Abstract
Background:
DNA methylation at the 5 position of cytosine (5mC) can be converted to 5-hydroxymethylcytosine (5hmC) by the ten–eleven translocation family. The loss of global levels of 5hmC has been regarded as a hallmark in various cancers. 5-hydroxymethylcytosine is distributed at protein-coding gene bodies and promoters; however, the role and distribution of 5hmC at long non-coding RNAs (lncRNAs) is not clear. We investigated the distribution and regulatory roles of 5hmC for lncRNAs in colorectal cancer (CRC).
Methods:
We integrated genome-wide profiles of 5hmC, 5mC, transcriptome and histone marks in CRC patients and examined the 5hmC-based clinical outcomes in patients.
Results:
5-hydroxymethylcytosine was distributed at lncRNA loci and positively correlated with lncRNA transcription. Dysreulated CRC lncRNAs were regulated by 5hmC directly or through abnormal activities of typical and super-enhancers and promoters modified by 5hmC. In addition, 5hmC was involved in long-range chromatin interactions at lncRNA loci. Finally, lncRNAs regulated by differential 5hmC marks were correlated with different clinical outcomes and tumour status in patients.
Conclusions:
5-hydroxymethylcytosine is critical in regulating the transcription of lncRNA and serve as novel biomarkers for clinical prognosis in CRC.
Similar content being viewed by others
Main
Colorectal cancer (CRC) is a major health problem associated with high morbidity and mortality worldwide (Jemal et al, 2011). Several critical genetic changes and pathways driving cancer initiation and progression have been uncovered, such as WNT, RAS, PI3K, TGF-B, p53 and mismatch repair proteins (Fearon, 2011; Cancer Genome Atlas Network, 2012). Epigenetic abnormalities have also been explored and proved to be essential in the pathogenesis of cancer. For example, aberrant DNA methylation at the 5 position of cytosine (5mC) occurs extensively in many types of cancer, exhibiting global hypomethylation and local hypermethylation at specific gene promoter CpG islands (Baylin and Jones, 2011; Dawson and Kouzarides, 2012). Such epigenetic changes were observed in CRC as well (Fearon, 2011; Vaiopoulos et al, 2014).
DNA methylation is dynamically regulated by DNA methyltransferases and the ten–eleven translocation (TET) family of 5mC hydroxylases. Ten–eleven translocation family proteins, including TET1, TET2 and TET3, can remove methylation in DNA and mediate the conversion of oxidised 5mC to 5-hydroxymethylcytosine (5hmC) in an α-ketoglutarate- and Fe(II)-dependent manner (Tahiliani et al, 2009; Ito et al, 2010). Further oxidation of 5hmC generates 5-formylcytosine and 5-carboxylcytosine (He et al, 2011; Ito et al, 2011). Notably, 5hmC is most abundant in self-renewing cells, pluripotent stem cells and adult neural cells (Kriaucionis and Heintz, 2009; Szwagierczak et al, 2010). Recent genome-wide studies in embryonic stem cells and developing T cells have revealed that 5hmC was enriched in protein-coding gene bodies, promoters and cell-type-specific enhancers (Pastor et al, 2011; Stroud et al, 2011; Tsagaratou et al, 2014; Kim et al, 2014a; Ichiyama et al, 2015). Cancers, however, exhibited strongly reduced 5hmC levels relative to the surrounding normal tissue (Jin et al, 2011). Several independent researchers have observed a reduction in global levels of 5hmC in CRC (Haffner et al, 2011; Li and Liu, 2011; Kudo et al, 2012; Zhang et al, 2013; Uribe-Lewis et al, 2015). However, the distribution of 5hmC at long non-coding RNAs (lncRNAs) remains to be elucidated.
Long non-coding RNA is longer than 200 nt and has little or no open reading frame. On the basis of the proximity between neighbouring transcripts, lncRNAs can be classified to five broad categories: sense, antisense, bidirectional, intronic and intergenic (Rinn and Chang, 2012). Accumulating evidence suggests that lncRNAs are involved in diverse physiological and pathological processes, such as cell growth, apoptosis, stem cell pluripotency, development and cancer biology (Guttman and Rinn, 2012; Ulitsky and Bartel, 2013; Hu et al, 2014). Epigenetic controls, such as DNA methylation (Yan et al, 2015) and histone modifications (Guttman et al, 2009; Prensner et al, 2011; Trimarchi et al, 2014; White et al, 2014), are essential for lncRNA transcription in cancers. Furthermore, the activities of enhancer- or promoter-affected transcription of some intergenic lncRNAs originate from these cis-elements (Marques et al, 2013).
To investigate the role of 5hmC in the transcriptional control of lncRNAs, we integrated genome-wide profiles of 5hmC, 5mC, transcriptome and several histone marks from patients with matched primary tumour and normal colon tissues. We showed that 5hmC was distributed at lncRNAs and positively correlated with lncRNA transcription. Dysreulated CRC lncRNAs were regulated by 5hmC directly or through abnormal activities of typical and super-enhancers and promoters modified by 5hmC. In addition, we found that 5hmC was involved in long-range chromatin interactions at lncRNA loci. Finally, lncRNAs regulated by differential 5hmC marks were correlated with different clinical outcomes and tumour status in patients. In conclusion, our findings revealed crucial roles for 5hmC in driving dysregulated transcriptional programmes of lncRNAs and contributed to the identification of novel biomarkers for prognosis evaluation in patients with CRC.
Materials and methods
Access to published data
Publicly available hmeDIP-seq and matched input data files for CRC were obtained from GSE47590. Methylcap-seq data sets for CRC were obtained from GSE39068. H3K4me1, H3K4me3, H3K27ac and matched input ChIP-seq data sets for CRC were obtained from GSE36204. RNA-seq data sets for CRC were obtained from GSE50760. Pol II ChIP-seq data sets for HCT116 cells were obtained from GSE60106 and GSE47677. GRO-seq data sets for HCT116 cells were obtained from GSE38140. ChIA-PET data for HCT116 cells were obtained from GSE33664.
hmeDIP-seq and methylcap-seq data processing
Each of the raw hmeDIP-seq data sets for four matched tumour and normal tissue samples and respective input data, as well as methylcap-seq data for eight matched tumour and normal tissue samples, were aligned to UCSC human genome build hg19 using Bowtie (Langmead et al, 2009). Duplicate reads were removed with picard-tools. Regions found to be enriched relative to input DNA were detected using the ‘findPeaks’ routine in HOMER (Heinz et al, 2010) with the ‘histone’ mode and default parameter values.
ChIP-seq data processing
Each of the raw H3K4me1, H3K4me3 and H3K27ac ChIP-seq data sets for tumour and normal tissue and input data was aligned to hg19 using Bowtie. Pol II ChIP-seq data sets for HCT116 cell were also aligned to hg19 using Bowtie. Enriched peaks or regions were identified by MACS (Zhang et al, 2008) and annotated by HOMER.
GRO-seq data processing
Three replicates of GRO-seq raw data for HCT116 colon cancer cells were aligned to hg19 using Bowtie. De novo transcript identification was performed using HOMER with the ‘groseq’ mode and default parameter values, which detected regions of continual GRO-Seq read coverage. For genome browse visualisation, the ‘makeUCSCfile’ command in HOMER was used to make a Bedgraph on each strand. The negative strand values were assigned negative values in the BedGraph file. The BedGraph values were then divided by the number of millions of mapped (309 772 394) reads to give values of reads per million per bp (RPM/bp) for each position in the genome. The two files (strands) were concatenated back together, and igvtools was used to create a TDF file for visualisation using the IGV browser (Robinson et al, 2011).
RNA-sequencing data analysis
All RNA-seq data were mapped to human reference genome hg19 using TopHat (Trapnell et al, 2009) with default parameters. The UCSC mRNA and lncRNA annotation files were merged into a single GTF file as a reference gene annotation file. The alignment BAM files were sorted and converted into SAM files with SAMtools, then subjected to read counting using the python package HTSeq (Anders et al, 2015). The read counts were converted into RPKM (Reads Per Kilobase of exon model per Million mapped reads).
Differentially epigenetic mark enrichment analysis
The alignment SAM files were converted into BAM files and sorted and indexed with SAMtools. Then the sorted BAM files were converted to BED files using BEDTools (Quinlan and Hall, 2010). diffReps (Shen et al, 2013) was used to quantitatively compare 5hmC, 5mC and H3K4me1 peaks in normal and tumour tissue using BED files. Peaks with FDR <0.05 and fold change >2 were considered as statistically significant.
Bidirectional enhancer RNA (eRNA) detection and correlation with gene transcription
For bidirectional eRNA analysis, we first searched the intergenic transcripts from GRO-seq. Paired eRNA peaks from opposite DNA strands were searched for, such that the plus strand peak centre had a greater genomic coordinate than the minus strand peak centre, and the distance between the 5′ ends of the peak pair was <1 kb. Then the two start sites of a plus transcript and a minus transcript were paired together, and their midpoint was defined as the centre of a bidirectional transcript.
Lists of genes in 5hmC- and non-5hmC-marked eRNA-associated genes in tumour were obtained by downloading the region-gene association file for each class from GREAT. Duplicate gene names in two region-gene association files were discarded, then merged to the normalised expression data. Boxplots of log10-transformed GRO-seq counts and RPKM RNA-seq values for genes were then generated in R. Statistical significance between groups was assessed by the Wilcoxon rank-sum test.
Patients and tissue samples
To validate the expression of lncRNAs, we recruited six pairs of colorectal cancer tissue and corresponding non-tumour tissue samples, all of which were obtained from patients who underwent surgical operations at Hospital No. 161 in 2014. All the participants were histologically confirmed to have colorectal cancer and did not receive any other therapy before enrolment. Informed consent was obtained from all the participants, and procedures used in this study were approved by the institutional review board at Hospital No. 161.
RNA extraction and qRT-PCR
All samples were immediately frozen with liquid nitrogen after surgical resection. Total RNA was isolated using Trizol Reagent (Invitrogen, Carlsbad, CA, USA). qPCR was performed using an iQ5 Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA) using SYBR Green Real-time PCR Master Mix (Toyobo Co., Ltd, Osaka, Japan) for the readout. All reactions were carried out in triplicate. Data were analysed using the 2−ΔΔCT method.
Results
Genome-wide distribution of 5hmC in CRC lncRNAs
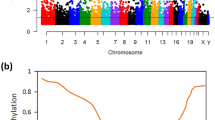
To explore the role of 5hmC in colon carcinogenesis, we analysed the hmeDIP-seq data from CRC patients (Uribe-Lewis et al, 2015). We also included methylcap-seq data sets that measure genome-wide 5mC for matched colon tumour and normal tissue (Simmer et al, 2012), as a direct comparison. In total, we identified 130 265 and 153 380 5hmC peaks, and 380 614 and 380 638 5mC peaks in tumour and normal colon. We first investigated the 5hmC distribution across long non-coding RNAs (lncRNAs) by plotting 5hmC and 5mC reads against UCSC lncRNAs across the whole gene body and 3 kb flanking regions distal to the TSS and TES. We showed that 5hmC was abundant at exons and depleted at the lncRNA TSSs but enriched at proximal sites downstream of TSSs and TESs in both tumour and normal colon (Figure 1A and B). The results showed a similar pattern for 5mC compared with 5hmC, except that the enrichment of 5mC at TSSs was observed in tumour (Figure 1A and B). The total number of peaks and read-density plot demonstrated a loss of 5hmC in tumour, which was consistent with the previously reported reduction of global levels of 5hmC in CRC (Haffner et al, 2011; Kudo et al, 2012). In contrast, 5mC modification levels in cancer and normal tissue seemed comparable, although a slight increase of 5mC in tumour was observed.
Comparison of genome-wide 5hmC and 5mC distribution in CRC lncRNAs.(A) Heat map representations of 5hmC and 5mC enrichment in UCSC lncRNAs across the whole gene body and 3 kb flanking region away from the TSS and TES. (B) The normalised tag density profiles for 5hmC and 5mC across lncRNA gene body ±3 kb flanking regions with 200 bp resolution are shown. (C) The distribution of 5hmC and 5mC modifications was analysed on the basis of location: promoter (within 1 kb upstream from the transcription start site), exon, intron, intergenic, 3′UTR, 5′UTR, DNA, ncRNA and CpG-Island. (D) Distributions of 5hmC and 5mC between lncRNAs and protein-coding genes.
Notably, we found sets of lncRNAs showed different patterns for 5hmC and 5mC. These lncRNAs formed distinct clusters through the enrichment of 5hmC, enrichment of 5mC or binary marks of both 5hmC and 5mC (Figure 1A), indicating that different potentially active DNA demethylation processes occurred in these clusters of lncRNAs. In addition, we analysed the distribution of all the 5hmC and 5mC peaks in the human genome in nine kinds of regions: promoter, exon, intron, intergenic, 3′UTR, 5′UTR, DNA, ncRNA and CpG-Island. In addition to the enrichment at promoters and exons, the vast majority of 5hmC and 5mC peaks are distributed at intron and intergenic regions (Figure 1C). We also compared the distributions of 5hmC and 5mC between lncRNAs and protein-coding genes. As is shown in Figure 1D, about 18% of 5hmC peaks and 23% of 5mC peaks enriched at lncRNA loci, with no significant difference between tumour and normal tissue.
The majority of CpG dinucleotides are methylated, whereas unmethylated CpGs are found primarily in those regions of DNA with a relatively high density of CpG (CpG islands) in mammals. We therefore explored the distribution of 5hmC and 5mC among CpG islands. Our results showed that 5hmC was depleted at the centre of CGIs in both tumour and normal colon, whereas 5mC was enriched at the centre of CGIs but depleted at proximal upstream and downstream regions with respect to the centre in tumours (Supplementary Figure S1).
Abnormal 5hmC associated with colon cancer lncRNA signatures
Although a global reduction was observed, locus-specific changes in 5hmC between tumour and normal colon remain unclear. Therefore, we set out to identify the differential modification of 5hmC regions in tumour relative to normal colon. In total, we found 1111 gain (up-5hmC) and 7687 loss (down-5hmC) sites (FDR <0.05 and fold change >2) in tumour (Figure 2A), reflecting the loss of 5hmC signal in CRC observed previously. For 5mC, we identified 4990 gain (up-5mC) and 3143 loss (down-5mC) sites (FDR <0.05 and fold change >2; Figure 2A), suggesting that changes in DNA methylation were mild compared with 5hmC. Functional interpretations using the genomic regions enrichment of annotations tool (GREAT) revealed that gained 5hmC regions were associated with MSigDB Perturbation terms of COLON_CANCER_MSI_UP, TP53_TARGETS_PHOSPHORYLATED and GRADE_COLON_CANCER_UP (FDR <0.05). Disease ontology and phenotype enrichment analysis also suggested these regions were associated with colon adenocarcinoma and abnormal colon morphology (FDR <0.05). In contrast, down-5hmC sites revealed that these 5hmC loss regions were associated with several pathways involved in colon cancer progression, including MAPK, NOTCH, TGF-beta, WNT, PI3K and RAS signalling (FDR <0.05).
Differential modifications for 5hmC between tumour and normal colon.(A) Heat maps and the normalised tag density profiles show the differential modifications for 5hmC and 5mC between tumour and normal colon. (B) Top enriched transcription factor (TF) motifs identified by de novo motif discovery at gain-5hmC (upper panel) and loss-5hmC (lower panel) loci. (C) Venn diagram showing the number of up-5hmC containing lncRNAs marked by down-5mC. (D) Venn diagram showing the number of down-5hmC containing lncRNAs marked by up-5mC. (E) Examples of up-5hmC (left panel) marked and down-5hmC (right panel) marked lncRNAs. (F) Boxplots of log10-transformed (RPKM) gene expression values for gain-5hmC- and loss-5hmC-associated lncRNAs. P values were determined by Wilcoxon rank-sum test with continuity correction.
Transcription factors typically regulate gene expression by binding cis-acting regulatory elements, including promoters and enhancers, and recruiting co-activators and RNA polymerase II (RNA Pol II) to target genes. We next performed de novo motif analysis on the 5hmC gain and loss regions to identify the enriched sequence elements. This analysis revealed 5hmC gain regions were highly enriched for motifs recognised by TFs, including TEAD, JUNB, ERG, AHR and NR2E3 (Figure 2B). 5-hydroxymethylcytosin loss regions were also significantly enriched for several TF motifs, such as HIF1A, YY1, HNF6, MYCN and TFAP2C (Figure 2B).
Because studies have suggested that 5hmC at gene bodies is positively correlated with protein-coding gene expression, we herein set out to investigate the relationship between 5hmC modification and lncRNA expression in colon cancer. By assigning the up-5hmC and down-5hmC peaks to their associated lncRNAs, we obtained 232 non-redundant lncRNAs marked with up-5hmC and 970 lncRNAs with down-5hmC (Supplementary Tables S1 and S2). Of these up-5hmC-associated lncRNAs, 26 also showed decreased 5mC levels, whereas 109 out of 970 down-5hmC-marked lncRNAs showed increased 5mC levels (Figure 2C and D). For example, upstream of TSS for TCONS_l2_00000584 displayed a gain of 5hmC and a loss of 5mC (Figure 2E). In contrast, the APCDD1L-AS1 locus showed a significant loss of 5hmC and gain of 5mC (Figure 2E). We analysed RNA-seq data (Kim et al, 2014b; Chen et al, 2016) in CRC to calculate expression levels for the up-5hmC and down-5hmC lncRNA list across normal, primary and liver metastatic cancer. Our results showed that up-5hmC-associated lncRNAs were expressed at a significantly higher level overall, relative to down-5hmC-associated lncRNAs (P<2.26e-16; Figure 2F). Thus, our results demonstrated that 5hmC was correlated with higher lncRNA levels in CRC.
Enrichment of 5hmC at typical and super-enhancers in colon cancer
The enrichment of 5hmC at enhancers in embryonic stem cells (Yu et al, 2012) and developing T cells (Tsagaratou et al, 2014; Ichiyama et al, 2015) but not in NPC and neurons (Hahn et al, 2013) suggests that the genomic distribution of 5hmC is cell and tissue specific. Thus, we integrated our 5hmC and 5mC profiles with genome-wide profiles of H3K4me1 and H3K27ac, two histone modifications that mark enhancers, to explore the enrichment of 5hmC at enhancer regions. Heat maps of 5mC, H3K4me3, H3K4me1 and H3K27ac patterns within 3 kb surrounding the 5hmC peaks generated six distinct clusters (Figure 3A). Of these clusters, C4 and C6 indicated 5hmC-associated proximal promoters as areas of H3K4me3 co-occurence. Importantly, we observed C3 representing 5hmC-marked distal enhancers, which exhibited highly enriched H3K4me1 and depleted H3K4me3 signals. In addition, this cluster was also enriched for H3K27ac, indicating that these enhancers can be further classified as active enhancers. We also found that 5mC was significantly depleted in these 5hmC-marked enhancers or promoters (Figure 3A and B). Collectively, these results suggest that 5hmC was enriched at active enhancers with 5mC depletion in CRC.
5-hydroxymethylcytosine is enriched at enhancer regions in colon cancer.(A) Heat maps showing 5hmC, 5mC, H3K4me3, H3K4me1 and H3K27ac patterns within 3 kb surrounding the 5hmC peaks. (B) The normalised tag density profiles for 5hmC, 5mC, H3K4me3, H3K4me1 and H3K27ac within 3 kb surrounding the 5hmC peaks. (C) Heat maps showing 5hmC and 5mC patterns within 3 kb surrounding the colon cancer super-enhancers. (D) The normalised tag density profiles for 5hmC within 3 kb surrounding the 5hmC colon cancer super-enhancers. (E, F) The normalised tag density profiles for 5hmC, 5mC, H3K4me3, H3K4me1 and H3K27ac within 3 kb surrounding the gain VELs and loss VELs.
The fact that enrichment of 5hmC at typical enhancers promoted us to further investigate whether it also enriched at super-enhancers, which consists of multiple enhancers in close genomic proximity (Hnisz et al, 2013; Loven et al, 2013; Whyte et al, 2013). We extracted the colon cancer specific super-enhancers coordinates and mapped 5hmC and 5mC to these elements. We observed that super-enhancers were marked with 5hmC, with depletion of 5mC (Figure 3C and D), indicating that super-enhancers were possibly regulated by 5hmC.
Recent findings have indicated that CRC displayed thousands of locus-specific loss- or gain-of-H3K4me1 marks called variant enhancer loci (VELs) compared with normal colonic crypts (Akhtar-Zaidi et al, 2012). This specific feature leads us to ask if 5hmC is associated with these VELs. To address this, we identified 15 216 up-H3K4me1 and 23 215 down-H3K4me1 sites (FDR <0.05 and fold change >2) between tumour and normal colonic crypts. We therefore defined the up-H3K4me1 sites as gain VELs and the down-H3K4me1 sites as loss VELs. By mapping our 5hmC profiles against the VELs, we found that a fraction of both gain and loss VELs are marked by 5hmC and high levels of H3K27ac (Figure 3E and F). We observed that 5hmC levels in gain VELs are higher than those in loss VELs, whereas 5mC was depleted in gain VELs but enriched at loss VELs, suggesting that 5hmC is associated with enhancer activity.
Given that lncRNAs could arise from enhancer-associated or promoter-associated elements (Marques et al, 2013), we explored the subclasses of lncRNAs regulated by 5hmC. We classified lncRNAs into enhancer- (elncRNA) and promoter-associated (plncRNA) groups according to chromatin status within 2 kb from each lncRNA TSS. As a result, 1513 lncRNAs were transcribed from 5hmC-marked enhancers, whereas 193 lncRNAs were from 5hmC-marked promoters. Further, we identified six lncRNAs associated with gain-5hmC-gain-VELs and one lncRNA associated with a gain-5hmC-gain promoter, whereas 15 lncRNAs were generated from loss-5hmC-loss-VELs and nine lncRNAs were generated from loss-5hmC-loss promoters. Taken together, these results indicated interaction between abnormal enhancer or promoter activities and that 5hmC modifications might drive dysregulated transcription of lncRNAs in CRC.
5hmC-marked enhancers display higher activity than non-5hmC-marked enhancers
Enhancers can also generate non-polyadenylated active bidirectional non-coding RNAs called enhancer RNAs (eRNAs) through the action of RNA polymerase II (pol II), which is considered a hallmark of functionally active enhancers (Lam et al, 2014). To characterise the regulatory relationship between 5hmC and eRNAs, we analysed genome-wide pol II binding and global nuclear run-on sequencing (GRO-seq) of nascent transcripts in colorectal cancer cells. First, we identified 32 821 intergenic H3K4me1 peaks, which we defined as intergenic enhancer loci. We then overlapped these intergenic enhancers with 5hmC profiles and obtained 8513 5hmC-marked and 25 215 non-5hmC-marked intergenic enhancers. After identifying pol II peaks and de novo transcripts, we found that 57 and 10.5% 5hmC-marked intergenic enhancers overlapped with at least one transcript and one pol II peak. In contrast, only 34 and 4.2% non-5hmC-marked intergenic enhancers overlapped with at least one transcript and one pol II peak.
Next, we identified 7763 intergenic bidirectional transcripts by searching both strands for paired peaks. We found that 5.2% 5hmC-marked intergenic enhancers and 4.7% non-5hmC-marked intergenic enhancers overlapped with those bidirectional transcripts, respectively. Although similar proportions of 5hmC-marked and non-5hmC-marked enhancers generating bidirectional transcripts were observed, we found that normalised GRO-seq read-density and pol II signals were more enriched at 5hmC-marked enhancers as compared with non-5hmC-marked enhancers (Figure 4A), suggesting that the transcriptional activity at 5hmC-marked enhancers was much higher than at non-5hmC-marked enhancers.
5-hydroxymethylcytosine is positively correlated with enhancer activity.(A) Average GRO-Seq reads and normalised pol II tag density profiles at 5hmC- and non-5hmC-marked enhancer regions. (B, C) Boxplots of log10-transformed GRO-seq reads and RNA-seq expression values (RPKM) for 5hmC-marked enhancers related genes and non-5hmC-marked enhancers related genes. (D, E) Biological process and signalling pathways enrichment analysis for genes targeted by 5hmC-associated eRNAs.
Given that eRNAs can promote the expression of proximal target genes (Kaikkonen et al, 2013; Lam et al, 2013; Li et al, 2013; Melo et al, 2013), we examined the transcriptional activity of putative target genes by quantifying the GRO-seq and RNA-sequencing (RNA-seq) read counts. Indeed, genes linked to higher eRNA production (Supplementary Table S3) at 5hmC-marked enhancers exhibited significantly higher GRO-seq and RNA-seq counts than genes linked to lower eRNA production (Supplementary Table S4) at non-5hmC enhancers (Figure 4B and C). Functional annotation of the genes targeted by 5hmC-associated eRNAs revealed that they were highly enriched for factors related to the immune process, cell cycle and signal transduction (Figure 4D). These genes were also implicated in critical signalling pathways closely related to colorectal cancer, including C-MYC, PI3K, NOTCH, P53 and WNT networks (Figure 4E). Collectively, our results indicated that enhancers marked with 5hmC displayed functional activity that might contribute to abnormal transcriptional programmes in colon cancer development.
5hmC is involved in long-range chromatin interactions in CRC
Higher-order chromosomal organisation represented by the physical interaction of promoters with distal regulatory elements is critical for transcriptional regulation. To test whether 5hmC is involved in chromatin interactions that regulate lncRNA transcription, we analysed ChIA-PET data sets targeting RNA pol II in colorectal cancer cells (Li et al, 2012), which provided high resolution for the detection of long-range interactions on a global scale. RNA pol II-mediated long-range chromatin interactomes in colorectal cancer contained 19 264 clusters with both intrachromosomal and interchromosomal interactions. Consistent with our hypothesis, 5hmC is highly enriched at these RNA pol II interaction sites in CRC (Figure 5A and B). Interestingly, 5mC was enriched at these sites but showed much weaker signal levels than 5hmC (Figure 5B). In addition, loss of 5hmC and gain of 5mC in tumour as compared with normal tissue was examined at the interaction loci (Supplementary Figure S2). The results indicated that epigenetic changes may affect chromatin interactions contributing to abnormal transcriptional regulation in cancer.
5-hydroxymethylcytosine is involved in RNA pol II-mediated chromatin interactome.(A) Circos map of the whole-genome pol II chromatin interactome (Interaction PET count >7) and 5hmC occupancies from chromosome 1 to chromosome X, generated using the OmicCircos package. Interactions are drawn in the innermost ring, followed by the 5hmC track (green) that shows the peak signals of 5hmC. Interchromosomal interactions are drown with blue and intrachromosomal interactions are drown with red. (B) The normalised tag density profiles for 5hmC, pol II, 5mC, H3K4me3, H3K4me1, H3K27ac and GRO-seq reads at chromatin interaction sites defined by ChIA-PET peaks. (C) Example showing enhancer–enhancer (left panel), promoter–promoter (middle panel) and promoter–enhancer (right panel) interaction at the lnRNA loci.
We next explored the histone modifications to determine which marks participate in long-range interactions. We found the enrichment of several active histone modification marks at RNA pol II interaction sites, including H3K4me3, H3K4me1 and H3K27ac, confirming that the interactions were highly associated with actively transcribed promoters and distal enhancers, as reflected by the enrichment of GRO-seq reads at these loci (Figure 5B).
We further identified three types of interactions at lncRNA loci: enhancer–promoter, enhancer–enhancer and promoter–promoter interactions. All were associated with 5hmC and corresponding histone modification signatures (Figure 5C). For example, one enhancer–enhancer interaction involved an enhancer located upstream of TCONS_00016184 that interacted with another enhancer 30 kb downstream of TCONS_00016491. Both the enhancers were marked by 5hmC and H3K4me1 and generated bidirectional transcripts. Furthermore, 5hmC was involved in promoter–promoter and promoter–enhancer interactions, as exemplified by observations that the promoter of TCONS_00009259 interacted with the promoter of coding gene PHF15 and that the promoter of TCONS_l2_00023557 interacted with the enhancer located within its intron. Interacting promoters and enhancers show high levels of transcriptional activity, as reflected by active histone mark occupation and transcription. Collectively, these observations suggest that 5hmC, together with several histone marks and transcription factors, established open chromatin structures that facilitate chromatin interactions that regulate lncRNA transcription.
Prognosis associated with lncRNAs regulated by 5hmC
To evaluate the clinical significance of dysregulated 5hmC, we next set out to determine the expressions of the 1202 lncRNAs marked with the gain or loss of 5hmC. We first explored the expression patterns of these lncRNAs using RNA-seq data. This allowed us to distinguish primary, metastatic and normal samples (Figure 6A). We also validated the expression of six lncRNAs in our six matched CRC samples using qPCR (Figure 6B), the results of which were consistent with 5hmC modification status.
Clinical evaluation of 5hmC-associated lncRNAs.(A) Heat map of lncRNAs expression pattern in primary, metastasis and normal colon. (B) qPCR validation of the differential 5hmC-associated lncRNAs. Replicates (n=6) of each sample were run and the Ct values averaged. All Ct values were normalised to β-actin. (C) Kaplan–Meier curves for overall survival time in patients with CRC according to expression of LINC00189. (D) Boxplot for MSI status in patients with CRC according to expression of ENSG00000182165.13. (E) Kaplan–Meier curves for overall survival time in patients with CRC according to expression of LOC286297. (F) Boxplot for tumour stage in patients with CRC according to expression of ENSG00000223561.2.
We then examined the prognostic value of those lncRNA signatures for TCGA through TANRIC (Li et al, 2015). Higher expression of gain-5hmC-marked lncRNA, LINC00189, was significantly correlated with poor overall patient survival (Figure 6C). We also observed the expression of another lncRNA, ENSG00000182165.13, was correlated with microsatellite instability (MSI; Figure 6D). In contrast, lower expression of loss-5hmC-marked lncRNA, LOC286297, was significantly correlated with poor overall patient survival (Figure 6E). The expression of ENSG00000223561.2 was correlated with tumour stage (Figure 6F). Taken together, these results suggest that lncRNAs had predictive and prognostic value in the management of CRC.
Discussion
Recent studies have shown that the global loss of 5hmC is a hallmark of various cancers, suggesting a potential role for 5hmC-mediated epigenetic regulation in cancer biology. Although genome-wide 5hmC maps have been generated for several cell types, the functional role of 5hmC remains limited. In this study, we systemically investigated the genome-wide patterns and functional relevance of 5hmC by integrating data resources, including 5hmC, transcriptome data, chromatin interactome data and several histone modifications. We provided the first report that in addition to previously reported protein-coding genes, 5hmC was also distributed at lncRNA loci and correlated with dysregulated lncRNAs. These findings together extend our knowledge about the epigenetic regulatory roles of 5hmC and cis-regulatory elements for controlling transcription in CRC.
Polyadenylated lncRNA is transcribed by RNA pol II and associated with epigenetic signatures that are common to protein-coding genes, such as H3K4me3, at the promoter and H3K36me3 throughout the gene body (Guttman et al, 2009). Previous work has shown enriched H3K4me3 at the TSS of lncRNA genes in cancers, including T-ALL, prostate cancer and lung cancer (Prensner et al, 2011; Trimarchi et al, 2014; White et al, 2014). Thus, our novel discovery that lncRNAs showed similar patterns to those of protein-coding genes in 5hmC distributions further indicated that lncRNAs share certain epigenetic control mechanisms with protein-coding genes. However, we observed that the average 5hmC signal for lncRNAs was lower than that observed for coding genes. As 5hmC at gene body is positively correlated with transcription, we suggest that the observation in previous studies that lncRNA was less actively transcribed than mRNA (Cabili et al, 2011; Trimarchi et al, 2014) could be partially explained by the lower-average 5hmC signal for lncRNAs.
We characterised thousands of locus-specific gain or loss of 5hmC regions, which were implicated in several pathways and transcription factor occupancy. TEAD family transcription factors, overpresented in gain-5hmC regions, are the major intracellular mediators of the Hippo-Yap pathway and TEAD-AP1 cooperation drives the expression of a unique core set of target genes, thereby directing cancer cell migration and invasion (Liu et al, 2016). Thus, we suggested that lncRNAs associated with the gain or loss 5hmC regions might be members of those signalling and regulated by the transcription factor.
To evaluate the clinical significance of differential modifications of 5hmC, we examined their associated lncRNA expression patterns in primary, metastatic tumour and normal colon. We showed that these lncRNAs exhibited distinct expression patterns in cancerous and normal tissue, whereas some metastatic tumours were similar to primary tumours with lncRNA expression changes, reflecting 5hmC changes across various tumour status. To further examine these lncRNA signatures in TCGA data using TANRIC, we mapped our lncRNAs to GENCODE V19 annotations. Among the mapped lncRNAs, some lncRNAs were correlated with distinct overall survival outcomes, tumour stage and MSI status. Thus, 5hmC as well as associated lncRNA signatures might serve as new diagnostic and prognostic prediction markers in CRC.
It has been suggested that several variants in LD could impact multiple enhancers and cooperatively affect target gene expression (Corradin et al, 2014). Here we obtained a list of the colon cancer-associated variant set, which included 18 clusters with 18 risk-associated SNPs and 568 linked SNPs that were in LD with each of the risk-associated SNPs. Of the 586 SNPs, 7.7% (45) were significantly mapped to 5hmC-marked regions (P=0.001), including 15 clusters. In addition, the vast majority of the remaining SNPs were located less than 10 kb from 5hmC regions. We also found that 5hmC regions containing SNPs exhibited a higher 5hmC signal level than control 5hmC sites not associated with colon cancer SNPs. We speculate that non-coding GWAS variants located within these types of enhancers could modulate gene expression, in some way, by affecting 5hmC-regulated enhancer activity. Notably, we also found CRC variants that mapped to a single 5hmC-marked enhancer at the SMAD7 locus, which contains a super-enhancer in CRC (Hnisz et al, 2013). A super-enhancer comprises large enhancer clusters that control cell identity (Hnisz et al, 2013; Whyte et al, 2013), can be defined by histone H3K27ac modification, and is associated with key oncogenes in several cancer cell types (Hnisz et al, 2013; Loven et al, 2013). In CRC, many cancer hallmark genes that function in the multistep process of tumour pathogenesis acquired super-enhancers (Hnisz et al, 2013). As a fraction of super-enhancers were occupied by 5hmC, we suggest that 5hmC is involved in the process of tumour pathogenesis driven by super-enhancers.
In conclusion, we have systemically investigated the genome-wide patterns and functional relevance of 5hmC in CRC. Our findings suggest potential roles for 5hmC in the regulation of tumour-specific promoters, enhancers and lncRNAs, thereby contributing to the pathogenesis of CRC, which will need to be taken into consideration in future studies of oncogenic transcriptional control in CRC.
Change history
28 February 2017
This paper was modified 12 months after initial publication to switch to Creative Commons licence terms, as noted at publication
References
Akhtar-Zaidi B, Cowper-Sal-lari R, Corradin O, Saiakhova A, Bartels CF, Balasubramanian D, Myeroff L, Lutterbaugh J, Jarrar A, Kalady MF, Willis J, Moore JH, Tesar PJ, Laframboise T, Markowitz S, Lupien M, Scacheri PC (2012) Epigenomic enhancer profiling defines a signature of colon cancer. Science 336 (6082): 736–739.
Anders S, Pyl PT, Huber W (2015) HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31 (2): 166–169.
Baylin SB, Jones PA (2011) A decade of exploring the cancer epigenome–biological and translational implications. Nat Rev Cancer 11 (10): 726–734.
Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, Rinn JL (2011) Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev 25 (18): 1915–1927.
Cancer Genome Atlas Network (2012) Comprehensive molecular characterization of human colon and rectal cancer. Nature 487 (7407): 330–337.
Chen X, Liu B, Yang R, Guo Y, Li F, Wang L, Hu H (2016) Integrated analysis of long non-coding RNAs in human colorectal cancer. Oncotarget 7 (17): 23897–23908.
Corradin O, Saiakhova A, Akhtar-Zaidi B, Myeroff L, Willis J, Cowper-Sal lari R, Lupien M, Markowitz S, Scacheri PC (2014) Combinatorial effects of multiple enhancer variants in linkage disequilibrium dictate levels of gene expression to confer susceptibility to common traits. Genome Res 24 (1): 1–13.
Dawson MA, Kouzarides T (2012) Cancer epigenetics: from mechanism to therapy. Cell 150 (1): 12–27.
Fearon ER (2011) Molecular genetics of colorectal cancer. Annu Rev Pathol 6: 479–507.
Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, Cabili MN, Jaenisch R, Mikkelsen TS, Jacks T, Hacohen N, Bernstein BE, Kellis M, Regev A, Rinn JL, Lander ES (2009) Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458 (7235): 223–227.
Guttman M, Rinn JL (2012) Modular regulatory principles of large non-coding RNAs. Nature 482 (7385): 339–346.
Haffner MC, Chaux A, Meeker AK, Esopi DM, Gerber J, Pellakuru LG, Toubaji A, Argani P, Iacobuzio-Donahue C, Nelson WG, Netto GJ, De Marzo AM, Yegnasubramanian S (2011) Global 5-hydroxymethylcytosine content is significantly reduced in tissue stem/progenitor cell compartments and in human cancers. Oncotarget 2 (8): 627–637.
Hahn MA, Qiu R, Wu X, Li AX, Zhang H, Wang J, Jui J, Jin SG, Jiang Y, Pfeifer GP, Lu Q (2013) Dynamics of 5-hydroxymethylcytosine and chromatin marks in mammalian neurogenesis. Cell Rep 3 (2): 291–300.
He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, Ding J, Jia Y, Chen Z, Li L, Sun Y, Li X, Dai Q, Song CX, Zhang K, He C, Xu GL (2011) Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science 333 (6047): 1303–1307.
Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK (2010) Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38 (4): 576–589.
Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-Andre V, Sigova AA, Hoke HA, Young RA (2013) Super-enhancers in the control of cell identity and disease. Cell 155 (4): 934–947.
Hu Y, Chen HY, Yu CY, Xu J, Wang JL, Qian J, Zhang X, Fang JY (2014) A long non-coding RNA signature to improve prognosis prediction of colorectal cancer. Oncotarget 5 (8): 2230–2242.
Ichiyama K, Chen T, Wang X, Yan X, Kim BS, Tanaka S, Ndiaye-Lobry D, Deng Y, Zou Y, Zheng P, Tian Q, Aifantis I, Wei L, Dong C (2015) The methylcytosine dioxygenase tet2 promotes DNA demethylation and activation of cytokine gene expression in T cells. Immunity 42 (4): 613–626.
Ito S, D’Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y (2010) Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature 466 (7310): 1129–1133.
Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y (2011) Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 333 (6047): 1300–1303.
Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D (2011) Global cancer statistics. CA Cancer J Clin 61 (2): 69–90.
Jin SG, Jiang Y, Qiu R, Rauch TA, Wang Y, Schackert G, Krex D, Lu Q, Pfeifer GP (2011) 5-Hydroxymethylcytosine is strongly depleted in human cancers but its levels do not correlate with IDH1 mutations. Cancer Res 71 (24): 7360–7365.
Kaikkonen MU, Spann NJ, Heinz S, Romanoski CE, Allison KA, Stender JD, Chun HB, Tough DF, Prinjha RK, Benner C, Glass CK (2013) Remodeling of the enhancer landscape during macrophage activation is coupled to enhancer transcription. Mol Cell 51 (3): 310–325.
Kim M, Park YK, Kang TW, Lee SH, Rhee YH, Park JL, Kim HJ, Lee D, Kim SY, Kim YS (2014a) Dynamic changes in DNA methylation and hydroxymethylation when hES cells undergo differentiation toward a neuronal lineage. Hum Mol Genet 23 (3): 657–667.
Kim SK, Kim SY, Kim JH, Roh SA, Cho DH, Kim YS, Kim JC (2014b) A nineteen gene-based risk score classifier predicts prognosis of colorectal cancer patients. Mol Oncol 8 (8): 1653–1666.
Kriaucionis S, Heintz N (2009) The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science 324 (5929): 929–930.
Kudo Y, Tateishi K, Yamamoto K, Yamamoto S, Asaoka Y, Ijichi H, Nagae G, Yoshida H, Aburatani H, Koike K (2012) Loss of 5-hydroxymethylcytosine is accompanied with malignant cellular transformation. Cancer Sci 103 (4): 670–676.
Lam MT, Cho H, Lesch HP, Gosselin D, Heinz S, Tanaka-Oishi Y, Benner C, Kaikkonen MU, Kim AS, Kosaka M, Lee CY, Watt A, Grossman TR, Rosenfeld MG, Evans RM, Glass CK (2013) Rev-Erbs repress macrophage gene expression by inhibiting enhancer-directed transcription. Nature 498 (7455): 511–515.
Lam MT, Li W, Rosenfeld MG, Glass CK (2014) Enhancer RNAs and regulated transcriptional programs. Trends Biochem Sci 39 (4): 170–182.
Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10 (3): R25.
Li G, Ruan X, Auerbach RK, Sandhu KS, Zheng M, Wang P, Poh HM, Goh Y, Lim J, Zhang J, Sim HS, Peh SQ, Mulawadi FH, Ong CT, Orlov YL, Hong S, Zhang Z, Landt S, Raha D, Euskirchen G, Wei CL, Ge W, Wang H, Davis C, Fisher-Aylor KI, Mortazavi A, Gerstein M, Gingeras T, Wold B, Sun Y, Fullwood MJ, Cheung E, Liu E, Sung WK, Snyder M, Ruan Y (2012) Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell 148 (1-2): 84–98.
Li J, Han L, Roebuck P, Diao L, Liu L, Yuan Y, Weinstein JN, Liang H (2015) TANRIC: an interactive open platform to explore the function of lncRNAs in cancer. Cancer Res 75 (18): 3728–3737.
Li W, Liu M (2011) Distribution of 5-hydroxymethylcytosine in different human tissues. J Nucleic Acids 2011: 870726.
Li W, Notani D, Ma Q, Tanasa B, Nunez E, Chen AY, Merkurjev D, Zhang J, Ohgi K, Song X, Oh S, Kim HS, Glass CK, Rosenfeld MG (2013) Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature 498 (7455): 516–520.
Liu X, Li H, Rajurkar M, Li Q, Cotton JL, Ou J, Zhu LJ, Goel HL, Mercurio AM, Park JS, Davis RJ, Mao J (2016) Tead and AP1 coordinate transcription and motility. Cell Rep 14 (5): 1169–1180.
Loven J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, Bradner JE, Lee TI, Young RA (2013) Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 153 (2): 320–334.
Marques AC, Hughes J, Graham B, Kowalczyk MS, Higgs DR, Ponting CP (2013) Chromatin signatures at transcriptional start sites separate two equally populated yet distinct classes of intergenic long noncoding RNAs. Genome Biol 14 (11): R131.
Melo CA, Drost J, Wijchers PJ, van de Werken H, de Wit E, Oude Vrielink JA, Elkon R, Melo SA, Leveille N, Kalluri R, de Laat W, Agami R (2013) eRNAs are required for p53-dependent enhancer activity and gene transcription. Mol Cell 49 (3): 524–535.
Pastor WA, Pape UJ, Huang Y, Henderson HR, Lister R, Ko M, McLoughlin EM, Brudno Y, Mahapatra S, Kapranov P, Tahiliani M, Daley GQ, Liu XS, Ecker JR, Milos PM, Agarwal S, Rao A (2011) Genome-wide mapping of 5-hydroxymethylcytosine in embryonic stem cells. Nature 473 (7347): 394–397.
Prensner JR, Iyer MK, Balbin OA, Dhanasekaran SM, Cao Q, Brenner JC, Laxman B, Asangani IA, Grasso CS, Kominsky HD, Cao X, Jing X, Wang X, Siddiqui J, Wei JT, Robinson D, Iyer HK, Palanisamy N, Maher CA, Chinnaiyan AM (2011) Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nat Biotechnol 29 (8): 742–749.
Quinlan AR, Hall IM (2010) BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26 (6): 841–842.
Rinn JL, Chang HY (2012) Genome regulation by long noncoding RNAs. Annu Rev Biochem 81: 145–166.
Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP (2011) Integrative genomics viewer. Nat Biotechnol 29 (1): 24–26.
Shen L, Shao NY, Liu X, Maze I, Feng J, Nestler EJ (2013) diffReps: detecting differential chromatin modification sites from ChIP-seq data with biological replicates. PloS One 8 (6): e65598.
Simmer F, Brinkman AB, Assenov Y, Matarese F, Kaan A, Sabatino L, Villanueva A, Huertas D, Esteller M, Lengauer T, Bock C, Colantuoni V, Altucci L, Stunnenberg HG (2012) Comparative genome-wide DNA methylation analysis of colorectal tumor and matched normal tissues. Epigenetics 7 (12): 1355–1367.
Stroud H, Feng S, Morey Kinney S, Pradhan S, Jacobsen SE (2011) 5-Hydroxymethylcytosine is associated with enhancers and gene bodies in human embryonic stem cells. Genome Biol 12 (6): R54.
Szwagierczak A, Bultmann S, Schmidt CS, Spada F, Leonhardt H (2010) Sensitive enzymatic quantification of 5-hydroxymethylcytosine in genomic DNA. Nucleic Acids Res 38 (19): e181.
Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, Rao A (2009) Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324 (5929): 930–935.
Trapnell C, Pachter L, Salzberg SL (2009) TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25 (9): 1105–1111.
Trimarchi T, Bilal E, Ntziachristos P, Fabbri G, Dalla-Favera R, Tsirigos A, Aifantis I (2014) Genome-wide mapping and characterization of Notch-regulated long noncoding RNAs in acute leukemia. Cell 158 (3): 593–606.
Tsagaratou A, Aijo T, Lio CW, Yue X, Huang Y, Jacobsen SE, Lahdesmaki H, Rao A (2014) Dissecting the dynamic changes of 5-hydroxymethylcytosine in T-cell development and differentiation. Proc Natl Acad Sci USA 111 (32): E3306–E3315.
Ulitsky I, Bartel DP (2013) lincRNAs: genomics, evolution, and mechanisms. Cell 154 (1): 26–46.
Uribe-Lewis S, Stark R, Carroll T, Dunning MJ, Bachman M, Ito Y, Stojic L, Halim S, Vowler SL, Lynch AG, Delatte B, de Bony EJ, Colin L, Defrance M, Krueger F, Silva AL, Ten Hoopen R, Ibrahim AE, Fuks F, Murrell A (2015) 5-hydroxymethylcytosine marks promoters in colon that resist DNA hypermethylation in cancer. Genome Biol 16 (1): 69.
Vaiopoulos AG, Athanasoula K, Papavassiliou AG (2014) Epigenetic modifications in colorectal cancer: molecular insights and therapeutic challenges. Biochim Biophys Acta 1842 (7): 971–980.
White NM, Cabanski CR, Silva-Fisher JM, Dang HX, Govindan R, Maher CA (2014) Transcriptome sequencing reveals altered long intergenic non-coding RNAs in lung cancer. Genome Biol 15 (8): 429.
Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, Young RA (2013) Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 153 (2): 307–319.
Yan X, Hu Z, Feng Y, Hu X, Yuan J, Zhao SD, Zhang Y, Yang L, Shan W, He Q, Fan L, Kandalaft LE, Tanyi JL, Li C, Yuan CX, Zhang D, Yuan H, Hua K, Lu Y, Katsaros D, Huang Q, Montone K, Fan Y, Coukos G, Boyd J, Sood AK, Rebbeck T, Mills GB, Dang CV, Zhang L (2015) Comprehensive genomic characterization of long non-coding RNAs across human cancers. Cancer Cell 28 (4): 529–540.
Yu M, Hon GC, Szulwach KE, Song CX, Zhang L, Kim A, Li X, Dai Q, Shen Y, Park B, Min JH, Jin P, Ren B, He C (2012) Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell 149 (6): 1368–1380.
Zhang LT, Zhang LJ, Zhang JJ, Ye XX, Xie AM, Chen LY, Kang JX, Cai C (2013) Quantification of the sixth DNA base 5-hydroxymethylcytosine in colorectal cancer tissue and C-26 cell line. Bioanalysis 5 (7): 839–845.
Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, Liu XS (2008) Model-based analysis of ChIP-Seq (MACS). Genome Biol 9 (9): R137.
Acknowledgements
We thank the respective authors for their contributions to generate the sequencing data, as well as all the people who participated in this study and helped us successfully complete the research. This work was supported by the National Natural Science Foundation of China (Grant No. 81372077).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License.
Supplementary Information accompanies this paper on British Journal of Cancer website
Rights and permissions
From twelve months after its original publication, this work is licensed under the Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/4.0/
About this article
Cite this article
Hu, H., Shu, M., He, L. et al. Epigenomic landscape of 5-hydroxymethylcytosine reveals its transcriptional regulation of lncRNAs in colorectal cancer. Br J Cancer 116, 658–668 (2017). https://doi.org/10.1038/bjc.2016.457
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/bjc.2016.457
Keywords
This article is cited by
-
Prevalence of human papillomavirus infection and associated factors among women attending cervical cancer screening in setting of Addis Ababa, Ethiopia
Scientific Reports (2024)
-
Dissection of transcriptomic and epigenetic heterogeneity of grade 4 gliomas: implications for prognosis
Acta Neuropathologica Communications (2023)
-
Epigenetic tumor heterogeneity in the era of single-cell profiling with nanopore sequencing
Clinical Epigenetics (2022)
-
Biological roles of RNA m5C modification and its implications in Cancer immunotherapy
Biomarker Research (2022)
-
DNA 5-hydroxymethylcytosine in pediatric central nervous system tumors may impact tumor classification and is a positive prognostic marker
Clinical Epigenetics (2021)