al, 2011). The COMMD family seems to be a tumour suppressor and has been shown to inhibit both NF-kB and HIF-mediated gene expression (de Bie et al, 2006). COMMD1 is downregulated and a potential novel prognostic factor in diffuse large B-cell lymphoma (Taskinen et al, 2014). COMMD5 level is decreased in many cancerous cell lines (Solban et al, 2000). Our data showed that COMMD10 was apparently downregulated in CRC tissues and low level of COMMD10 was relevant to CRC patients’ worse overall survival. Moreover, the expressions of FMNL2, COMMD10 and p65 were highly linked in CRC tissues.
In summary, our study reveals that FMNL2 targets COMMD10 for ubiquitin degradation and subsequently activates NF-κB signalling pathway by increasing the nuclear accumulation of p65 in CRC cells. COMMD10 is necessary for FMNL2-induced proliferation, invasion and metastasis of CRC. Downregulation of COMMD10 is associated with the survival and clinical stage of CRC patients. Our study provides a new mechanism to understand the development of CRC and that will be a challenge for future studies and a potential, novel therapeutic target for clinical CRC patients.
Change history
10 October 2017
This paper was modified 12 months after initial publication to switch to Creative Commons licence terms, as noted at publication
References
Baldwin AJ (1996) The NF-kappa B and I kappa B proteins: new discoveries and insights. Annu Rev Immunol 14: 649–681.
Bartuzi P, Hofker MH, van de Sluis B (2013) Tuning NF-κB activity: a touch of COMMD proteins. Biochim Biophys Acta (BBA) 1832: 2315–2321.
Brenner H, Kloor M, Pox CP (2014) Colorectal cancer. Lancet 383: 1490–1502.
de Bie P, van de Sluis B, Burstein E, Duran KJ, Berger R, Duckett CS, Wijmenga C, Klomp LW (2006) Characterization of COMMD protein-protein interactions in NF-kappaB signalling. Biochem J 398: 63–71.
Faix J, Grosse R (2006) Staying in shape with formins. Dev Cell 10: 693–706.
Gardberg M, Heuser VD, Koskivuo I, Koivisto M, Carpen O (2016) FMNL2/FMNL3 formins are linked with oncogenic pathways and predict melanoma outcome. J Pathol Clin Res 2: 41–52.
Gkiafi Z, Panayotou G (2011) Comparative proteomic analysis implicates COMMD proteins as Epstein-Barr virus targets in the BL41 Burkitt’s lymphoma cell line. J Proteome Res 10: 2959–2968.
Hu S, Xie Z, Onishi A, Yu X, Jiang L, Lin J, Rho HS, Woodard C, Wang H, Jeong JS, Long S, He X, Wade H, Blackshaw S, Qian J, Zhu H (2009) Profiling the human protein-DNA interactome reveals ERK2 as a transcriptional repressor of interferon signaling. Cell 139: 610–622.
Karin M, Lin A (2002) NF-kappaB at the crossroads of life and death. Nat Immunol 3: 221–227.
Kitzing TM, Wang Y, Pertz O, Copeland JW, Grosse R (2010) Formin-like 2 drives amoeboid invasive cell motility downstream of RhoC. Oncogene 29: 2441–2448.
Li H, Burstein E (2014) COMMD1 regulates inflammation and colitis-associated cancer progression. OncoImmunology 3: e947891.
Li S, Ning H, Ye Y, Wei W, Guo R, Song Q, Liu L, Liu Y, Na L, Niu Y, Chu X, Feng R, Moustaid-Moussa N, Li Y, Sun C (2017) Increasing extracellular Ca2+ sensitizes TNF-alpha-induced vascular cell adhesion molecule-1 (VCAM-1) via a TRPC1/ERK1/2/NFkappaB-dependent pathway in human vascular endothelial cells. Biochim Biophys Acta 1864 (10): 1566–1577.
Li Y, Zhu X, Zeng Y, Wang J, Zhang X, Ding YQ, Liang L (2010) FMNL2 enhances invasion of colorectal carcinoma by inducing epithelial-mesenchymal transition. Mol Cancer Res 8: 1579–1590.
Liang L, Guan J, Zeng Y, Wang J, Li X, Zhang X, Ding Y (2011) Down-regulation of formin-like 2 predicts poor prognosis in hepatocellular carcinoma. Hum Pathol 42: 1603–1612.
Liang L, Li X, Zhang X, Lv Z, He G, Zhao W, Ren X, Li Y, Bian X, Liao W, Liu W, Yang G, Ding Y (2013) MicroRNA-137, an HMGA1 target, suppresses colorectal cancer cell invasion and metastasis in mice by directly targeting FMNL2. Gastroenterology 144: 624–635.e4.
Maine GN, Burstein E (2007a) COMMD proteins and the control of the NF kappa B pathway. Cell Cycle 6: 672–676.
Maine GN, Burstein E (2007b) COMMD proteins: COMMing to the scene. Cell Mol Life Sci 64: 1997–2005.
Moody SE, Perez D, Pan TC, Sarkisian CJ, Portocarrero CP, Sterner CJ, Notorfrancesco KL, Cardiff RD, Chodosh LA (2005) The transcriptional repressor Snail promotes mammary tumor recurrence. Cancer Cell 8: 197–209.
Papacleovoulou G, Critchley HO, Hillier SG, Mason JI (2011) IL1alpha and IL4 signalling in human ovarian surface epithelial cells. J Endocrinol 211: 273–283.
Peladeau C, Heibein A, Maltez MT, Copeland SJ, Copeland JW (2016) A specific FMNL2 isoform is up-regulated in invasive cells. BMC Cell Biology 17: 32.
Ren XL, He GY, Li XM, Men H, Yi LZ, Lu GF, Xin SN, Wu PX, Li YL, Liao WT, Ding YQ, Liang L (2016) MicroRNA-206 functions as a tumor suppressor in colorectal cancer by targeting FMNL2. J Cancer Res Clin 142: 581–592.
Ruiz DPV, Bystrup S, Martinez-Cardus A, Pluvinet R, Sumoy L, Howells L, James MI, Iwuji C, Manzano JL, Layos L, Buges C, Abad A, Martinez-Balibrea E (2016) Curcumin mediates oxaliplatin-acquired resistance reversion in colorectal cancer cell lines through modulation of CXC-Chemokine/NF-kappaB signalling pathway. Sci Rep 6: 24675.
Schwartzberg PL (2007) Formin the way. Immunity 26: 139–141.
Siegel RL, Miller KD, Jemal A (2016) Cancer statistics, 2016. Cancer J Clin 66: 7–30.
Solban N, Jia HP, Richard S, Tremblay S, Devlin AM, Peng J, Gossard F, Guo DF, Morel G, Hamet P, Lewanczuk R, Tremblay J (2000) HCaRG, a novel calcium-regulated gene coding for a nuclear protein, is potentially involved in the regulation of cell proliferation. J Biol Chem 275: 32234–32243.
Song L, Gong H, Lin C, Wang C, Liu L, Wu J, Li M, Li J (2012) Flotillin-1 promotes tumor necrosis factor-α receptor signaling and activation of NF-κB in esophageal squamous cell carcinoma cells. Gastroenterology 143: 995–1005.e12.
Taskinen M, Louhimo R, Koivula S, Chen P, Rantanen V, Holte H, Delabie J, Karjalainen-Lindsberg ML, Bjorkholm M, Fluge O, Pedersen LM, Fjorden K, Jerkeman M, Eriksson M, Hautaniemi S, Leppa S (2014) Deregulation of COMMD1 is associated with poor prognosis in diffuse large B-cell lymphoma. PLoS One 9: e91031.
van de Sluis B, Mao X, Zhai Y, Groot AJ, Vermeulen JF, van der Wall E, van Diest PJ, Hofker MH, Wijmenga C, Klomp LW, Cho KR, Fearon ER, Vooijs M, Burstein E (2010) COMMD1 disrupts HIF-1alpha/beta dimerization and inhibits human tumor cell invasion. J Clin Invest 120: 2119–2130.
van De Sluis B, Rothuizen J, Pearson PL, van Oost BA, Wijmenga C (2002) Identification of a new copper metabolism gene by positional cloning in a purebred dog population. Hum Mol Genet 11: 165–173.
Volpato M, Perry SL, Marston G, Ingram N, Cockbain AJ, Burghel H, Mann J, Lowes D, Wilson E, Droop A, Randerson-Moor J, Coletta PL, Hull MA (2016) Changes in plasma chemokine C-C motif ligand 2 levels during treatment with eicosapentaenoic acid predict outcome in patients undergoing surgery for colorectal cancer liver metastasis. Oncotarget 7: 28139–28150.
Wang Y, Arjonen A, Pouwels J, Ta H, Pausch P, Bange G, Engel U, Pan X, Fackler OT, Ivaska J, Grosse R (2015) Formin-like 2 promotes beta1-integrin trafficking and invasive motility downstream of PKCalpha. Dev Cell 34: 475–483.
Xia Y, Shen S, Verma IM (2014) NF-kappaB, an active player in human cancers. Cancer Immunol Res 2: 823–830.
Yeh DW, Chen YS, Lai CY, Liu YL, Lu CH, Lo JF, Chen L, Hsu LC, Luo Y, Xiang R, Chuang TH (2016) Downregulation of COMMD1 by miR-205 promotes a positive feedback loop for amplifying inflammatory- and stemness-associated properties of cancer cells. Cell Death Differ 23: 841–852.
Zhu XL, Liang L, Ding YQ (2008) Overexpression of FMNL2 is closely related to metastasis of colorectal cancer. Int J Colorectal Dis 23: 1041–1047.
Zhu XL, Zeng YF, Guan J, Li YF, Deng YJ, Bian XW, Ding YQ, Liang L (2011) FMNL2 is a positive regulator of cell motility and metastasis in colorectal carcinoma. J Pathol 224: 377–388.
Zoubeidi A, Ettinger S, Beraldi E, Hadaschik B, Zardan A, Klomp LW, Nelson CC, Rennie PS, Gleave ME (2010) Clusterin facilitates COMMD1 and I-kappaB degradation to enhance NF-kappaB activity in prostate cancer cells. Mol Cancer Res 8: 119–130.
Acknowledgements
We thank Professor Reddy for editing the English writing. This work was supported by the National Key R&D Program of China (2017YFC1309002), National Basic Research Program of China (973 Program, 2015CB554002), Key project of National Natural Science Fund (Guangdong Province NSFC-joint fund, U1201226), National Natural Science Foundation of China (81672821, 81272759); National Natural Youth Science Foundation of China (81302151).
Author contributions
We thank master Yang SS, Li XM and Yang M carried out experiments. Dr Hu JL took on the statistical analysis. Professor Liao WT and Ding YQ gave assistance in collecting tissue samples. Yang SS, Li XM and Yang M contributed equally to this work. Professor Liang L and Guan J conceived experiments and analysed the data. All authors were involved in writing the paper and had final approval of the submitted and published versions.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License.
Supplementary Information accompanies this paper on British Journal of Cancer website
Rights and permissions
From twelve months after its original publication, this work is licensed under the Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/4.0/
About this article
Cite this article
Yang, S., Li, X., Yang, M. et al. FMNL2 destabilises COMMD10 to activate NF-κB pathway in invasion and metastasis of colorectal cancer. Br J Cancer 117, 1164–1175 (2017). https://doi.org/10.1038/bjc.2017.260
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/bjc.2017.260
Keywords
This article is cited by
-
LINC00839 promotes malignancy of liver cancer via binding FMNL2 under hypoxia
Scientific Reports (2022)
-
FMNL2 suppresses cell migration and invasion of breast cancer: a reduction of cytoplasmic p27 via RhoA/LIMK/Cofilin pathway
Cell Death Discovery (2022)
-
Mesenchymal Stem Cell-derived Extracellular Vesicles Transmitting MicroRNA-34a-5p Suppress Tumorigenesis of Colorectal Cancer Through c-MYC/DNMT3a/PTEN Axis
Molecular Neurobiology (2022)
-
Transcriptional analysis of the expression, prognostic value and immune infiltration activities of the COMMD protein family in hepatocellular carcinoma
BMC Cancer (2021)
-
Expression profile and bioinformatics analysis of COMMD10 in BALB/C mice and human
Cancer Gene Therapy (2020)