Abstract
Background:
Identifying informative prognostic biomarkers for oral tongue squamous cell carcinoma (OTSCC) is of great importance in order to better predict tumour behaviour and to guide treatment planning. Here, we summarise existing evidence regarding immunohistochemical prognostic biomarkers for OTSCC.
Methods:
A systematic search of the literature was performed using the databases of Scopus, Ovid Medline, Web of Science and Cochrane Library. All studies which had investigated the prognostic significance of immunohistochemical biomarkers in OTSCC during the period from 1985 to 2015 were retrieved. For the five most often evaluated biomarkers a random-effects meta-analysis on overall survival was performed, including those studies that provided the necessary statistical results.
Results:
A total of 174 studies conducted during the last three decades were found, and in these 184 biomarkers were evaluated for the prognostication of OTSCC. The five biomarkers most frequently assessed were p53, Ki-67, p16, VEGFs and cyclin D1. In the meta-analyses, the most promising results of the prognostic power for OTSCC were obtained for cyclin D1. For studies of VEGF A and C the results were equivocal, but the pooled analysis of VEGF A separately showed it to be a useful prognosticator for OTSCC. There was no sufficient evidence to support p53, Ki-67 and p16 as prognostic biomarkers for OTSCC. Limitations in the quality of the published studies (e.g., small cohorts, lack of compliance with REMARK guidelines) are widespread.
Conclusions:
Numerous biomarkers have been presented as useful prognosticators for OTSCC, but the quality of the conduct and reporting of original studies is overall unsatisfactory which does not allow reliable conclusions. The value of two biomarkers (VEGF-A and cyclin D1) should be validated in a multicentre study setting following REMARK guidelines.
Similar content being viewed by others
Main
Oral tongue squamous cell carcinoma (OTSCC) is the most common malignancy of the oral cavity. OTSCC is increasing in incidence (Patel et al, 2011; Ng et al, 2016), and has an aggressive clinical behaviour with a relatively poor prognosis (Bello et al, 2010a). The 5-year relative survival rate was 63% in a recent report from the Netherlands (van Dijk et al, 2016). During 2017, almost 16 400 new cases of tongue cancer, and 2400 deaths are projected to occur due to this cancer in the United States (Siegel et al, 2017).
Predicting the outcome of OTSCC patients is important when planning treatment. In early stages (cT1–T2) of OTSCC, which are expected to have favourable prognosis, the cancer-related mortality affects about 19% of patients (Almangush et al, 2015). This indicates the need for better prognostic tools. Identification of robust prognostic biomarkers that could accurately predict the behaviour of OTSCC will aid in the selection of appropriate treatment strategies.
Recent research in the field of molecular pathology has introduced thousands of tumour biomarkers, which are associated with the progression and/or prognosis of different cancers. Many of these biomarkers have been evaluated for their prognostic power in OTSCC. However, the use of any molecular biomarker for OTSCC in daily practice has not yet been approved, although several biomarkers have been presented as promising prognosticators that could provide added value upon the classical ones such as stage, tumour grade and depth of invasion.
The task of the current systematic review of literature was to retrieve original studies that have examined the prognostic value of immunohistochemical biomarkers of OTSCC and to meta-analyse the studies of the most repeatedly reported biomarkers. We also summarise the current understanding of the topic and highlight the main shortcomings of the published studies to improve future research in this field.
Materials and methods
Search strategy
A search strategy combining the terms (tongue) AND (cancer* OR squamous cell carcinoma* OR neoplas* OR tumo*) AND (prognos* OR predict* OR surviv* OR recur* OR mortal* OR metasta*) AND (immunohisto* OR protei* OR marke* OR biomark*) was developed. The search terms were entered into Scopus, Ovid Medline, Web of Science and Cochrane Library (1985–2015).
In advanced search, the following search fields were included: title, abstract, subject heading, and keyword. The Preferred Reporting Items for Systematic Review and Meta-Analysis (PRISMA) were utilised (Moher et al, 2009).
Screening
Two independent researchers (AA & IH) examined the retrieved hits, and discarded duplicated ones. We also excluded unrelated studies through careful browsing of the title and/or abstract of each publication. Existing review articles for prognostic biomarkers of tongue cancer (Ferrari et al, 2009; Bello et al, 2010a, 2010b) were screened for papers missed in the search strategy.
Data extraction
For relevant articles, we retrieved information about the name of the first author, country, year of publication, number of patients and the immunohistochemical biomarker/s examined. For those biomarkers which were reported repeatedly, further data including the primary antibody used and its dilution, unadjusted and adjusted analyses, statistical results reported (estimated hazard ratio (HR), 95% confidence interval (CI) and P-value) were retrieved when available.
Exclusion criteria
-
1
Studies in languages other than English.
-
2
Data based on animal samples.
-
3
Studies on cancers other than SCC, or on rare histological variants of SCC.
-
4
Studies including samples from other subsites of the oral cavity, and studies that mixed samples from the oral tongue and the base of the tongue.
-
5
Studies that did not report the prognostic value of the biomarker (i.e., studies that only reported association between the biomarker and classical parameters, such as stage and grade, but did not provide results for the association between the biomarker and survival outcomes).
Quality assessment
The guidelines from Reporting Recommendations for Tumor Marker Prognostic Studies (REMARK) (McShane et al, 2005; Altman et al, 2012b) were used to evaluate the quality of studies that were eligible for the meta-analyses of the five most often reported biomarkers. The selected guidelines taken from the REMARK criteria are summarised in Table 1.
Statistical analysis
A meta-analysis on overall survival (OS) was performed for each of the five selected biomarkers, including only those original studies which provided an estimate of the HR and the associated 95% CI for the contrast of interest. As most studies provided only unadjusted or ‘univariate’ estimates of HR, the meta-analysis was primarily based on them. However, in case only an estimated HR was reported which was adjusted for various other prognostic factors using multiple Cox regression, such a ‘multivariate’ estimate was entered to the analysis. Depending on the biomarker the direction of HR contrast was either positive vs negative (high vs low; applied for p53, Ki-67, VEGF and cyclin D1) or negative vs positive (low vs high; applied for p16). Where an individual study reported the HR estimate in the opposite direction, the inverse HR and CI were calculated to obtain results concordant with other studies. HR and CIs were transformed into log(HR) and its standard error (s.e.). Pooled estimates of the HR were computed both by a fixed-effect and by a random-effects model based on the generic inverse variance approach (Schwartzer et al, 2015). In the random-effects analysis the between-studies variance was obtained by the Sidik–Jonkmann method (Sidik and Jonkmann, 2005). The I2 statistic and tau-squared, the estimated heterogeneity variance, were used to measure the heterogeneity of HRs between studies. Analyses were performed and forest plots created using the functions ‘metagen’ and ‘forest’ contained in package ‘meta’ (Schwarzer, 2007) of the R environment (R Core Team, 2016).
Results
Search results
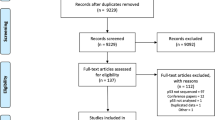
Our search retrieved 1817 hits, and 771 of these were relevant for our study (Figure 1). A total of 174 studies remained that evaluated a total of 184 biomarkers for OTSCC. Of these, 32 biomarkers (17.4%) were reported at least in three studies (Figure 2). Of the latter, five biomarkers including p53, Ki-67, p16, vascular endothelial growth factors (VEGFs) and cyclin D1 were reported more often than any other (Tables 2, 3, 4, 5, 6; Supplementary Tables 1–5).
A clear majority (86%) of the studies reported at least one biomarker as a promising prognosticator for OTSCC based on obtaining a ‘statistically significant’ result for an association of that biomarker with at least one outcome variable considered. Most of the studies on these five biomarkers were based on quite small cohorts (<100 patients), and in the analyses of the outcome they commonly mixed early and late stage cancers (Tables 2, 3, 4, 5, 6; Supplementary Tables 1–5). It was also common to exclude a biomarker from an adjusted analysis using Cox regression, when the biomarker turned out to be statistically ‘non-significant’ in an unadjusted analysis, but biomarkers ‘significant’ in an unadjusted analysis were often included in an adjusted analysis. Many authors, though, presented the results based on unadjusted analysis only.
Most of the publications of those frequently studied biomarkers did not fulfil completely REMARK guidelines (Altman et al, 2012b). We used the selected criteria mentioned in Table 1 to evaluate the most often studied biomarkers, on which estimated HRs and their 95% CI were reported for at least one survival endpoint in a number of studies, and more than one study analysed the OS to be included in our meta-analyses (Tables 2, 3, 4, 5, 6). We particularly noted that guideline no. 5 (related to statistics) was not fulfilled in several studies (e.g., adjusted analysis using multiple Cox regression was not conducted), and guideline no. 1 (related to the patient series) was sometimes ignored by the authors (e.g., medical treatment applied to the patients was not well-explained). In addition, guideline no. 6 (related to classical prognostic factors) was not fulfilled properly in many studies (e.g., the relationship between biomarker/s and classical prognostic factors were not reported) (Tables 2, 3, 4, 5, 6).
Results of meta-analyses
The results of the meta-analyses on OS for the five most often studied biomarkers, each being evaluated in at least two studies that reported necessary statistical data, are summarised in Figures 3 and 4. The number of eligible studies was very small ranging from two for cyclin D1 to six for p16, the remaining biomarkers having three studies each. Essential heterogeneity in the HRs across the individual studies was observed for Ki-67, VEGFs and p16, whereas the pertinent measures (I2 and tau-squared) had very low values for p53 and cyclin D1. The point estimates and the error margins of the pooled HR from the fixed effect model and from the random effects model were very similar for the two last ones, whereas for the three first ones the 95% CI of the pooled HR was substantially wider when based on the random effects model as expected. Thus, it is reasonable to focus on the results of the random effects model. On the basis of the available data there was not sufficient evidence for p53, Ki-67 and p16 to be informative prognostic biomarkers. The two available studies on cyclin D1 suggest that this biomarker could be a useful prognosticator worth further evaluation. For cyclin D1 the pooled HR estimate was 2.86 (95% CI from 1.34 to 6.08). As regards VEGFs, the results were mixed with a very wide CI for HR from the random effects model. However, two of the VEGFs studies analysed VEGF-A and one VEGF-C. A positive or high VEGF-C value was shown to be associated with an improved prognosis (Morita et al, 2014), which was in sharp contrast with the two other studies analysing the expression of VEGF-A, both indicating a much worse survival for a positive VEGF-A value. When the VEGF-C study was excluded from the meta-analysis, the pooled estimate for HR was 7.34 (95% CI from 2.32 to 23.22), which provides rather strong evidence for VEGF-A being a useful prognostic biomarker.
Discussion
Molecular biomarkers may highlight biological differences between cancers and help to prognosticate patient outcome. During the last three decades (1985–2015), more than one hundred molecular biomarkers (identified by immunohistochemistry) were introduced as prognosticators for OTSCC. Five biomarkers including p53, Ki-67, p16, VEGFs and cyclin D1 were most often reported, and their biology has been reviewed elsewhere (Oliveira and Ribeiro-Silva, 2011; Wang et al, 2013). According to the findings of the present meta-analysis, the prognostic usefulness of VEGF-A and cyclin D1 is worth further evaluation. On the other hand, for p53, Ki-67 and p16 there appears to be no sufficient evidence for any prognostic value for OTSCC.
The evaluation of molecular biomarkers in different subsites of the oral cavity is common in literature. However, variations in the immunohistochemical staining results reflect variations in proteomic (and genomic) properties of SCC between different oral subsites. For example, various immunohistological biomarkers analysed in OTSCC and buccal carcinoma samples did not associate with survival in OTSCC, whereas some of them were prognostic in buccal carcinoma (Sathyan et al, 2006; Trivedi et al, 2011). The histological structures and the carcinogenesis are different in buccal mucosa and in oral tongue. Similarly, the base portion and the oral (mobile) portion of the tongue have differences in the etiopathogenesis of the cancer. In the base of the tongue, HPV is commonly linked with the cancer, whereas the virus is rarely founded in mobile tongue. However, still several studies combine oral and base of the tongue SCC samples, or they do not specify for which part of the tongue the analyses were done (Supplementary Tables 1, 2, 4 and 5). To avoid the effect of tumour heterogeneity, we included in the present meta-analyses studies in which the authors defined their cohort as OTSCC. Our meta-analysis and conclusions are based on those studies which included previously untreated, surgically resected, primary OTSCC.
Almost all of the most reported biomarkers (except VEGFs, which contribute to tumour angiogenesis) reflect important growth-related properties of the cancer cells. However, non-neoplastic cells of the tumour stroma, including fibroblasts, endothelial cells and inflammatory cells, seem also to have a critical role in cancer progression (Marsh et al, 2011). Accordingly, biomarkers of the stromal microenvironment might even have a greater impact on prognosis than biomarkers related to tumour cells (Marsh et al, 2011). However, biomarkers related to tumour microenvironment such as Activin A (Kelner et al, 2015) are not yet widely studied in OTSCC. The only tumour microenvironment biomarker that was reported repeatedly in OTSCC, is the cancer-associated fibroblast identified by a α-smooth muscle actin antibody (Supplementary Table 6). Further studies should also focus on the evaluation of promising biomarkers of tumour microenvironment, such as fibronectin and tenascin-C (Sundquist et al, 2017), in addition to biomarkers related to cancer cells.
Notably, 152 biomarkers (83%) have been studied only once or twice, so it is not possible to reach trustworthy conclusions on the basis of such limited evidence. About 86% of the studies claimed to have found at least one biomarker to have prognostic value. Negative or ‘non-significant’ prognostic finding as the only result was not widely published (about 14%). This might well indicate a fair amount of publication bias as noted by Soland and Brusevold (2013). However, the articles which reported new promising biomarkers have also usually evaluated the prognostic significance of previously known biomarkers (e.g., Ki-67, p53, and p16). This approach helps to validate previously published data and allows the accumulation of evidence for known biomarkers. Relationships between biomarkers and clinicopathologic manifestations, other than survival, have also been studied in several publications (Albert et al, 2012; Wang et al, 2012). However, even though it is important to understand such relationships, clinically the most relevant information is provided by proper survival analysis of the tested biomarker.
REMARK guidelines (Altman et al, 2012b) have suggested items to be reported in prognostic studies of tumour markers. Tables 2, 3, 4, 5, 6 show that many studies on p53, Ki-67, p16, VEGFs and cyclin D1 did not follow REMARK criteria accurately. This indicates shortcomings in the reporting of the biomarkers tested in these studies, and subsequently limits the possibilities to reach definitive conclusions of their usefulness.
To date, none of the most often reported biomarkers can be recommended as prognosticators valid for clinical use. This may be related to the mixing of early and late stages of OTSCC in the same analysis, the small sizes cohorts or numbers of events seen in many studies. In addition, antibodies supplied by different manufacturers, variable staining conditions, and different cut-off values were also seen. All of these factors might affect the biomarker results published.
Even though it is highly desirable to apply multiple Cox regression or similar predictive modelling (‘multivariate’ analysis) to adjust for important classical prognostic factors (like TNM stage and histologic grade) and relevant patient characteristics (like age), limited cohort size does not allow conducting a regression analysis of good predictive performance, and might affect the validity of the estimation results (Ogundimu et al, 2016). Moreover, numerous studies which included small cohorts (and small numbers of events) have reported regression analyses with many variables, and such studies may run the risk of overfitting. A rule of thumb of 10 outcome events for each prognostic term involved in multiple Cox regression is widely advocated (Peduzzi et al, 1995; Ogundimu et al, 2016), but in many instances at least 20 events per term would be needed for reliable modelling (Ogundimu et al, 2016). It seems that no consideration has been devoted to this issue in most of the studies applying Cox regression. Another common statistical shortcoming was to report the P value only and to lean on it when making judgments of the prognostic value of a biomarker depending on whether the result was ‘significant’ (when P<0.05) or ‘non-significant’ (when P⩾0.05). Such statistical malpractice meant that many studies were not eligible for inclusion in the current meta-analyses; and may have caused false inferences in some studies, thus it should be avoided in future studies. More useful results can be obtained by reporting the estimated HR, preferably an adjusted one, and its CI, as is recommended by REMARK guidelines (Altman et al, 2012a). When predicting mortality, it was quite common to report OS but not cancer-specific mortality. Analysis by cause of death would, however, provide more relevant prognostic information than that of OS only (Läärä et al, 2016). For better statistical analysis, guidelines for improving statistical reporting (Greenland et al, 2016) should be followed, the authors should involve a statistician, and journals should assign an experienced statistician as one of their reviewers.
Recently, the digital evaluation of biomarkers has been suggested to facilitate the evaluation and to avoid inter- and intra-observer variability (Bouzin et al, 2016). So far only a few studies have used this technique in the prognostication of OTSCC (Hannen et al, 2001, 2002), and none of the studies in the meta-analysis had applied digital scorings. In the future, image analysis will most likely be more commonly applied also for OTSCC scorings.
Regarding the global distribution of the studies, the Japanese population was widely represented in studies of p53, Ki-67, VEGFs and cyclin D1 (Table 7). However, the studies included in our meta-analyses (Figures 3 and 4) were conducted in a different population (marked with an asterisk in Table 7).
The primary treatment was surgical OTSCC resection in all of the publications included in the meta-analyses. However, one of the p16 studies included cases with either surgical excision or external beam radiotherapy for patients unwilling/unfit for surgery (Ramshankar et al, 2014). None of the studies in the meta-analyses mentioned any preoperative therapy, which could have led to molecular changes in the resected tumour tissue. Neck dissection, radiotherapy and/or chemotherapy are used for selected cases of OTSCC, especially those in advanced stages. Such variations in treatment modalities must have an impact on the prognosis, as well as on the prognostic analyses. Unfortunately, therefore a limitation in our meta-analyses is that the cases included were not homogenous with regard to neck dissection or other adjuvant therapies. The results of our meta-analyses must, in any event, be interpreted with due caution because of various shortcomings in the published studies and their reporting. First, for each biomarker very few original studies were available that provided the necessary statistical data. Second, essential heterogeneity was evident within and across individual studies with respect to the spectrum of patients, staining methods, and so on. Third, the numbers of patients and outcome events were mostly small implying poor statistical precision. Fourth, only crude HR estimates, unadjusted for classical prognostic factors were typically reported, leaving room for an unknown amount of bias in the assessment of the prognostic power of the pertinent marker due to uncontrolled confounding.
Conclusion
The identification of biomarkers may increase the possibility of detecting cancers with high risk for poor prognosis. Here, for the first time, we systematically reviewed the literature for immunohistochemical prognostic biomarkers for OTSCC. On the basis of our meta-analysis VEGF-A and cyclin D1 may be useful prognostic biomarkers for OTSCC. Therefore, these biomarkers, in addition to the new biomarker candidates, should be further evaluated carefully following the REMARK criteria using large, preferably multicentre, cohorts of OTSCC cases separated into early and late stages.
Change history
05 September 2017
This paper was modified 12 months after initial publication to switch to Creative Commons licence terms, as noted at publication
References
Adduri R Sr, Kotapalli V, Gupta NA, Gowrishankar S, Srinivasulu M, Ali MM, Rao S, Uppin SG, Nayak UK, Dhagam S, Chigurupati MV, Bashyam MD (2014) P53 nuclear stabilization is associated with FHIT loss and younger age of onset in squamous cell carcinoma of oral tongue. BMC Clin Pathol 14: 37.
Albert S, Hourseau M, Halimi C, Serova M, Descatoire V, Barry B, Couvelard A, Riveiro ME, Tijeras-Raballand A, de Gramont A, Raymond E, Faivre S (2012) Prognostic value of the chemokine receptor CXCR4 and epithelial-to-mesenchymal transition in patients with squamous cell carcinoma of the mobile tongue. Oral Oncol 48 (12): 1263–1271.
Almangush A, Bello IO, Coletta RD, Makitie AA, Makinen LK, Kauppila JH, Pukkila M, Hagstrom J, Laranne J, Soini Y, Kosma VM, Koivunen P, Kelner N, Kowalski LP, Grenman R, Leivo I, Läärä E, Salo T (2015) For early-stage oral tongue cancer, depth of invasion and worst pattern of invasion are the strongest pathological predictors for locoregional recurrence and mortality. Virchows Arch 467 (1): 39–46.
Altman DG, McShane LM, Sauerbrei W, Taube SE (2012a) Reporting recommendations for tumor marker prognostic studies (REMARK): explanation and elaboration. BMC Med 10: 51.
Altman DG, McShane LM, Sauerbrei W, Taube SE (2012b) Reporting Recommendations for Tumor Marker Prognostic Studies (REMARK): explanation and elaboration. PLoS Med 9 (5): e1001216.
Bello IO, Soini Y, Salo T (2010a) Prognostic evaluation of oral tongue cancer: means, markers and perspectives (I). Oral Oncol 46 (9): 630–635.
Bello IO, Soini Y, Salo T (2010b) Prognostic evaluation of oral tongue cancer: means, markers and perspectives (II). Oral Oncol 46 (9): 636–643.
Bello IO, Vered M, Dayan D, Dobriyan A, Yahalom R, Alanen K, Nieminen P, Kantola S, Läärä E, Salo T (2011) Cancer-associated fibroblasts, a parameter of the tumor microenvironment, overcomes carcinoma-associated parameters in the prognosis of patients with mobile tongue cancer. Oral Oncol 47 (1): 33–38.
Bockelman C, Hagström J, Mäkinen LK, Keski-Santti H, Häyry V, Lundin J, Atula T, Ristimäki A, Haglund C (2011) High CIP2A immunoreactivity is an independent prognostic indicator in early-stage tongue cancer. Br J Cancer 104 (12): 1890–1895.
Bouzin C, Saini ML, Khaing KK, Ambroise J, Marbaix E, Gregoire V, Bol V (2016) Digital pathology: elementary, rapid and reliable automated image analysis. Histopathology 68 (6): 888–896.
Bova RJ, Quinn DI, Nankervis JS, Cole IE, Sheridan BF, Jensen MJ, Morgan GJ, Hughes CJ, Sutherland RL (1999) Cyclin D1 and p16INK4A expression predict reduced survival in carcinoma of the anterior tongue. Clin Cancer Res 5 (10): 2810–2819.
Ferrari D, Codeca C, Fiore J, Moneghini L, Bosari S, Foa P (2009) Biomolecular markers in cancer of the tongue. J Oncol 2009: 412908.
Goto M, Tsukamoto T, Inada K, Mizoshita T, Ogawa T, Terada A, Hyodo I, Shimozato K, Hasegawa Y, Tatematsu M (2005) Loss of p21WAF1/CIP1 expression in invasive fronts of oral tongue squamous cell carcinomas is correlated with tumor progression and poor prognosis. Oncol Rep 14 (4): 837–846.
Greenland S, Senn SJ, Rothman KJ, Carlin JB, Poole C, Goodman SN, Altman DG (2016) Statistical tests, P values, confidence intervals, and power: a guide to misinterpretations. Eur J Epidemiol 31 (4): 337–350.
Hannen EJ, van der Laak JA, Kerstens HM, Cuijpers VM, Hanselaar AG, Manni JJ, de Wilde PC (2001) Quantification of tumour vascularity in squamous cell carcinoma of the tongue using CARD amplification, a systematic sampling technique, and true colour image analysis. Anal Cell Pathol 22 (4): 183–192.
Hannen EJ, van der Laak JA, Manni JJ, Freihofer HP, Slootweg PJ, Koole R, de Wilde PC (2002) Computer assisted analysis of the microvasculature in metastasized and nonmetastasized squamous cell carcinomas of the tongue. Head Neck 24 (7): 643–650.
Harris SL, Thorne LB, Seaman WT, Hayes DN, Couch ME, Kimple RJ (2011) Association of p16(INK4a) overexpression with improved outcomes in young patients with squamous cell cancers of the oral tongue. Head Neck 33 (11): 1622–1627.
Hogmo A, Kuylenstierna R, Lindholm J, Nathansson A, Auer G, Munck-Wikland E (1998) Nuclear DNA content and p53 overexpression in stage I squamous cell carcinoma of the tongue compared with advanced tongue carcinomas. Mol Pathol 51 (5): 268–272.
Hwa JS, Kwon OJ, Park JJ, Woo SH, Kim JP, Ko GH, Seo JH, Kim RB (2015) The prognostic value of immunohistochemical markers for oral tongue squamous cell carcinoma. Eur Arch Otorhinolaryngol 272 (10): 2953–2959.
Imayama N, Yamada S, Yanamoto S, Naruse T, Matsushita Y, Takahashi H, Seki S, Fujita S, Ikeda T, Umeda M (2015) FOXC2 expression is associated with tumor proliferation and invasion potential in oral tongue squamous cell carcinoma. Pathol Oncol Res 21 (3): 783–791.
Kelner N, Rodrigues PC, Bufalino A, Fonseca FP, Santos-Silva AR, Miguel MC, Pinto CA, Leme AF, Graner E, Salo T, Kowalski LP, Coletta RD (2015) Activin A immunoexpression as predictor of occult lymph node metastasis and overall survival in oral tongue squamous cell carcinoma. Head Neck 37 (4): 479–486.
Läärä E, Korpi JT, Pitkänen H, Alho OP, Kantola S (2016) Competing risks analysis of cause-specific mortality in patients with oral squamous cell carcinoma. Head Neck 39 (1): 56–62.
Lim AM, Do H, Young RJ, Wong SQ, Angel C, Collins M, Takano EA, Corry J, Wiesenfeld D, Kleid S, Sigston E, Lyons B, Fox SB, Rischin D, Dobrovic A, Solomon B (2014) Differential mechanisms of CDKN2A (p16) alteration in oral tongue squamous cell carcinomas and correlation with patient outcome. Int J Cancer 135 (4): 887–895.
Marsh D, Suchak K, Moutasim KA, Vallath S, Hopper C, Jerjes W, Upile T, Kalavrezos N, Violette SM, Weinreb PH, Chester KA, Chana JS, Marshall JF, Hart IR, Hackshaw AK, Piper K, Thomas GJ (2011) Stromal features are predictive of disease mortality in oral cancer patients. J Pathol 223 (4): 470–481.
McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM Statistics Subcommittee of the NCIEWGoCD (2005) REporting recommendations for tumour MARKer prognostic studies (REMARK). Br J Cancer 93 (4): 387–391.
Mineta H, Miura K, Ogino T, Takebayashi S, Misawa K, Ueda Y (2002) Vascular endothelial growth factor (VEGF) expression correlates with p53 and ki-67 expressions in tongue squamous cell carcinoma. Anticancer Res 22 (2B): 1039–1044.
Moher D, Liberati A, Tetzlaff J, Altman DG Group P (2009) Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Br Med J 339: b2535.
Morita Y, Morita N, Hata K, Nakanishi M, Kimoto N, Omata T, Nakamura Y, Yoneda T (2014) Cyclooxygenase-2 expression is associated with vascular endothelial growth factor-c and lymph node metastasis in human oral tongue cancer. Oral Surg Oral Med Oral Pathol Oral Radiol 117 (4): 502–510.
Ng JH, Iyer NG, Tan MH, Edgren G (2016) Changing epidemiology of oral squamous cell carcinoma of the tongue: a global study. Head Neck 39 (2): 297–304.
Ogundimu EO, Altman DG, Collins GS (2016) Adequate sample size for developing prediction models is not simply related to events per variable. J Clin Epidemiol 76: 175–182.
Oliveira LR, Ribeiro-Silva A (2011) Prognostic significance of immunohistochemical biomarkers in oral squamous cell carcinoma. Int J Oral Maxillofac Surg 40 (3): 298–307.
Patel SC, Carpenter WR, Tyree S, Couch ME, Weissler M, Hackman T, Hayes DN, Shores C, Chera BS (2011) Increasing incidence of oral tongue squamous cell carcinoma in young white women, age 18 to 44 years. J Clin Oncol 29 (11): 1488–1494.
Peduzzi P, Concato J, Feinstein AR, Holford TR (1995) Importance of events per independent variable in proportional hazards regression analysis. II. Accuracy and precision of regression estimates. J Clin Epidemiol 48 (12): 1503–1510.
Petera J, Sirak I, Laco J, Kasaova L, Tucek L, Dolezalova H (2015) High-dose-rate brachytherapy in early oral cancer with close or positive margins. Brachytherapy 14 (1): 77–83.
R Core Team (2016) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing: Vienna, Austria, Available at https://www.R-project.org/.
Ramshankar V, Soundara VT, Shyamsundar V, Ramani P, Krishnamurthy A (2014) Risk stratification of early stage oral tongue cancers based on HPV status and p16 immunoexpression. Asian Pac J Cancer Prev 15 (19): 8351–8359.
Sakata K, Someya M, Nagakura H, Nakata K, Oouchi A, Takagi M, Hareyama M (2008) Brachytherapy for oral tongue cancer: an analysis of treatment results with various biological markers. Jpn J Clin Oncol 38 (6): 402–407.
Sathyan KM, Sailasree R, Jayasurya R, Lakshminarayanan K, Abraham T, Nalinakumari KR, Abraham EK, Kannan S (2006) Carcinoma of tongue and the buccal mucosa represent different biological subentities of the oral carcinoma. J Cancer Res Clin Oncol 132 (9): 601–609.
Schwartzer G, Carpenter JR, Rücker G (2015) Meta-Analysis with R. Springer: New York.
Schwarzer G (2007) Meta: an R package for meta-analysis. R News 7 (3): 40–45.
Shao Z, Zhang WF, Chen XM, Shang ZJ (2008) Expression of EphA2 and VEGF in squamous cell carcinoma of the tongue: correlation with the angiogenesis and clinical outcome. Oral Oncol 44 (12): 1110–1117.
Sidik K, Jonkmann JF (2005) Simple heterogeneity variance estimation for meta-analysis. J R Stat Soc 54 (2): 367–384.
Siegel RL, Miller KD, Jemal A (2017) Cancer statistics, 2017. CA Cancer J Clin 67 (1): 7–30.
Soland TM, Brusevold IJ (2013) Prognostic molecular markers in cancer—quo vadis? Histopathology 63 (3): 297–308.
Sundquist E, Kauppila JH, Veijola J, Mroueh R, Lehenkari P, Laitinen S, Risteli J, Soini Y, Kosma VM, Sawazaki-Calone I, Macedo CC, Bloigu R, Coletta RD, Salo T (2017) Tenascin-C and fibronectin expression divide early stage tongue cancer into low- and high-risk groups. Br J Cancer 116 (5): 640–648.
Trivedi TI, Tankshali RA, Goswami JV, Shukla SN, Shah PM, Shah NG (2011) Identification of site-specific prognostic biomarkers in patients with oral squamous cell carcinoma. Neoplasma 58 (3): 217–226.
van Dijk BA, Brands MT, Geurts SM, Merkx MA, Roodenburg JL (2016) Trends in oral cavity cancer incidence, mortality, survival and treatment in the Netherlands. Int J Cancer 139 (3): 574–583.
Wang A, Liu X, Sheng S, Ye H, Peng T, Shi F, Crowe DL, Zhou X (2009) Dysregulation of heat shock protein 27 expression in oral tongue squamous cell carcinoma. BMC Cancer 9: 167.
Wang C, Liu X, Huang H, Ma H, Cai W, Hou J, Huang L, Dai Y, Yu T, Zhou X (2012) Deregulation of Snai2 is associated with metastasis and poor prognosis in tongue squamous cell carcinoma. Int J Cancer 130 (10): 2249–2258.
Wang H, Sun R, Lin H, Hu WH (2013) P16INK4A as a surrogate biomarker for human papillomavirus-associated oropharyngeal carcinoma: consideration of some aspects. Cancer Sci 104 (12): 1553–1559.
Acknowledgements
This article was supported by grants of Finnish Dental Society (Alhadi Almangush and Ilkka Heikkinen), the Helsinki University Hospital Research Fund (Antti Mäkitie and Tuula Salo), the Maritza and Reino Salonen Foundation (Ilmo Leivo), the Sigrid Juselius Foundation and the Finnish Cancer Society (Tuula Salo).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License.
Supplementary Information accompanies this paper on British Journal of Cancer website
Rights and permissions
From twelve months after its original publication, this work is licensed under the Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/4.0/
About this article
Cite this article
Almangush, A., Heikkinen, I., Mäkitie, A. et al. Prognostic biomarkers for oral tongue squamous cell carcinoma: a systematic review and meta-analysis. Br J Cancer 117, 856–866 (2017). https://doi.org/10.1038/bjc.2017.244
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/bjc.2017.244
Keywords
This article is cited by
-
A preliminary study of IgG4 expression and its prognostic significance in oral squamous cell carcinoma
BMC Cancer (2024)
-
The Significance of Systemic Inflammatory Markers in Prognosis of Head and Neck Squamous Cell Cancers
Indian Journal of Otolaryngology and Head & Neck Surgery (2024)
-
Identify and validate circadian regulators as potential prognostic markers and immune infiltrates in head and neck squamous cell carcinoma
Scientific Reports (2023)
-
Relationship Between Worst Pattern of Invasion and Extranodal Extension in Oral Tongue Squamous Cell Carcinomas
Head and Neck Pathology (2023)
-
CT radiomics nomogram for prediction of the Ki-67 index in head and neck squamous cell carcinoma
European Radiology (2022)