Abstract
Background:
NF-κB signalling appears deregulated in breast tumours. The purpose of this study was to determine whether the non-canonical NF-κB pathway, is activated in oestrogen receptor positive (ER+) breast cancer, to identify any correlation between its activity and the clinico-pathological phenotype and to explore whether NF-κB2 and RelB subunits and/or any of their target genes might be used as a predictive marker.
Methods:
Two independent cohorts of ER+ early breast cancer patients treated with adjuvant endocrine therapy were included in the study. Activation of RelB and NF-κB2 subunits was determined in a training set of 121 patients by measuring DNA-binding activities in nuclear extracts from fresh frozen specimens by an ELISA-based assay. Samples of 15 ER− breast cancer patients were also included in the study. In a large validation cohort of 207 patients, nuclear immunostaining of RelB and NF-κB2 on formalin-fixed paraffin-embedded specimens was performed. Statistical correlation within clinico-pathological factors, disease-free survival (DFS) and overall survival (OS) was evaluated. Publicly available gene expression and survival data have been interrogated aimed to identify target genes.
Results:
Activation of NF-κB2 and RelB was found in 53.7 and 49.2% of the 121 ER+ tumours analysed, with similar levels to ER− breast tumours analysed in parallel for comparisons. In the validation cohort, we obtained a similar proportion of cases with activation of NF-κB2 and RelB (59.9 and 32.4%), with a 39.6% of co-activation. Multiplexing immunofluorescence in breast cancer tissue confirmed an inverse spatial distribution of ER with NF-κB2 and RelB nuclear expression in tumour cells. Interestingly, NF-κB2 and RelB mRNA expression was inversely correlated with ER gene (ESR1) levels (P<0.001, both) and its activation was significantly associated with worse DFS (P=0.005 and P=0.035, respectively) in ER+ breast cancer. Moreover, the co-activation of both subunits showed a stronger association with early relapse (P=0.002) and OS (P=0.001). Finally, higher expression of the non-canonical NF-κB target gene myoglobin was associated with a poor outcome in ER+ breast cancer (DFS, P<0.05).
Conclusions:
The non-canonical NF-κB pathway activation is inversely associated with oestrogen receptor expression in ER+ breast cancer and predicts poor survival in this subgroup. The myoglobin gene expression has been identified as a possible surrogate marker of the non-canonical NF-κB pathway activation in these tumours.
Similar content being viewed by others
Main
Breast cancer is a leading cause of death for women in industrialised countries with a 10% lifetime risk of disease and over 1 million new cases annually. The oestrogen receptor positive (ER+) breast cancers suppose the 80% of all breast cancers. Despite the success of anti-estrogenic therapies, resistance still appears in patients and constitute the main obstacle in their management. Although the molecular mechanisms behind anti-oestrogen resistance are yet to be fully elucidated, evidences link treatment resistance with activation of the nuclear factor-κB (NF-κB) pathway (Zhou et al, 2005a). Moreover, NF-κB affects ER expression and activity in breast cancer through multiple mechanisms (Sas et al, 2012).
NF-κB transcription factors are either homodimers or heterodimers of five proteins: RelA, c-Rel, RelB, NF-κB2 (p52) and p50 and they are tightly regulated. In unstimulated normal cells, inactive NF-κB exists in the cytoplasm bound to inhibitory proteins. The phosphorylation and degradation of the inhibitory molecule that is induced by a specific signal causes the subsequent release and nuclear shuttling of NF-κB dimers to induce the expression of genes. Each NF-κB dimer selectively regulates a set of target genes involved in cell proliferation and survival, inflammation and innate immunity (Saccani et al, 2003). In tumour tissues, NF-κB signalling pathway is constitutively active (Ben-Neriah and Karin, 2011) and has been implied in tumorogenesis and resistance mechanisms towards treatment (Basseres and Baldwin, 2006; Inoue et al, 2007; Prasad et al, 2010).
There are two major NF-κB activation pathways, the canonical and the non-canonical that differ by the stimuli that activate them, the kinases complexes implicated as well as the inhibitory NF-κB proteins. The canonical pathway used mainly p65 and p50 subunits, and IκBs as inhibitory protein. Instead, the non-canonical pathway involves NF-κB2 and RelB subunits, and the NF-κB-inducing kinase (NIK) and IKKα kinases. RelB is synthetised in mature form (Millet et al, 2013) and NF-κB2 as a precursor form named p100, which also function as IκB-like molecules (Mise-Omata et al, 2014). As p100 preferentially interacts with RelB (Fusco et al, 2008; Solan et al, 2002), the processing of p100 not only generates NF-κB2 but also causes NF-κB2/RelB translocation (Sun, 2011) and regulation of responsive target genes (Wang et al, 2012).
The vast majority of studies on NF-κB in cancer have focused on p65 and p50 subunits and little information about the non-canonical pathway activity is available. The purpose of this study was to evaluate the implication of NF-κB2 and RelB subunits in ER+ breast cancer patients using tumour samples from two cohorts of patients that were analysed either by DNA-binding ELISA or immunohistochemistry. We have also aimed to correlate non-canonical NF-κB activation with clinico-pathological characteristics and to evaluate their potential prognostic significance.
Materials and methods
Clinical samples
The training cohort included fresh-frozen tissues specimens (n=121) obtained from chemo-naive surgically resected ER+ breast cancer patients and diagnosed in the Hospital Clínico de Valencia. During surgery, tumour fragments were collected in sterile containers, frozen and stored at −80°C. Samples of 15 ER− breast cancer patients were also included in the study. The clinico-pathological characteristics of these breast tissues are summarised in Table 1. The validation cohort included formalin-fixed paraffin-embedded (FFPE) tissues of 207 ER+ breast cancer patients from Biobanks of Hospital del Mar (Barcelona), Fundación Jimenez Diaz (Madrid) and Hospital Clinico de Valencia/INCLIVA, treated with tamoxifen. The clinico-pathological characteristics of these samples are summarised in Table 2. The study was approved by the Institutional Ethics Committee of the three institutions and all subjects gave written informed consent to enter into the study.
Tissue quality determination and tissue microarray construction
Frozen tissues were embedded in optimal cutting temperature compound and sectioned at a 10-μm thickness with a cryostat, mounted onto microscope slides and stained with haematoxylin and eosin. Stained sections of the cases were reviewed, and the areas of the tumour were marked on the slides. Samples that contained more than 70% malignant cells were selected for inclusion in the study. From each FFPE tissue block, three tumour representative cores (1 mm in diameter) were sampled from each marked area in the donor block and mounted on a recipient paraffin block using a TMA workstation (T1000 Chemicon/Merck Millipore, Temecula, CA, USA), as previously described (Rojo et al, 2009).
Determination of NF-κB subunits activation by immunohistochemistry
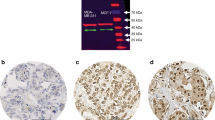
Four microns tissue sections from TMAs were placed on plus charged glass slides. After deparaffinization, heat antigen retrieval was performed in pH9 EDTA-based buffer (Dako, Carpinteria, CA, USA). Endogenous peroxidase was blocked and slides were incubated with primary antibodies previously described by us (Tapia et al, 2007) to NF-κB2 (K27, SC298) and RelB (C19, SC226), both from Santa Cruz Biotechnologies (Dallas, TX, USA) for 60 min, followed by appropriate anti-Ig horseradish peroxidase-conjugated polymer (Flex+, Dako) using a Dako Autostainer, and visualised with 3,3′-diaminobenzidine. Sections incubated with non-immunised serum were used as negative controls and breast tumours with a known expression of markers were used as positive control (Tapia et al, 2007). The interpretation of immunohistochemical staining was based on the percentage and intensity of the immunoreactive tumour cells with nuclear expression of markers in each core, and the final values of the positive tumour cells were evaluated as the mean of the immunoreactivity in three cores. The staining intensity was visually scored and stratified as follows: 0 (negative), 1 (weak, if it was a blush), 2 (intermediate) and 3 (strong staining). A semiquantitative HistoScore (H-score) was calculated by estimation of the percentage of tumour cells positively stained with low, medium or high staining intensity. The final score was determined after applying a weighting factor to each estimate. The following formula was used: H-score=(low%) × 1+(medium%) × 2+(high%) × 3 and the results ranged from 0 to 300. All of the slides were evaluated independently by two experienced pathologists (FR and JF) blinded to clinico-pathological data. Expression cutoffs for each marker was defined using ROC curves.
Determination of DNA-binding activity of NF-κB subunits
Nuclear protein extracts were obtained from frozen breast cancer tissue using Nuclear Extract Kit (Active Motif, Carlsbad, CA, USA) according to the manufacturer’s instructions. The protein concentration was determined using Bradford Reagent assay (Sigma-Aldrich, St Louis, MO, USA). NF-κB activity was measured in nuclear protein extracts by the TransAM NF-κB Kit (Active Motif), an ELISA-based method designed to specifically detect and quantify NF-κB subunits activation, with high sensitivity and reproducibility.
Briefly, the NF-κB TransAM kit contains a 96-well plate with immobilised oligonucleotides containing the NF-κB consensus (5′-GGGACTTTCC-3′) sequence for NF-κB. Nuclear extracts are added to each well and the active form of NF-κB that is present in the extract binds specifically to the oligonucleotide coated on the plate. Then specific antibodies for the activated forms of NF-κB2 and RelB subunits of NF-κB are added. These antibodies recognise epitopes on either NF-κB2 or RelB that are accessible only when NF-κB is activated and bound to its target DNA. A secondary horseradish peroxidase (HRP)-conjugated antibody and chromogenic substrate is added. Colorimetric readout is quantified by spectrophotometry at 450 nm with a reference wavelength of 655 nm. DNA-binding specificity was assessed by competition assays using NF-κB wild type and mutated consensus oligonucleotides. The median value of the analysed tumours for each subunit was taken as cutoff of activation.
Immunofluorescence staining of human breast carcinomas
Immunofluorescence analysis of RelB and NF-κB2 was performed on two sequential formalin-fixed paraffin-embedded 3 μm tissue sections from breast carcinoma specimens placed on positively charged glass slides (two per specimen) according to our previous experience (Tapia et al, 2007). After deparaffinization, antigen retrieval was performed by incubation in Tris-EDTA buffer, pH 9.0 (Dako), in a heated (97 °C) water bath for 40 min. The tumour sections were immersed in TBS/5% BSA for 10 min to block non-specific binding. For co-localisation experiments on the same cells, double immunofluorescence techniques were used. Formalin-fixed paraffin-embedded samples were incubated with a mixture of anti-NF-κB2 rabbit (1 : 100 dilution), anti-RelB mouse (1 : 200 dilution) and anti-ER mouse (clone 1D5, 1 : 1 dilution, Dako) and rabbit (clone SP1, 1 : 1 dilution, Ventana, Tucson, AZ, USA) antibodies. Secondary detection antibodies were Alexa 488 coupled goat antirabbit IgG and Alexa 555 coupled goat antimouse IgG (1 : 1000 dilutions) (Lifetechnologies, Grand Island, NY, USA) generating green fluorescence and red fluorescence for polyclonal and monoclonal antibodies, respectively. Sections were counterstained with 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI, Vysis, Downers Grove, IL, USA) to visualise cell nuclei. All incubations were performed at room temperature. Fluorescence assays were performed using a Dako Autostainer. Staining was evaluated by a pathologist (FR) using a Leica fluorescence DM2000 microscope. Pictures were obtained by Leica Application Suite v2.8.1 and colour deconvolution and image composition was performed using ImageJ software (http://rsb.info.nih.gov/ij).
In silico data analysis of publicly available genomic data sets related to breast cancer
Normalised expression (RNA-seq RPKMs) data of breast tumours in the TCGA cohort and clinical data of patients of the same cohort were downloaded from synapse (syn300013) and TCGA Data Portal (https://tcga-data.nci.nih.gov/tcga/), respectively. These data were used for the analysis of the expression levels of RelB and NF-κB2 genes in both ER− and ER+ breast tumours. The expression and clinical data of breast cancer patients in the GSE3494 data set (PMC1197273), that includes a total of 251 tumours (Miller et al, 2005) were downloaded from the GEO data repository (Edgar et al, 2002). These data were exploited to build prediction models for ER+ patient’s survival using NF-κB-driven selected genes. The expression data of the latter data set was gene-wise mean-centred before the analysis.
Statistical analysis
Statistical analysis was carried out with the program SPSS version 22.0 (SPSS, Inc., Chicago, IL, USA). Receiver operating curve (ROC) was used to determine the optimal cutoff point based on relapse endpoint for RelB and NF-κB2 expression by immunohistochemistry. To analyse correlations between NF-κB2 and RelB expression and clinico-pathological variables we used χ2 test (Fisher exact test), based on bimodal distribution of data. Disease-free survival (DFS) was considered from date of diagnosis to date of any primary, regional or distant recurrence of the infiltrating carcinoma, as well as appearance of a secondary tumour or death. Overall survival (OS) was defined as the time from the date of surgery to the date of death from any cause or last follow-up. Univariate analysis for DFS and OS curves were constructed based on Kaplan–Meier life-table method and analysed using log-rank test of equality. Multivariate survival analysis was calculated using Cox proportion model. Graphical summaries of the distributions were made using boxplots. All the statistical tests were conducted at two sided 0.05 level of significance. This work was carried out in accordance with Reporting Recommendations for Tumour Marker Prognostic Studies (REMARK) guidelines.
Results
Non-canonical NF-κB pathway is activated in ER+ breast cancer with low expression of the oestrogen receptor gene
We assessed the involvement of RelB and NF-κB2 in ER+ breast cancer. Very few studies have analysed NF-κB non-canonical pathway and breast cancer (Wang et al, 2007), demonstrating only an inverse correlation between RelB and ER mRNA expression based on publicly available data. Because NF-κB acts as a dimer, we decided to recapitulate whether the levels of expression of both RelB and NF-κB2 genes correlate with the ER gene (ESR1) expression in breast tumours. To that end, we obtained normalised RNA-seq data of tumours in the breast cancer TCGA cohort from synapse (syn300013) and clinical data of the patients in this cohort from the TCGA Data Portalhttps://tcga-data.nci.nih.gov/tcga/. RNA-seq data and clinical annotations were then merged by mapping patients and tumours barcodes. According to the clinical observations, the cohort included 489 ER+ and 177 ER− breast tumour samples with expression data for comparison. The 489 ER+ tumours were further divided in four subsets corresponding to the quartiles of gene expression of ESR1. Our analysis confirmed that RelB mRNA levels correlate inversely with ER status in primary breast cancer (P<0.001). Moreover, detailed quartile-based analysis of expression level showed an inverse association between ER and RelB expression levels (P<0.001) supporting that low ER+ tumours have a phenotype more similar to the ER− breast cancer. This behaviour is also true for NF-κB2 mRNA in human primary breast cancers (P<0.001 for both subunits) (Figure 1).
Because these data is at gene expression level, we decided to explore whether differences on RelB and NF-κB2 protein activation does exist between ER− and ER+ patients. For this analytical validation, we used an available set of fresh-frozen breast tissues of 121 ER+ and 15 ER− breast cancer patients blinded to clinical data. We analysed nuclear extracts prepared by a commercial ELISA-like assay for detecting DNA-binding activity of NF-κB. Demographic data of the patients are shown in Table 1. Box plot in Figure 2 shows that RelB and NF-κB2 are active in similar percentages in ER- and ER+ tumours (53.7% NF-κB2 activation in ER+ and 60% in ER-; 49.2% RelB activation in ER+ and 50% in ER-). In these series, no significant correlation was found between RelB or NF-κB2 activation and clinico-pathological characteristics of the patients at the time of diagnosis, including age, tumour size, nodal status, histological type and histological grade (data not shown).
DNA-binding activity (activation) of NF- κ B2 and RelB proteins in ER+ and ER− breast tumours. NF-κB DNA binding for the two NF-κB family members was evaluated using the ELISA-based TransAM NF-κB Family kit (Active Motif). The read absorbance at 450 nm of lysis buffer added with the NF-κB antibody dilution was used as blank to normalise the samples measure.
Because of these results, we then plan to study in more detail the clinical consequences of the non-canonical NF-κB pathway activation in ER+ breast cancer. For that, we use a second larger cohort (n=207) of ER+ breast cancer patients with clinical follow-up, assaying RelB and NF-κB2 expression by IHC on FFPE tumour samples. Demographic and clinical characteristics of the patients are shown in Table 2. RelB and NF-κB2 subunits were present diffusely in the cytoplasm and in a proportion of nuclei of neoplastic cells, homogeneously distributed in tumours. Weak expression was also detected in the nucleus of some stromal cells, including fibroblasts and lymphocytes (Figure 3A). This nuclear expression of RelB was present in 108 (32.4%) breast tumours, and NF-κB2 in 124 (59.9%) cases. In tumours with detected nuclear expression of NF-κB2 and RelB, multiplexing immunofluorescence confirmed the significant co-localisation of both subunits up to 73% of tumour cells (Figure 3B) (P=0.002); only 9% of cells expressed only RelB and 11% of cells, only NF-κB2 subunit. Accordingly to previous results, we attempted to correlate the ER expression levels by immunohistochemistry with the nuclear detection of NF-κB2 and RelB in breast tumours. Considering those tumour samples (184 from the total of 207) from which the information of % of cells expressing ER was available, we divided into quartiles the cohort (Figure 3C). The ER expression was inversely correlated with the expression in the nucleus of RelB and NF-κB2 (P<0.001). Finally, multiplexing immunofluorescence for ER, NF-κB2 and RelB determined an inverse spatial distribution of ER and NF-κB2, and ER and RelB nuclear expression in tumour cells (Figure 3D), detecting lower expression or absence of ER in tumour cells with detectable NF-κB2 and RelB nuclear expression.
Immunohistochemical staining for nuclear NF- κ B2 and RelB proteins expression in ER+ breast tumours. (A) Representative image of breast cancer tissue sections with immunohistochemistry staining for RelB and NF-κB2 proteins. Breast cancer cells evidence intense nuclear immunoreactivity for both NF-κB family members. Images of random fields were taken at a magnification of × 100. Scale bars, 25 μm. (B) Representative image of co-localisation RelB, and NF-κB2 proteins by immunofluorescence. Sections were stained with antibodies against RelB (red) and NF-κB2 (green); nuclei were visualised with DAPI (blue). Merged frames indicate co-localisation (yellow). Scale bars, 25 μm. (C) Expression of NF-κB2 and RelB protein in ER+ breast tumours according to expression levels of the receptor (P<0.001). (D) Multiplexing immunofluorescence for ER and NF-κB2, and for ER and RelB in sequential tissue sections. Scale bars, 25 μm.
In this second cohort of breast cancer patients, we detected associations between nuclear NF-κB2 expression with lymph node involvement (P=0.028) and relapse of the disease (P=0.022). In addition, RelB nuclear expression was correlated with relapse (P=0.038). Finally, the co-expression of RelB and NF-κB2 in breast tumours was correlated with relapse (P=0.015). No additional significant correlations were found between RelB or NF-κB2 and the rest of clinico-pathological variables analysed (data not shown).
NF-κB2 and RelB activation are associated with poor prognosis in ER+ breast cancer
Then, we investigate the association between activation of NF-κB2 and RelB subunits in tumour cells and the outcome of the patients (Figure 4A). In these series, nuclear NF-κB2 and expression correlated with DFS (P=0.005, HR=2.1, 95% CI: 0.9–4.5) and a trend towards worse OS (P=0.056, HR=2.3, 95% CI: 0.7–8.1) in ER+ population. As expected, nuclear RelB expression was also associated with DFS (P=0.035, HR=2.1, 95% CI: 1.1–4.1) and a trend towards worse OS (P=0.078, HR=1.6, 95% CI: 0.7–4.2) in same patients. Supporting these data, we analysed the prognosis of the co-expression of RelB and NF-κB2 in ER+ breast cancer patients (Figure 4B). The presence of both subunits strongly correlated with DFS (P=0.002, HR=2.8, 95% CI: 1.2–6.7) and OS (P=0.001, HR=2.4, 95% CI: 0.8–7.3).
Finally, in the multivariate survival analysis, RelB and NF-κB2 co-expression in ER+ tumours samples retained this significant association with worse DFS (P=0.005, HR=5.5, 95% CI: 1.6–15.8) and OS (P=0.018, HR=4.8, 95% CI: 1.1–12.1) in ER+ breast cancer cases. This association is deeply related to patients survival that each subunit separately suggesting that the heterodimer NF-κB2/RelB is more important in the response to treatment that the formation of the homodimers.
NF-κB2 /RelB expression, target gene expressions and clinical outcomes
Our data results related the activation of the heterodimer RelB/NF-κB2 with poor survival in the subgroup of breast cancer patients with ER positivity. In order to go further and to analyse whether we could found not only a prognostic significance of nuclear NF-κB2 and RelB but also of its target gene products in ER+ breast cancer patients. We exploited bioinformatics to evaluate the correlation between NF-κB2 and RelB gene expression and survival in ER+ patients from public databases. For our studies, we selected those genes whose levels correlated with the levels of NF-κB2 and RelB of RE+ in the GSE3494 data set (PMC1197273) and data set (Miller et al, 2005) from the published target genes of NF-κB (Peri et al, 2013). Among the genes analysed, we found that the expression of the myoglobin (MB) gene (NM_203377) was coherent with RelB expression values (P-value<0.01), and confirmed that MB gene expression was associated in the same way to poor outcome in ER+ breast cancer patients (Figure 5). In tumour samples, the expression (mRNA) of MB was not significantly coherent with those of nor p50 subunits (Rel A: P-value=0.39; p50: P-value=0.07). Although it is known that extensive cross-regulation between the canonical and non-canonical signalling axes seems to exist (Carracedo et al, 2012), a recent work reinforces the idea of MB as a possible surrogate of the activation of the non-canonical NF-κB pathway (Iwamoto et al, 2012). This study provides gene expression data (GSE30670) on MDA-MB-231 cells with Rel A or RelB depletion achieved by siRNA transfection. MB gene was downregulated by the down-regulation of RelB, and did not change by the down-regulation of RelA. Overall we have assumed that MB depends on RelB and discriminates between ER+ patients.
Discussion
Approximately 80% of breast cancers express ER. Once activated by 17B-estradiol, ER dimerises and translocates into the nucleus where it binds to DNA and regulates the expression of different target genes (Levin, 2005). The administration of endocrine therapy targeting ER has significantly improved the survival in ER+ breast cancer patients (Early Breast Cancer Trialists’ Collaborative G, 2005). However, efficacy of endocrine therapy is limited by initial de novo and acquired resistance mechanisms, including the deregulation of components of the ER pathway, changes in the expression or action of the ER itself (Zhao and Ramaswamy, 2014), alterations in cell cycle by maintained/high cyclin-D1 and cyclin-E1 expression, alterations in survival signalling molecules, or adaptive upregulation of alternate signalling pathways (Garcia-Becerra et al, 2012). However, definition of the specific genetic lesions and molecular processes that determine clinical endocrine resistance is incomplete (Musgrove and Sutherland, 2009; Tormo et al, 2015) and therefore candidate molecular biomarkers of endocrine response are not yet available. Another signal transduction pathway that might contribute to the development of endocrine resistance is NF-κB. A regulated activation pattern of NF-κB pathway is essential for normal development of the mammary gland, but an aberrant constant increase in NF-κB activity is implicated in breast cancer. NF-κB is able to affect ER pathway by multiple and complex mechanisms (Sas et al, 2012; Yde et al, 2012; Oida et al, 2014), including downregulation of ER expression (Frasor et al, 2014). Several studies have demonstrated an inverse correlation between ER expression and NF-κB pathway (Nakshatri et al, 1997; Biswas et al, 2004; Van Laere et al, 2006), supporting a predominant NF-κB activation in ER− breast cancer (Biswas et al, 2000). In addition, using publicly available gene expression data, a highly significant negative correlation between a gene NF-κB activation signature expression (Annunziata et al, 2007) and the ER activity has been demonstrated (Sas et al, 2012). Similarly, the levels of NF-κB DNA binding (Zhou et al, 2005a) or the expression of target genes (Van Laere et al, 2007) are also inversely correlated with ER expression. Association between ER levels and the risk of recurrence exists (Harvey et al, 1999), and the ability of NF-κB to repress ER might be clinically relevant. However, this clinical significance of NF-κB in breast cancer has been investigated mainly by studying p65 and p50 of the classical pathway. Constitutive NF-κB has been observed in a subset of ER+ tumours that respond poorly to tamoxifen or aromatase inhibitors, and is associated with shortened DFS and increased metastatic recurrence (Nakshatri et al, 1997; Zhou et al, 2005b; Jones et al, 2011; Kubo et al, 2013). Then, the clinical relevance of non-canonical NF-κB pathway represented mainly by NF-κB2 and RelB subunits has been lest inquired (Cogswell et al, 2000; Wang et al, 2007). To our knowledge, no studies relating non-canonical NF-κB pathway and survival in breast cancer have been reported.
Several studies have showed that heterogeneity exist within ER+ tumours. Our group and others have provided evidences that low ER+ tumours are more similar to the ER− than to usual ER+ tumours (Carracedo et al, 2012; Iwamoto et al, 2012). Although the ASCO/CAP guidelines recommend that ER status should be considered positive if 1% or more of tumour cells demonstrate positive nuclear staining (Hammond et al, 2010), recent clinical data suggest that low expression of ER is associated with a downregulation of ESR1-associated gene signature and worse outcome, that is, highly ER-positive breast tumours (Cheang et al, 2009; Prat et al, 2013). In fact, luminal B HER2-subtype is less responsive to endocrine therapy and is associated with poor survival (Sorlie et al, 2001), exhibiting low expression of ER and ER-related genes compared with luminal A tumours (Sorlie et al, 2001; Creighton, 2012).
In this work, in order to ascertain whether the non-canonical NF-κB has a role in ER+ patient’s outcome, we first based our analysis in mining public genomic databases, confirming that RelB and NF-κB2 mRNA levels correlate inversely with ER status in primary breast cancer. An inverse correlation between constitutive RelB expression and ER has been described in breast (Wang et al, 2009). The high RelB expression found in ER− tumours has been attributed in part to the fact that the ER signalling represses the RelB synthesis (Wang et al, 2007). Moreover, RelB reciprocally inhibits ER synthesis by inducing the expression of a called Blimp1 protein that in turn mediates ER repression (Wang et al, 2009). Nuclear expression of RelB is associated with more aggressive breast cancer (Mineva et al, 2009). Similarly, NF-κB2 overexpression in mammary epithelium has been associated with tumorigenesis in animal models (Connelly et al, 2007), but have not shown apparent correlation with ER status in human breast tumours (Cogswell et al, 2000). Then, we confirmed the non-canonical NF-κB pathway activation, based on RelB and NF-κB2 in a large cohort of ER+ breast cancer patients. Two methods were used to analyse both NF-κB2 and RelB subunits activation in ER+ breast cancer, by a commercial ELISA assay, which has an immobilised oligonucleotide encoding an NF-κB consensus site that only binds the active form of NF-κB-specific subunit, and second, by evaluation of nuclear expression of NF-κB2 and RelB subunits as a surrogate of activation by IHC. Non-canonical NF-κB activation was independently associated with poor survival in ER+ breast cancer patients.
It is believed that non-canonical NF-κB promotes a more invasive phenotype by induction of several genes and some authors suggest that resistance to therapy may proceed from the RelB promotes survival (Mineva et al, 2009). Among a list of reported NF-κB genes (Green et al, 2010), we have found that overexpression of the MB gene is associated with worse survival in ER+ patients.Myoglobin is a cytoplasmic oxygen-binding hemoprotein that was thought to be solely present in both cardiac and skeletal muscle cells, and has a role in coping with chronic hypoxia. MB has been found to be overexpressed in a variety of cancers, including breast cancer, and seems to participate actively in carcinogenesis, although its role is not clear in tumour development and progression (Flonta et al, 2009; Oleksiewicz et al, 2011). Some studies have shown that MB expression correlated significantly with a positive ER alpha status and a better prognosis (Kristiansen et al, 2010), but it has been also suggested that solid tumours could take advantage of MB to manage with hypoxic environmental conditions associated with neoplastic growth (Flonta et al, 2009), confirmed by the description of co-expression of MB with markers of tumour hypoxia (Bicker et al, 2015), or that MB expression has been associated with worse outcome in NSCLC (Oleksiewicz et al, 2011).
Translational relevance
Conventional endocrine therapy is not always efficient in the treatment of ER+ breast cancer patients. There is not an efficient way to discriminate groups with different survival in ER+ breast cancer patients (Beelen et al, 2012) and predictive markers of response are needed. We explored the relationship between the activation of the non-canonical RelB and NF-κB2 subunits, and the survival in this breast cancer subtype. Activation levels of NF-κB2 and RelB predict outcome in ER+ breast cancer, but analysis of the co-expression more reliably predicts survival. Our findings suggest the activation of RelB and NF-κB2 subunits as new predictive markers of response in ER+ breast cancers patients. Applications of this study are that increased expression of NF-κB2 and RelB was associated with poor survival in ER+ breast cancer patients and MB expression in this subgroup of luminal tumours might be a surrogate marker of non-canonical NF-κB2 pathway activation.
Change history
26 July 2016
This paper was modified 12 months after initial publication to switch to Creative Commons licence terms, as noted at publication
References
Annunziata CM, Davis RE, Demchenko Y, Bellamy W, Gabrea A, Zhan F, Lenz G, Hanamura I, Wright G, Xiao W, Dave S, Hurt EM, Tan B, Zhao H, Stephens O, Santra M, Williams DR, Dang L, Barlogie B, Shaughnessy JD Jr, Kuehl WM, Staudt LM (2007) Frequent engagement of the classical and alternative NF-kappaB pathways by diverse genetic abnormalities in multiple myeloma. Cancer Cell 12: 115–130.
Basseres DS, Baldwin AS (2006) Nuclear factor-kappaB and inhibitor of kappaB kinase pathways in oncogenic initiation and progression. Oncogene 25: 6817–6830.
Beelen K, Zwart W, Linn SC (2012) Can predictive biomarkers in breast cancer guide adjuvant endocrine therapy? Nat Rev Clin Oncol 9: 529–541.
Ben-Neriah Y, Karin M (2011) Inflammation meets cancer, with NF-kappaB as the matchmaker. Nat Immunol 12: 715–723.
Bicker A, Brahmer AM, Meller S, Kristiansen G, Gorr TA, Hankeln T (2015) The distinct gene regulatory network of myoglobin in prostate and breast cancer. PLoS One 10: e0142662.
Biswas DK, Cruz AP, Gansberger E, Pardee AB (2000) Epidermal growth factor-induced nuclear factor kappa B activation: a major pathway of cell-cycle progression in estrogen-receptor negative breast cancer cells. Proc Natil Acad Sci USA 97: 8542–8547.
Biswas DK, Shi Q, Baily S, Strickland I, Ghosh S, Pardee AB, Iglehart JD (2004) NF-kappa B activation in human breast cancer specimens and its role in cell proliferation and apoptosis. Proc Natil Acad Sci USA 101: 10137–10142.
Carracedo A, Salido M, Corominas JM, Rojo F, Ferreira BI, Suela J, Tusquets I, Corzo C, Segura M, Espinet B, Cigudosa JC, Arumi M, Albanell J, Serrano S, Sole F (2012) Are ER+PR+ and ER+PR- breast tumors genetically different? A CGH array study. Cancer Genet 205: 138–146.
Cogswell PC, Guttridge DC, Funkhouser WK, Baldwin AS Jr (2000) Selective activation of NF-kappa B subunits in human breast cancer: potential roles for NF-kappa B2/p52 and for Bcl-3. Oncogene 19: 1123–1131.
Connelly L, Robinson-Benion C, Chont M, Saint-Jean L, Li H, Polosukhin VV, Blackwell TS, Yull FE (2007) A transgenic model reveals important roles for the NF-kappa B alternative pathway (p100/p52) in mammary development and links to tumorigenesis. J Biol Chem 282: 10028–10035.
Creighton CJ (2012) The molecular profile of luminal B breast cancer. Biologics 6: 289–297.
Cheang MC, Chia SK, Voduc D, Gao D, Leung S, Snider J, Watson M, Davies S, Bernard PS, Parker JS, Perou CM, Ellis MJ, Nielsen TO (2009) Ki67 index, HER2 status, and prognosis of patients with luminal B breast cancer. J Natl Cancer Inst 101: 736–750.
Early Breast Cancer Trialists’ Collaborative G (2005) Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: an overview of the randomised trials. Lancet 365: 1687–1717.
Edgar R, Domrachev M, Lash AE (2002) Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30: 207–210.
Flonta SE, Arena S, Pisacane A, Michieli P, Bardelli A (2009) Expression and functional regulation of myoglobin in epithelial cancers. Am J Pathol 175: 201–206.
Frasor J, El-Shennawy L, Stender JD, Kastrati I (2014) NFkappaB affects estrogen receptor expression and activity in breast cancer through multiple mechanisms. Mol Cell Endocrinol 418 (Pt 3): 235–239.
Fusco AJ, Savinova OV, Talwar R, Kearns JD, Hoffmann A, Ghosh G (2008) Stabilization of RelB requires multidomain interactions with p100/p52. J Biol Chem 283: 12324–12332.
Garcia-Becerra R, Santos N, Diaz L, Camacho J (2012) Mechanisms of resistance to endocrine therapy in breast cancer: focus on signaling pathways, miRNAs and genetically based resistance. Int J Mol Sci 14: 108–145.
Green JA, Elkington PT, Pennington CJ, Roncaroli F, Dholakia S, Moores RC, Bullen A, Porter JC, Agranoff D, Edwards DR, Friedland JS (2010) Mycobacterium tuberculosis upregulates microglial matrix metalloproteinase-1 and -3 expression and secretion via NF-kappaB- and Activator Protein-1-dependent monocyte networks. J Immunol 184: 6492–6503.
Hammond ME, Hayes DF, Dowsett M, Allred DC, Hagerty KL, Badve S, Fitzgibbons PL, Francis G, Goldstein NS, Hayes M, Hicks DG, Lester S, Love R, Mangu PB, McShane L, Miller K, Osborne CK, Paik S, Perlmutter J, Rhodes A, Sasano H, Schwartz JN, Sweep FC, Taube S, Torlakovic EE, Valenstein P, Viale G, Visscher D, Wheeler T, Williams RB, Wittliff JL, Wolff AC (2010) American Society of Clinical Oncology/College Of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. J Clin Oncol 28: 2784–2795.
Harvey JM, Clark GM, Osborne CK, Allred DC (1999) Estrogen receptor status by immunohistochemistry is superior to the ligand-binding assay for predicting response to adjuvant endocrine therapy in breast cancer. J Clin Oncol 17: 1474–1481.
Inoue J, Gohda J, Akiyama T, Semba K (2007) NF-kappaB activation in development and progression of cancer. Cancer Sci 98: 268–274.
Iwamoto T, Booser D, Valero V, Murray JL, Koenig K, Esteva FJ, Ueno NT, Zhang J, Shi W, Qi Y, Matsuoka J, Yang EJ, Hortobagyi GN, Hatzis C, Symmans WF, Pusztai L (2012) Estrogen receptor (ER) mRNA and ER-related gene expression in breast cancers that are 1% to 10% ER-positive by immunohistochemistry. J Clin Oncol 30: 729–734.
Jones RL, Rojo F, A'Hern R, Villena N, Salter J, Corominas JM, Servitja S, Smith IE, Rovira A, Reis-Filho JS, Dowsett M, Albanell J (2011) Nuclear NF-kappaB/p65 expression and response to neoadjuvant chemotherapy in breast cancer. J Clin Pathol 64: 130–135.
Kristiansen G, Rose M, Geisler C, Fritzsche FR, Gerhardt J, Luke C, Ladhoff AM, Knuchel R, Dietel M, Moch H, Varga Z, Theurillat JP, Gorr TA, Dahl E (2010) Endogenous myoglobin in human breast cancer is a hallmark of luminal cancer phenotype. Br J Cancer 102: 1736–1745.
Kubo M, Kanaya N, Petrossian K, Ye J, Warden C, Liu Z, Nishimura R, Osako T, Okido M, Shimada K, Takahashi M, Chu P, Yuan YC, Chen S (2013) Inhibition of the proliferation of acquired aromatase inhibitor-resistant breast cancer cells by histone deacetylase inhibitor LBH589 (panobinostat). Breast Cancer Res Treat 137: 93–107.
Levin ER (2005) Integration of the extranuclear and nuclear actions of estrogen. Mol Endocrinol 19: 1951–1959.
Miller LD, Smeds J, George J, Vega VB, Vergara L, Ploner A, Pawitan Y, Hall P, Klaar S, Liu ET, Bergh J (2005) An expression signature for p53 status in human breast cancer predicts mutation status, transcriptional effects, and patient survival. Proc Natl Acad Sci USA 102: 13550–13555.
Millet P, McCall C, Yoza B (2013) RelB: an outlier in leukocyte biology. J Leukoc Biol 94: 941–951.
Mineva ND, Wang X, Yang S, Ying H, Xiao ZX, Holick MF, Sonenshein GE (2009) Inhibition of RelB by 1,25-dihydroxyvitamin D3 promotes sensitivity of breast cancer cells to radiation. J Cell Physiol 220: 593–599.
Mise-Omata S, Obata Y, Doi TS (2014) p100, a precursor of NF-kappaB2, inhibits c-Rel and reduces the expression of IL-23 in dendritic cells. Biochem Biophys Res Commun 453: 332–337.
Musgrove EA, Sutherland RL (2009) Biological determinants of endocrine resistance in breast cancer. Nat Rev Cancer 9: 631–643.
Nakshatri H, Bhat-Nakshatri P, Martin DA, Goulet RJ Jr., Sledge GW Jr. (1997) Constitutive activation of NF-kappaB during progression of breast cancer to hormone-independent growth. Mol Cell Biol 17: 3629–3639.
Oida K, Matsuda A, Jung K, Xia Y, Jang H, Amagai Y, Ahn G, Nishikawa S, Ishizaka S, Jensen-Jarolim E, Matsuda H, Tanaka A (2014) Nuclear factor-kB plays a critical role in both intrinsic and acquired resistance against endocrine therapy in human breast cancer cells. Sci Rep 4: 4057.
Oleksiewicz U, Daskoulidou N, Liloglou T, Tasopoulou K, Bryan J, Gosney JR, Field JK, Xinarianos G (2011) Neuroglobin and myoglobin in non-small cell lung cancer: expression, regulation and prognosis. Lung Cancer 74: 411–418.
Peri S, Devarajan K, Yang DH, Knudson AG, Balachandran S (2013) Meta-analysis identifies NF-kappaB as a therapeutic target in renal cancer. PloS One 8: e76746.
Prasad S, Ravindran J, Aggarwal BB (2010) NF-kappaB and cancer: how intimate is this relationship. Mol Cell Biochem 336: 25–37.
Prat A, Cheang MC, Martin M, Parker JS, Carrasco E, Caballero R, Tyldesley S, Gelmon K, Bernard PS, Nielsen TO, Perou CM (2013) Prognostic significance of progesterone receptor-positive tumor cells within immunohistochemically defined luminal A breast cancer. J Clin Oncol 31: 203–209.
Rojo F, Gonzalez-Navarrete I, Bragado R, Dalmases A, Menendez S, Cortes-Sempere M, Suarez C, Oliva C, Servitja S, Rodriguez-Fanjul V, Sanchez-Perez I, Campas C, Corominas JM, Tusquets I, Bellosillo B, Serrano S, Perona R, Rovira A, Albanell J (2009) Mitogen-activated protein kinase phosphatase-1 in human breast cancer independently predicts prognosis and is repressed by doxorubicin. Clin Cancer Res 15: 3530–3539.
Saccani S, Pantano S, Natoli G (2003) Modulation of NF-kappaB activity by exchange of dimers. Mol Cell 11: 1563–1574.
Sas L, Lardon F, Vermeulen PB, Hauspy J, Van Dam P, Pauwels P, Dirix LY, Van Laere SJ (2012) The interaction between ER and NFkappaB in resistance to endocrine therapy. Breast Cancer Res 14: 212.
Solan NJ, Miyoshi H, Carmona EM, Bren GD, Paya CV (2002) RelB cellular regulation and transcriptional activity are regulated by p100. J Biol Chem 277: 1405–1418.
Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, Thorsen T, Quist H, Matese JC, Brown PO, Botstein D, Lonning PE, Borresen-Dale AL (2001) Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA 98: 10869–10874.
Sun SC (2011) Non-canonical NF-kappaB signaling pathway. Cell Res 21: 71–85.
Tapia MA, Gonzalez-Navarrete I, Dalmases A, Bosch M, Rodriguez-Fanjul V, Rolfe M, Ross JS, Mezquita J, Mezquita C, Bachs O, Gascon P, Rojo F, Perona R, Rovira A, Albanell J (2007) Inhibition of the canonical IKK/NF kappa B pathway sensitizes human cancer cells to doxorubicin. Cell Cycle 6: 2284–2292.
Tormo E, Pineda B, Serna E, Guijarro A, Ribas G, Fores J, Chirivella E, Climent J, Lluch A, Eroles P (2015) MicroRNA profile in response to doxorubicin treatment in breast cancer. J Cell Biochem 116: 2061–2073.
Van Laere SJ, Van der Auwera I, Van den Eynden GG, Elst HJ, Weyler J, Harris AL, van Dam P, Van Marck EA, Vermeulen PB, Dirix LY (2006) Nuclear factor-kappaB signature of inflammatory breast cancer by cDNA microarray validated by quantitative real-time reverse transcription-PCR, immunohistochemistry, and nuclear factor-kappaB DNA-binding. Clin Cancer Res 12: 3249–3256.
Van Laere SJ, Van der Auwera I, Van den Eynden GG, van Dam P, Van Marck EA, Vermeulen PB, Dirix LY (2007) NF-kappaB activation in inflammatory breast cancer is associated with oestrogen receptor downregulation, secondary to EGFR and/or ErbB2 overexpression and MAPK hyperactivation. Br J Cancer 97: 659–669.
Wang B, Parobchak N, Rosen T (2012) RelB/NF-kappaB2 regulates corticotropin-releasing hormone in the human placenta. Mol Endocrinol 26: 1356–1369.
Wang X, Belguise K, Kersual N, Kirsch KH, Mineva ND, Galtier F, Chalbos D, Sonenshein GE (2007) Oestrogen signalling inhibits invasive phenotype by repressing RelB and its target BCL2. Nat Cell Biol 9: 470–478.
Wang X, Belguise K, O'Neill CF, Sanchez-Morgan N, Romagnoli M, Eddy SF, Mineva ND, Yu Z, Min C, Trinkaus-Randall V, Chalbos D, Sonenshein GE (2009) RelB NF-kappaB represses estrogen receptor alpha expression via induction of the zinc finger protein Blimp1. Mol Cell Biol 29: 3832–3844.
Yde CW, Emdal KB, Guerra B, Lykkesfeldt AE (2012) NFkappaB signaling is important for growth of antiestrogen resistant breast cancer cells. Breast Cancer Res Treat 135: 67–78.
Zhao M, Ramaswamy B (2014) Mechanisms and therapeutic advances in the management of endocrine-resistant breast cancer. World J Clin Oncol 5: 248–262.
Zhou Y, Eppenberger-Castori S, Eppenberger U, Benz CC (2005a) The NFkappaB pathway and endocrine-resistant breast cancer. Endocr Relat Cancer 12 (Suppl 1): S37–S46.
Zhou Y, Eppenberger-Castori S, Marx C, Yau C, Scott GK, Eppenberger U, Benz CC (2005b) Activation of nuclear factor-kappaB (NFkappaB) identifies a high-risk subset of hormone-dependent breast cancers. Int J Biochem Cell Biol 37: 1130–1144.
Acknowledgements
This work was supported by RD12/0036/0070 (A. Ll), RD12/0036/0051 (J.A.), PI12/01421 and PI15/01617 (AL), PI12/00680 and PI15/00146 (JA), PI12/01552 and PI15/00934 (FR) and 2014 SGR 740 grants. AGP is supported by a Ramon y Cajal grant by the Spanish Ministry of Economy. JA is recipient of intensification program ISCIII/FEDER. We thank Fundacion Cellex for a generous donation to the Oncology Service of Hospital del Mar.
Author contributions
FR carried out the analysis of data, participated in the design and coordination, and in the elaboration of the draft of the manuscript. AG carried out the analysis of the publicly available gene expression and survival data to identify ER expression and target genes. JF carried out the ELISA-based assay determinations and analysis of results. MJN, JF and OB selected patients of the training cohort, build the formalin-fixed paraffin-embedded and performed the reading of immunostaining. IC, SZ and JM performed selection of Fundación Jimenez Díaz samples and the immunostaining. IT carried out the selection of Hospital del Mar patients and participated in the elaboration of the manuscript. LS and SS carried out the TMA constructions, and MAS and IG performed the immunostaining of Hospital del Mar and Hospital Clínico de Valencia samples. JA and AL selected patients coordinated the study and participate in the elaboration of the manuscript. AR and PE designed and coordinated the study, analysed the data and participate in the elaboration of the draft of the manuscript. All authors read and approved the final manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License.
Rights and permissions
From twelve months after its original publication, this work is licensed under the Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/4.0/
About this article
Cite this article
Rojo, F., González-Pérez, A., Furriol, J. et al. Non-canonical NF-κB pathway activation predicts outcome in borderline oestrogen receptor positive breast carcinoma. Br J Cancer 115, 322–331 (2016). https://doi.org/10.1038/bjc.2016.204
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/bjc.2016.204
Keywords
This article is cited by
-
RelB sustains endocrine resistant malignancy: an insight of noncanonical NF-κB pathway into breast Cancer progression
Cell Communication and Signaling (2020)
-
Differential expression of p52 and RelB proteins in the metastatic and non-metastatic groups of uveal melanoma with patient outcome
Journal of Cancer Research and Clinical Oncology (2019)
-
The nuclear transcription factor RelB functions as an oncogene in human lung adenocarcinoma SPC-A1 cells
Cancer Cell International (2018)
-
IΚΚε cooperates with either MEK or non-canonical NF-kB driving growth of triple-negative breast cancer cells in different contexts
BMC Cancer (2018)