Abstract
Background:
Adjuvant chemotherapy offered to treat colon cancer is based on the TNM staging system, which often fails due to molecular heterogeneity and undefined molecular mechanisms independent of TNM. Therefore, identification of markers to better predict therapeutic option and outcome is needed. In this study we have characterised the clinical association of CCR6 with colon cancer and defined CCR6-mediated molecular pathway.
Methods:
Immunohistochemistry, RT-qPCR, western blot and FACS were used to determine expression of CCR6 and/or EMT markers in colon tissues/cells. BrdU assay and trans-well system were used to determine cell proliferation, migration and invasion in response to CCL20.
Results:
CCR6 was higher in cancer cases compared to normal adjacent tissue and expression was associated with nodal status and distant metastasis. Similarly, CCR6 expression was higher in cells derived from node-positive cases and highest expression was in cells derived from metastatic cases. Significant changes in EMT markers, that is, E-cadherin, vimentin, β-catenin, N-cadherin, α-SMA, SNAILl and ZEB1 were observed in response to CCL20 along with decreased proliferation, increased migratory and invasive potential.
Conclusions:
Results suggest CCR6 as a potential therapeutic target as well as biomarker in addition to nodal status for predicting therapeutic option.
Similar content being viewed by others
Main
Colon cancer is the third most common cancer worldwide, attributing to 49 700 deaths (26 100 males and 23 600 females) in 2015 in US alone (Siegel et al, 2015). Though the disease can be completely cured if diagnosed early, increasing deaths and poor survival of colon cancer patients are mainly due to metastasis and rapid progression. In addition to this, adjuvant chemotherapy offered to patients with positive lymph nodes or distant metastasis often fails to achieve optimal therapeutic outcomes. This poor therapeutic outcome of adjuvant chemotherapy is primarily due to limitation of TNM classification, used to predict if patient is candidate for adjuvant chemotherapy, which are often inaccurate because biological characteristics and predictors of tumour behaviour are not included in this assessment (Baxter et al, 2005; Sarli et al, 2005; Lee et al, 2007; Morris et al, 2007). Therefore, understanding and defining the molecular signature on colon cancer cell which dictates metastatic process and aggressiveness is needed to improve the disease outcome.
Cancer metastasis is a multi-step process beginning with morphological changes from compact to a more migratory phenotype, dissemination of primary cancerous cells, finally culminating into distant homing. The process of transition from less-motile epithelial- to more-motile spindle-shaped mesenchymal phenotype, known as epithelial-to-mesenchymal (EMT), is complex and correlates with disease progression. EMT involves downregulation of genes responsible for maintaining the sessile nature of epithelial cells with simultaneous increase in expression of genes that trigger migration and invasion (Kim et al, 2009; Thiery et al, 2009). Specifically, the level of proteins involved in various adherens junctions and cytoskeleton reorganisation are regulated in a coordinated manner. These proteins include, but are not limited to, E-cadherin, N-cadherin, β-catenin, vimentin, α-SMA and occludin. Transcriptional regulators, for example, SNAIL1 and ZEB1, indirectly control this process (Lee et al, 2006). Unravelling factors that induce EMT would greatly aid in the development of diagnostic and prognostic markers, as well as better therapeutics for colon cancer.
Studies from our laboratory and others have shown that chemokine–chemokine receptor interaction supports metastatic process in many cancers including colon cancer (Singh et al, 2004b; Sarli et al, 2005; Kakinuma and Hwang, 2006; Singh et al, 2009b; El-Haibi et al, 2011; Singh et al, 2011; Balkwill, 2012; Wyler et al, 2014; Mir et al, 2015). Particularly, CCR6 and CCL20 are highly expressed in a variety of human cancers including colorectal cancer (Dellacasagrande et al, 2003; Kimsey et al, 2004; Ghadjar et al, 2006; Rubie et al, 2006; Ghadjar et al, 2008; Kirshberg et al, 2011; Frick et al, 2013). CCL20, also known as liver activation-regulated chemokine or macrophage-infiltrating factor protein-3α, is the sole ligand for CCR6 (Baba et al, 1997). CCR6–CCL20 interaction has been shown to be involved in several inflammatory and autoimmune diseases, like inflammatory bowel disease (Kaser et al, 2004), psoriasis (Harper et al, 2009), multiple sclerosis (Yamazaki et al, 2008; Reboldi et al, 2009) and rheumatoid arthritis (Hirota et al, 2007). Reports suggest a direct correlation between chronic inflammation and neoplastic transformation leading to metastatic progression (Kuper et al, 2000; Wang et al, 2009), but the underlying molecular mechanisms responsible for this transformation remain elusive. In the present study, we show that increased CCR6 expression correlates with advanced colon cancer. In addition to this, we show the biological significance of CCR6–CCL20 axis in colon cancer and also demonstrate involvement of this chemokine in EMT transition, which is known to be involved in poor outcome of colon cancer.
Materials and Methods
Tissue specimens
Tissue microarray (TMA) was purchased from commercial source (AccuMax/ISU; Abxis Co. Ltd., San Diego, CA, USA) under the IRB guidelines. This study falls under exemption-4 (E4) category, because the specimens and/or data that have already been collected by the company and the patients information were not disclosed. Only information available with tissue was stage and grade. TMA contains adenocarcinoma (AC) tissues (45 cancer cases with 1 mm duplets from each case; n=90) and adjacent normal tissues (n=8). AC on TMA had no regional lymph node metastasis (N0: 24 spots in duplicate); single lymph node (N1: 13 spots in duplicate); two lymph nodes, (N2: 3 spots in duplicate) as well metastasis to distant sites (M1: 5 spots in duplicate).
Immunohistochemistry
Colon TMA, containing neoplastic and normal adjacent tissues, were stained for CCR6. Briefly, slides were incubated at 60 °C for 1 h, de-paraffinized by three changes in xylene. Sections were rehydrated for 5 min each in decreasing concentrations of ethanol (100%, 95% and 70%) and washed in de-ionised water. The slide was then incubated with 3% H2O2 in PBS for 3 min for blocking endogenous peroxidase, rinsed three times with de-ionised water followed by Tris-buffer (pH 7.6). Biotin–Avidin blocking was performed at RT for 15 min using BioGenex kit, (BioGenex; San Ramon, CA, USA). Sections were blocked with 3% normal mouse serum for 30 min at RT and then incubated with 25 μg ml−1 anti-human CCR6 antibody (MAB195, R&D Systems, Minneapolis, MN, USA) overnight at 4 °C in a humidity chamber followed by incubation with multi-species link (BioGenex) at RT for 20 min. Next, sections were washed and incubated with multi-species label (BioGenex) for 20 min at RT. After washing the label, CCR6 immunohistochemical reaction colour was developed with 3,3′-diaminobenzidine (DAB) substrate kit (Vector Laboratories, Burlingame, CA, USA) for 10 s. Counterstaining was done with hematoxylin (Sigma-Aldrich, St Louis, MO, USA). Subsequently, sections were dehydrated using 70%, 95% and absolute alcohol followed by three changes in xylene for 1 min each. The slide was finally mounted with Permount (Sigma-Aldrich). The slide was scanned using TissueFAXS and analysed by HistoQuest software (TissueGnostics GmBH, Vienna, Austria). Briefly, hematoxylin staining was used as a master marker for cellular identification. The range of intensities of CCR6 staining and master marker (hematoxylin) were set by autodetection feature of the software, and ring mask algorithm was used to measure the colour intensity of CCR6 staining at the surface of each cell. Identical settings were used to analyse all the images and intensity measurements were represented as CCR6-positive counts per unit area (mm2). Well-qualified pathologists further validated the results.
Cell lines and cell culture
Duke’s type C colon cancer cell lines (CCL221 and CCL225) derived from the node-positive AC patients and Duke’s type D colon cancer cell lines (CCL222 and CCL224) derived from the AC patients with distant metastasis were used for this study. Cell lines were cultured in RPMI-1640 supplemented with 10% FBS (HyClone, Pittsburgh, PA, USA), 100 μg ml−1 of streptomycin and 100 U ml−1 of penicillin (HyClone). Normal colon epithelial cells (CRL-1790) were cultured in EMEM supplemented with 10% FBS. All cell lines were maintained at 37 °C with 5% CO2.
RNA isolation and RT-qPCR analysis
To determine the effect of CCR6–CCL20 interaction on EMT, colon cancer cells (2.5 × 106 cells) were seeded in T-25 flasks. After overnight culture cells were treated with CCL20 (100 ng ml−1) for different time intervals (15 min, 30 min, 1 h, 2 h, 4 h, 6 h and 24 h). Untreated cells were used as negative control. Total cellular RNA was isolated using Tri-Reagent (Sigma-Aldrich), precipitated and re-suspended in RNA Secure (Ambion, Life technologies, New York, NY, USA). Next, cDNA was generated by reverse transcribing 1 μg total RNA using Verso kit (Thermo Fisher Scientific, Grand Island, NY, USA). Transcripts of EMT markers (E-cadherin, N-cadherin, β-catenin, α-SMA, Vimentin, SNAIL1 and ZEB1) and CCR6 were quantitified by RT-qPCR using iQ SYBR-Green Supermix (Bio-Rad, Hercules, CA, USA; Mir et al, 2015). Absolute copy number of each target was calculated by means of a standard curve, and results are represented as copies of target gene per 106 copies of 18S rRNA or fold change in expression with respect to controls. All RT-qPCR experiments were done in duplicates and repeated three times to validate the results.
Western blot analysis
The relative expression of EMT markers was further confirmed by western blot analyses. Cells were treated with 100 ng ml−1 CCL20 and cell lysates were prepared using protease inhibitor cocktail containing RIPA lysis buffer (Thermo Fisher Scientific, Waltham, MA USA) at different time points (30 min, 1 h, 2 h, 4 h and 6 h) after CCL20 treatment. Proteins isolated from untreated cells were used as control. Proteins were resolved using 10% SDS-PAGE and transferred electrophoretically onto PVDF membranes. The membrane was blocked using 5% skimmed milk for 1 h at RT and then probed for specific proteins using antibody against specific EMT markers purchased from Cell Signalling Technology (CST Inc., Danvers, MA, USA). Chemiluminescent substrate ECL kit (Thermo Fisher Scientific) was used to develop the bands using manufacturer’s protocol, and GAPDH was used as the loading control. The immuno-reactive bands were visualised using ImageQuant LAS4010 imager (GE Healthcare Bio-Sciences, Pittsburgh, PA, USA).
Flow cytometry
Surface expression of CCR6 in colon cancer cell lines was quantified by flow cytometry. Briefly, cells (106) were incubated at 4 °C for 30 min with FITC-conjugated mouse anti-human CCR6 antibody or corresponding isotype control (R&D Systems). After staining, cells were washed with FACS buffer (2% FBS in PBS) and fixed with 2% PFA in PBS for 10 min at RT. Fixative was removed and cells were suspended in 500 μl FACS buffer. Immunofluorescence was acquired using Guava flow cytometer (Millipore, Billerica, MA, USA) and analysed using FlowJo 10.0.6 software (Treestar Inc., Ashland, OR, USA).
Cell proliferation assay
Colon cancer cells were seeded at a density of 2 × 104 per 100 μl per well in 96-well plates. After overnight incubation, cells were stimulated with different concentrations (0–200 ng ml−1) of recombinant CCL20 (Peprotech, Rocky Hill, NJ, USA). BrdU labelling and detection Kit III (Roche Diagnostics Corporation, Indianapolis, IN, USA) was used to determine the effect of CCL20 on colon cancer cell proliferation according to the manufacturer’s protocol. Briefly, 10 μ M BrdU labelling reagent was added to the cells and incorporated BrdU was detected using peroxidase-labeled monoclonal anti-BrdU-POD after 24 h. The bound conjugate was visualised with the soluble chromogenic ABTS substrate (2,2′-azino-bis(3-ethylbenzothiazoline-6-sulphonic acid)) and measured at 405 nm with a reference wavelength at 492 nm. All concentrations were tested in triplicates and the experiment was repeated three times.
Migration and invasion assays
BD Biocoat tumour migration and Matrigel tumour invasion chambers were used for migration and invasion studies, respectively. The inserts were activated with serum-free bicarbonate-based medium (DMEM) for 2 h at 37 °C with 5% CO2. Medium was carefully removed after rehydration without disturbing the matrigel matrix layer. Colon cancer cells (1.0 × 105) with or without blocking CCR6 using 1.0 μg ml−1 anti-human CCR6 antibody (MAB19, R&D Systems) were added to the top chamber of inserts and 100 ng ml−1 CCL20 (Peprotech) was added in the bottom chamber as chemo-attractant. Cells were then allowed to migrate/invade under chemotactic gradient of CCL20 overnight in a CO2 incubator at 37 °C. Non-migrated/invaded cells from top chamber were removed with a cotton swab. Cells at the bottom surface of the insert were fixed with 100% methanol for 2 min, stained for 2 min with 1% Toluidine Blue (Thermo Fisher Scientific), and rinsed twice with de-ionised water. Migrated/invaded cells were counted under the microscope at 40 × magnification. All experiments were repeated three times to validate the results.
Statistics
Unpaired non-parametric Mann–Whitney’s U-test was used to compare CCR6 immunostaining in AC tissue sections with adjacent normal tissues derived from colon cancer patients using GraphPad Prism 6 software (GraphPad Software, Inc., La Jolla, CA, USA). Results were declared significant when the P-value<0.05. Analysis of CCR6 staining was assessed in a semi-quantitative manner by scoring the immunoreactivity as a function of per cent CCR6-positive cells as well as CCR6 immuno-intensity, both derived using HistoQuest software (TissueGnostics GmBH). Percentage of colon cancer cells stained positive for CCR6 were scored on the scale of 1 to 4: ‘1’ for<25%, ‘2’ for 25 to 50%, ‘3’ for 51 to 75% and ‘4’ for>75% positively stained cells (Table 1). Similar scoring grades were assigned based on differential immunostaining intensity, where ‘0’ represents no immunostaining, ‘1’ for weak staining (DAB intensity 5 to 50), ‘2’ for moderate staining (DAB intensity=51 to 100), ‘3’ for high staining (DAB intensity=101 to 200) and ‘4’ for very high staining (DAB intensity>200). The mean composite score was calculated by summation of intensity score multiplied by CCR6-positive cells percentage score. Significance of correlation of CCR6 expression to disease progression was estimated using GraphPad prism contingency table analysis. Probabilities were computed by performing chi-square test with confidence interval set at 95%. Comparison of CCR6 expression between colon cancer cell lines derived from AC patients and normal colon epithelial cells was performed by ordinary non-parametric one-way analysis of variance (ANOVA, multiple comparison with control group). Mean fluorescence intensities of CCR6 surface expression in colon cancer cells were compared with normal colon cells using unpaired t-test. Comparative analysis for differential expression of EMT markers between CCL20-treated and untreated cells were done using unpaired multiple t-test, where results were considered significant when α<0.05. Multiple t-tests using Holm–Sidak method with α=5% were performed for BrdU proliferation, migration and invasion assays to determine the statistical significance. All in vitro experiments were repeated multiple times.
Results
CCR6 expression in colon cancer tissue and cell lines correlates with disease progression
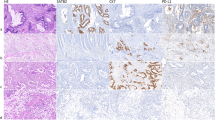
Colon AC tissues were immuno-stained for CCR6 and mean composite scores were found to be higher in tissue section from metastatic cases (M1) compared with N0/N1/N2 and normal adjacent tissues (Table 1). When CCR6 immunostaining was calculated as DAB-positive signals per mm2 of total membrane surface area of tissue sections, expression of CCR6 was significantly higher in cancer tissues (**P-value<0.01), as compared with adjacent normal tissue (Figure 1A and B). CCR6 expression was further compared between non-metastatic cases vs cases with lymph node-positive or with distant metastasis. Expression of CCR6 was higher in cases with lymph node-positive and distant metastasis (****P-value<0.0001) compared with non-metastatic cases (N0= 3.59 × 107 counts per mm2). Further, expression of CCR6 was higher in cases with distant metastasis (M1: 1.35 × 108 counts per mm2) when compared with node-positive cases (N1: 1.11 × 108 counts per mm2, N2: 1.21 × 108), and it is important to mention that expression of CCR6 correlates with number of positive nodes (Figure 1C), which was not separated well by mean composite score (Table 1).
CCR6 expression in colon cancer tissue. (A) Immuno-intensity of CCR6 (brown) in normal adjacent tissue and AC tissue with no nodal involvement (N0), nodal involvement (N1 and N2) and distant metastasis (M1). Digital images were acquired using 20 × -objective of TissueFAXS and CCR6 expression was quantified using HistoFAXS analysis software. (B) CCR6 expression in adjacent normal tissue and AC tissues is represented as DAB-positive counts per mm2 area. **P-value<0.01 between normal adjacent and AC. (C) Cases were further stratified according to nodal involvement (N0, N1 and N2) and distant metastasis (M1), and CCR6 expression in each subtype is presented as box plot, showing the minimum and maximum values. Median value of CCR6 expression is represented by the line in the box plots. *P-value<0.05 between adjacent normal vs N0 and ****P-value<0.0001 between adjacent normal vs N1/N2/M1 cases. Significant increase (****P-value<0.0001) was observed when non-metastatic cases (N0) were compared with cases with regional and distant metastasis. Mann–Whitney’s U-test was used to determine statistical significance between each group.
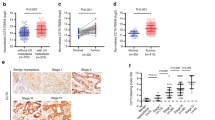
To further elucidate the biological significance of CCR6 in colon cancer tissue, its expression was determined in Duke’s type C (CCL221 and CCL225) and Duke’s type D (CCL222 and CCL224) cell lines derived from colon AC patient with regional lymph node and distant metastasis, respectively. Similar to tissue expression, CCR6 surface protein (Figure 2A and B) and mRNA levels (Figure 2C) were significantly higher (****P-value<0.0001) in cancer cell lines compared to normal colon epithelial cell line (CRL-1790). Flow cytometry analysis revealed increase in surface expression of CCR6 in colon cancer cells (Figure 2A) with respect to isotype control as shown by shift in histogram. However, CCR6 signal was not changed in case of normal colon cells CRL-1790 (Figure 2A) as observed by overlapping CCR6 and isotype control histograms. Mean fluorescence intensities of CCR6 surface expression were significantly higher (***P-value<0.001) in colon cancer cells as compared with normal colon cells (Figure 2B). CCR6 expression at mRNA levels was also significantly higher (*P-value<0.05) in cell line derived from patient with distant metastasis compared to that derived from patient with regional lymph node metastasis (Figure 2C).
CCR6 expression in colon cancer cell lines. Normal colon epithelial (CRL-1790), Duke’s type C (CCL221) and type D (CCL224) cells were stained with FITC-conjugated anti-CCR6 antibodies or isotype antibodies and data was acquired using Guava Flow cytometer and analysed with FlowJo. (A) Surface expression of CCR6. Solid histogram show CCR6 expression and open histogram represents isotype control. (B) Mean fluorescence intensity of CCR6 surface staining with respect to the isotype control in respective cell lines. ***P-value<0.001 between normal colon cells vs colon cancer cells. Copies of CCR6 transcript per million copies of 18S rRNA are shown in C. Values represent the mean±s.e.m. from three independent experiments. ****P-value<0.0001 difference in CCR6 expression in colon cancer cell lines compared with the normal colon cells.
CCR6–CCL20 interaction promotes EMT in colon cancer cells
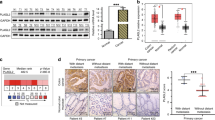
EMT supports metastasis and has a negative impact on disease and therapeutic outcome. Hence, we evaluated the effect of CCR6–CCL20 interaction on EMT markers. Reduction in E-cadherin protein was observed 1 h after CCL20 treatment in all cell lines and reduction was continued until 6 h (Figure 3). N-cadherin, β-catenin and vimentin expression increased 30 min after CCL20 treatment of Duke’s type C and D cell lines compared with untreated cells. For CCL225 cells, the increase in these markers occurred 1 h after CCL20 treatment. Further, increase in α-SMA and SNAIL was observed 4 h after CCL20 treatment in colon cancer cells compared with untreated cells. α-SMA expression in CCL225 cells increased 6 h after CCL20 treatment. Similar expression pattern after CCL20 treatment was observed at mRNA level. In addition, transcripts of another mesenchymal marker, ZEB1, were elevated in colon cancer cells 4 h after CCL20 treatment (Figure 3). These results clearly demonstrate the role of CCR6–CCL20 interaction in EMT induction in colon cancer.
Effect of CCR6–CCL20 interactions on EMT. Transcript levels of E-cadherin, N-cadherin, β-catenin, Vimentin, α-SMA, SNAIL1, ZEB1 in untreated (open box) and CCL20-treated (solid box) cells were quantified by qRT-PCR and normalised with 18S rRNA. Relative fold change, using ΔΔCt method was calculated with untreated samples as control. Fold change in E-cadherin, N-cadherin, β-catenin, Vimentin, α-SMA, SNAIL1, ZEB1 mRNA transcripts in colon cancer cell lines: CCL221 (A), CCL222 (B), CCL224 (C) and CCL225 (D) are shown on left side. Statistical significance of change in mRNA level of EMT marker in CCL20-treated cells compared with untreated cells are indicated as *P-value<0.05, **P-value<0.001 and ***P-value<0.0001. Western blot analysis was used to confirm mRNA expression results at protein level. GAPDH was used as loading control. Western blot images of EMT markers are shown at right side in A to D.
CCR6-activation affects proliferation, migration and invasion in colon cancer cells
Proliferation of all colon cancer cell lines decreased in a dose-dependent manner after CCL20 stimulation. Maximum reduction in proliferation was observed at 24 h following CCL20 treatment (Figure 4A). There was a major decrease (20–35%) in proliferation in colon cancer cells treated with CCL20 compared with untreated samples (Figure 4B). The effect of CCR6–CCL20 axis on colon cancer cell migration and invasion was characterised using trans-well migration and invasion chambers using CCL20 as a chemo-attractant. Colon cancer cell lines showed higher invasive and migratory potential toward CCL20 gradients, compared to respective untreated cells, which was significantly inhibited after CCR6 blockade (Figure 5).
CCR6–CCL20 interaction inhibits proliferation of colon cancer cells. (A) BrDU incorporation assay was used to determine proliferation in untreated (open box) and CCL20-treated (solid box) colon cancer cells. Significant (***P-value<0.001 and **P-value<0.01) differences in proliferation rate of CCL20-treated cells compared with the untreated cells are shown. (B) Percentage decrease in proliferation of colon cancer cells after CCL20 treatment compared with untreated cells. Significant (**P-value<0.00001, ***P-value<0.000001 and ****P-value<0.0000001) decrease in proliferation of CCL20-treated cells compared with untreated cells is shown.
CCR6–CCL20 interaction mediates migration and invasion. Migratory (A) and invasive (B) potential of colon cancer cells was tested without chemo-attractant (open box), CCL20 as a chemo-attractant (solid box) and using CCL20 as chemo-attractant after blocking CCR6 (hashed box). Cells were counted in three random fields and data is presented as mean±s.d., n=3. Asterisks indicate significant differences in migration and invasion between untreated and CCL20-treated cell lines (**P-value<0.01, ***P-value<0.001, and ****P-value<0.0001). Significant differences in migration and invasion between CCL20-treated and anti-CCR6-treated cell lines are shown (##P-value<0.01, ###P-value<0.001 and ####P-value<0.0001).
Discussion
Current adjuvant chemotherapies are inadequate in achieving optimal therapeutic response in colon cancer patients with lymph node metastasis. The TNM classification used to predict whether a patient is a good candidate for adjuvant chemotherapy is often inaccurate because biological characteristics and predictors of tumour behaviour are not part of this assessment (Baxter et al, 2005; Sarli et al, 2005; Lee et al, 2007; Morris et al, 2007). Higher CCR6 expression in tumour tissues derived from AC patients compared with normal adjacent colon tissue strongly suggests potential role of CCR6 in etiopathogenesis of colon cancer. Higher expression of CCR6 in node-positive cases compared with N0 and highest expression in metastatic cases further suggests CCR6 is potential marker for disease aggressiveness. The receptor could be used as an independent biomarker for making decision of adjuvant chemotherapy, as well as potential adjuvant or neoadjuvant therapeutic target.
To establish the biological significance of CCR6 in colon cancer, its expression was evaluated in colon AC cell lines derived from patients with positive regional lymph node and distant metastasis. Higher expression of CCR6 in AC cells compared with normal colon cells and highest CCR6 expression in cells derived from patient with distant metastasis compared with those derived from regional lymph node-positive corroborates with the clinical significance of CCR6 at cellular level.
To achieve metastatic goal tumour cells escape from primary site, which requires invasion, intravasation, systemic transport and colonisation at secondary site. Coordinated interaction of various signalling pathways dictates EMT that supports these metastatic processes (Lee et al, 2006; Kim et al, 2009; Thiery et al, 2009). Of these, chemokine signalling significantly contributes to this process (Kakinuma and Hwang, 2006; Balkwill, 2012; Wyler et al, 2014). Epithelial-cadherin (E-cadherin) is a calcium-dependent cell–cell adhesion glycoprotein responsible for maintaining mechanical rigidity of epithelium (van Roy and Berx, 2008). Downregulation of E-cadherin is essential for loss of adhesion and escape. Loss of E-cadherin and cancer progression through EMT has been well-established (Kim et al, 2002; Jeanes et al, 2008).
Interaction of E-cadherin intracellular domain with β-catenin is required for maintaining architectural rigidity of cell (Scott and Yap, 2006; Daugherty and Gottardi, 2007). Hence, decrease in E-cadherin and increase in β-catenin observed in our study following CCL20 treatment implicates potential role of CCR6–CCL20 axis in EMT, which also accords with the studies showing activation of EMT transcription factors by nuclear translocation of elevated β-catenin (Gottardi et al, 2001; Stockinger et al, 2001; Kim et al, 2002). SNAIL1 is another transcription factor and an important mesenchymal marker, which was elevated in CCL20-treated colon cancer cells. This EMT regulator recruits G9α-methyl transferase facilitating DNA methylation of the promoter region of E-cadherin (Dong et al, 2012; Lin et al, 2014) and downregulates E-cadherin (Batlle et al, 2000; Cano et al, 2000; Yokoyama et al, 2001) to promote the invasive phenotype in cancer cells (Yokoyama et al, 2001; Blanco et al, 2002).
E-A zinc-finger box-binding homeobox transcription factor 1 (ZEB1) is also overexpressed in colon cancer cells following CCL20 treatment. ZEB1 is a repressor of E-cadherin that can transcriptionally activate several mesenchymal genes like vimentin that facilitate EMT (Guaita et al, 2002; Pena et al, 2005; Spoelstra et al, 2006; Witta et al, 2006; Peinado et al, 2007; Liu et al, 2008). We showed that vimentin was upregulated in colon cancer cells after CCL20 treatment. This ubiquitous mesenchymal protein protects cells against mechanical stress and maintains cellular integrity (Satelli and Li, 2011). It modulates the invasiveness of tumour cells by regulating the E-cadherin/β-catenin complex (Wei et al, 2008). Overexpression of vimentin correlates with invasive potential and tumour cell motility (McInroy and Maatta, 2007; Zhao et al, 2008; Kim et al, 2009; Zhu et al, 2011). In addition to ZEB1, vimentin could also be induced by SNAIL1 (Olmeda et al, 2007). Alpha-smooth muscle actin (α-SMA) was also upregulated in CCL20-treated colon cancer cells and is known to control cell motility by maintaining cytoskeletal structures (Lambrechts et al, 2004). Gene silencing experiments showed α-SMA regulates FAK and c-MET, which in turn influence the metastatic potential of cancer cells (Lee et al, 2013). Although there was a difference in activation time and duration in sustaining the expression of EMT protein in different colon cancer cells, change was consistent. This further suggests inclusion of increased CCR6–CCL20, in addition to the nodal status, as molecular signature while determining therapeutic regime for colon cancer.
Cell proliferation fuels tumour growth and this critical event substantially influences tumour fate and therapeutic outcomes. Our data show significant inhibition in cell proliferation following CCL20 treatment. This inhibition in cell proliferation could be due to the observed role of CCR6–CCL20 axis in promoting mesenchymal phenotype, which are slow proliferating compared with epithelial phenotype (Evdokimova et al, 2009; Liu et al, 2014). Furthermore, cancer cells utilise various migratory and invasive mechanisms in response to their external environment including EMT. Hence, decoding the molecular determinants, which support migratory and invasive potential of colon cancer cell, may provide new therapeutic option. Our previous published data on various cancers show that chemokines and their corresponding receptor interactions mediate cell migration and invasion (Singh et al, 2004a, 2004b, 2009a; Johnson et al, 2010; Johnson-Holiday et al, 2011; Gupta et al, 2014; Mir et al, 2015). Hence, our data showing higher migratory and invasive potential of colon cancer cells derived from node-positive and metastatic colon cancer in response to chemotactic gradient of CCL20 suggests a role of CCR6–CCL20 axis in colon cancer cell migration, and emphasises the potential of CCR6 as a therapeutic target.
In conclusion, higher expression of CCR6 in colon cancer tissues and cell lines compared with normal adjacent tissue and normal colon cell, as well as association of expression status with nodal involvement and distant metastasis implies its important role in colon cancer progression. Activation of EMT markers and inhibition of colon cancer cell proliferation following CCL20 stimulation, and high migratory and invasive potential towards CCL20 gradient implicates association of CCR6–CCL20 with colon cancer progression. Overall these cellular and molecular process affected by CCR6–CCL20 are involved in cancer progression and poor therapeutic response. Hence, inclusion of CCR6 expression status with TNM may serve as a better predictor of disease and therapeutic outcome than TNM grading alone; although this would require validation in larger sample size. In addition to this our study provides rationale for CCR6-directed drug designing against this disease.
Change history
14 June 2016
This paper was modified 12 months after initial publication to switch to Creative Commons licence terms, as noted at publication
References
Baba M, Imai T, Nishimura M, Kakizaki M, Takagi S, Hieshima K, Nomiyama H, Yoshie O (1997) Identification of CCR6, the specific receptor for a novel lymphocyte-directed CC chemokine LARC. J Biol Chem 272 (23): 14893–14898.
Balkwill FR (2012) The chemokine system and cancer. J Pathol 226 (2): 148–157.
Batlle E, Sancho E, Franci C, Dominguez D, Monfar M, Baulida J, Garcia De Herreros A (2000) The transcription factor snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat Cell Biol 2 (2): 84–89.
Baxter NN, Virnig DJ, Rothenberger DA, Morris AM, Jessurun J, Virnig BA (2005) Lymph node evaluation in colorectal cancer patients: a population-based study. J Natl Cancer Inst 97 (3): 219–225.
Blanco MJ, Moreno-Bueno G, Sarrio D, Locascio A, Cano A, Palacios J, Nieto MA (2002) Correlation of Snail expression with histological grade and lymph node status in breast carcinomas. Oncogene 21 (20): 3241–3246.
Cano A, Perez-Moreno MA, Rodrigo I, Locascio A, Blanco MJ, del Barrio MG, Portillo F, Nieto MA (2000) The transcription factor snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat Cell Biol 2 (2): 76–83.
Daugherty RL, Gottardi CJ (2007) Phospho-regulation of Beta-catenin adhesion and signaling functions. Physiology 22: 303–309.
Dellacasagrande J, Schreurs OJ, Hofgaard PO, Omholt H, Steinsvoll S, Schenck K, Bogen B, Dembic Z (2003) Liver metastasis of cancer facilitated by chemokine receptor CCR6. Scand J Immunol 57 (6): 534–544.
Dong C, Wu Y, Yao J, Wang Y, Yu Y, Rychahou PG, Evers BM, Zhou BP (2012) G9a interacts with Snail and is critical for Snail-mediated E-cadherin repression in human breast cancer. J Clin Invest 122 (4): 1469–1486.
El-Haibi CP, Singh R, Sharma PK, Singh S, Lillard JW Jr (2011) CXCL13 mediates prostate cancer cell proliferation through JNK signalling and invasion through ERK activation. Cell Prolif 44 (4): 311–319.
Evdokimova V, Tognon C, Ng T, Sorensen PH (2009) Reduced proliferation and enhanced migration: two sides of the same coin? Molecular mechanisms of metastatic progression by YB-1. Cell Cycle 8 (18): 2901–2906.
Frick VO, Rubie C, Kolsch K, Wagner M, Ghadjar P, Graeber S, Glanemann M (2013) CCR6/CCL20 chemokine expression profile in distinct colorectal malignancies. Scand J Immunol 78 (3): 298–305.
Ghadjar P, Coupland SE, Na IK, Noutsias M, Letsch A, Stroux A, Bauer S, Buhr HJ, Thiel E, Scheibenbogen C, Keilholz U (2006) Chemokine receptor CCR6 expression level and liver metastases in colorectal cancer. J Clin Oncol 24 (12): 1910–1916.
Ghadjar P, Loddenkemper C, Coupland SE, Stroux A, Noutsias M, Thiel E, Christoph F, Miller K, Scheibenbogen C, Keilholz U (2008) Chemokine receptor CCR6 expression level and aggressiveness of prostate cancer. J Cancer Res Clin Oncol 134 (11): 1181–1189.
Gottardi CJ, Wong E, Gumbiner BM (2001) E-cadherin suppresses cellular transformation by inhibiting beta-catenin signalling in an adhesion-independent manner. J Cell Biol 153 (5): 1049–1060.
Guaita S, Puig I, Franci C, Garrido M, Dominguez D, Batlle E, Sancho E, Dedhar S, De Herreros AG, Baulida J (2002) Snail induction of epithelial to mesenchymal transition in tumor cells is accompanied by MUC1 repression and ZEB1 expression. J Biol Chem 277 (42): 39209–39216.
Gupta P, Sharma PK, Mir H, Singh R, Singh N, Kloecker GH, Lillard JW Jr, Singh S (2014) CCR9/CCL25 expression in non-small cell lung cancer correlates with aggressive disease and mediates key steps of metastasis. Oncotarget 5 (20): 10170–10179.
Harper EG, Guo C, Rizzo H, Lillis JV, Kurtz SE, Skorcheva I, Purdy D, Fitch E, Iordanov M, Blauvelt A (2009) Th17 cytokines stimulate CCL20 expression in keratinocytes in vitro and in vivo: implications for psoriasis pathogenesis. J Invest Dermatol 129 (9): 2175–2183.
Hirota K, Yoshitomi H, Hashimoto M, Maeda S, Teradaira S, Sugimoto N, Yamaguchi T, Nomura T, Ito H, Nakamura T, Sakaguchi N, Sakaguchi S (2007) Preferential recruitment of CCR6-expressing Th17 cells to inflamed joints via CCL20 in rheumatoid arthritis and its animal model. J Exp Med 204 (12): 2803–2812.
Jeanes A, Gottardi CJ, Yap AS (2008) Cadherins and cancer: how does cadherin dysfunction promote tumor progression? Oncogene 27 (55): 6920–6929.
Johnson EL, Singh R, Singh S, Johnson-Holiday CM, Grizzle WE, Partridge EE, Lillard JW Jr (2010) CCL25-CCR9 interaction modulates ovarian cancer cell migration, metalloproteinase expression, and invasion. World J Surg Oncol 8: 62.
Johnson-Holiday C, Singh R, Johnson E, Singh S, Stockard CR, Grizzle WE, Lillard JW Jr (2011) CCL25 mediates migration, invasion and matrix metalloproteinase expression by breast cancer cells in a CCR9-dependent fashion. Int J Oncol 38 (5): 1279–1285.
Kakinuma T, Hwang ST (2006) Chemokines, chemokine receptors, and cancer metastasis. J Leukoc Biol 79 (4): 639–651.
Kaser A, Ludwiczek O, Holzmann S, Moschen AR, Weiss G, Enrich B, Graziadei I, Dunzendorfer S, Wiedermann CJ, Murzl E, Grasl E, Jasarevic Z, Romani N, Offner FA, Tilg H (2004) Increased expression of CCL20 in human inflammatory bowel disease. J Clin Immunol 24 (1): 74–85.
Kim K, Lu Z, Hay ED (2002) Direct evidence for a role of beta-catenin/LEF-1 signaling pathway in induction of EMT. Cell Biol Int 26 (5): 463–476.
Kim MA, Lee HS, Lee HE, Kim JH, Yang HK, Kim WH (2009) Prognostic importance of epithelial-mesenchymal transition-related protein expression in gastric carcinoma. Histopathology 54 (4): 442–451.
Kimsey TF, Campbell AS, Albo D, Wilson M, Wang TN (2004) Co-localization of macrophage inflammatory protein-3alpha (Mip-3alpha) and its receptor, CCR6, promotes pancreatic cancer cell invasion. Cancer J 10 (6): 374–380.
Kirshberg S, Izhar U, Amir G, Demma J, Vernea F, Beider K, Shlomai Z, Wald H, Zamir G, Shapira OM, Peled A, Wald O (2011) Involvement of CCR6/CCL20/IL-17 axis in NSCLC disease progression. PloS one 6 (9): e24856.
Kuper H, Adami HO, Trichopoulos D (2000) Infections as a major preventable cause of human cancer. J Intern Med 248 (3): 171–183.
Lambrechts A, Van Troys M, Ampe C (2004) The actin cytoskeleton in normal and pathological cell motility. Int J Biochem Cell Biol 36 (10): 1890–1909.
Lee HW, Park YM, Lee SJ, Cho HJ, Kim DH, Lee JI, Kang MS, Seol HJ, Shim YM, Nam DH, Kim HH, Joo KM (2013) Alpha-smooth muscle actin (ACTA2) is required for metastatic potential of human lung adenocarcinoma. Clin Cancer Res 19 (21): 5879–5889.
Lee HY, Choi HJ, Park KJ, Shin JS, Kwon HC, Roh MS, Kim C (2007) Prognostic significance of metastatic lymph node ratio in node-positive colon carcinoma. Ann Surg Oncol 14 (5): 1712–1717.
Lee JM, Dedhar S, Kalluri R, Thompson EW (2006) The epithelial-mesenchymal transition: new insights in signaling, development, and disease. J Cell Biol 172 (7): 973–981.
Lin Y, Dong C, Zhou BP (2014) Epigenetic regulation of EMT: the Snail story. Curr Pharm Des 20 (11): 1698–1705.
Liu S, Cong Y, Wang D, Sun Y, Deng L, Liu Y, Martin-Trevino R, Shang L, McDermott SP, Landis MD, Hong S, Adams A, D'Angelo R, Ginestier C, Charafe-Jauffret E, Clouthier SG, Birnbaum D, Wong ST, Zhan M, Chang JC, Wicha MS (2014) Breast cancer stem cells transition between epithelial and mesenchymal states reflective of their normal counterparts. Stem Cell Rep 2 (1): 78–91.
Liu Y, El-Naggar S, Darling DS, Higashi Y, Dean DC (2008) Zeb1 links epithelial-mesenchymal transition and cellular senescence. Development 135 (3): 579–588.
McInroy L, Maatta A (2007) Down-regulation of vimentin expression inhibits carcinoma cell migration and adhesion. Biochem Biophys Res Commun 360 (1): 109–114.
Mir H, Singh R, Kloecker GH, Lillard JW Jr, Singh S (2015) CXCR6 expression in non-small cell lung carcinoma supports metastatic process via modulating metalloproteinases. Oncotarget 6 (12): 9985–9998.
Morris EJ, Maughan NJ, Forman D, Quirke P (2007) Who to treat with adjuvant therapy in Dukes B/stage II colorectal cancer? The need for high quality pathology. Gut 56 (10): 1419–1425.
Olmeda D, Moreno-Bueno G, Flores JM, Fabra A, Portillo F, Cano A (2007) SNAI1 is required for tumor growth and lymph node metastasis of human breast carcinoma MDA-MB-231 cells. Cancer Res 67 (24): 11721–11731.
Peinado H, Olmeda D, Cano A (2007) Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat Rev Cancer 7 (6): 415–428.
Pena C, Garcia JM, Silva J, Garcia V, Rodriguez R, Alonso I, Millan I, Salas C, de Herreros AG, Munoz A, Bonilla F (2005) E-cadherin and vitamin D receptor regulation by SNAIL and ZEB1 in colon cancer: clinicopathological correlations. Hum Mol Genet 14 (22): 3361–3370.
Reboldi A, Coisne C, Baumjohann D, Benvenuto F, Bottinelli D, Lira S, Uccelli A, Lanzavecchia A, Engelhardt B, Sallusto F (2009) C-C chemokine receptor 6-regulated entry of TH-17 cells into the CNS through the choroid plexus is required for the initiation of EAE. Nat Immunol 10 (5): 514–523.
Rubie C, Oliveira V, Kempf K, Wagner M, Tilton B, Rau B, Kruse B, Konig J, Schilling M (2006) Involvement of chemokine receptor CCR6 in colorectal cancer metastasis. Tumour Biol 27 (3): 166–174.
Sarli L, Bader G, Iusco D, Salvemini C, Mauro DD, Mazzeo A, Regina G, Roncoroni L (2005) Number of lymph nodes examined and prognosis of TNM stage II colorectal cancer. Eur J Cancer 41 (2): 272–279.
Satelli A, Li S (2011) Vimentin in cancer and its potential as a molecular target for cancer therapy. Cell Mol Life Sci 68 (18): 3033–3046.
Scott JA, Yap AS (2006) Cinderella no longer: alpha-catenin steps out of cadherin's shadow. J Cell Sci 119 (Pt 22): 4599–4605.
Siegel RL, Miller KD, Jemal A (2015) Cancer statistics, 2015. CA Cancer J Clin 65 (1): 5–29.
Singh R, Stockard CR, Grizzle WE, Lillard JW Jr, Singh S (2011) Expression and histopathological correlation of CCR9 and CCL25 in ovarian cancer. Int J Oncol 39 (2): 373–381.
Singh S, Singh R, Sharma PK, Singh UP, Rai SN, Chung LW, Cooper CR, Novakovic KR, Grizzle WE, Lillard JW Jr (2009a) Serum CXCL13 positively correlates with prostatic disease, prostate-specific antigen and mediates prostate cancer cell invasion, integrin clustering and cell adhesion. Cancer Lett 283 (1): 29–35.
Singh S, Singh R, Singh UP, Rai SN, Novakovic KR, Chung LW, Didier PJ, Grizzle WE, Lillard JW Jr (2009b) Clinical and biological significance of CXCR5 expressed by prostate cancer specimens and cell lines. Int J Cancer 125 (10): 2288–2295.
Singh S, Singh UP, Grizzle WE, Lillard JW Jr (2004a) CXCL12-CXCR4 interactions modulate prostate cancer cell migration, metalloproteinase expression and invasion. Lab Invest 84 (12): 1666–1676.
Singh S, Singh UP, Stiles JK, Grizzle WE, Lillard JW Jr (2004b) Expression and functional role of CCR9 in prostate cancer cell migration and invasion. Clin Cancer Res 10 (24): 8743–8750.
Spoelstra NS, Manning NG, Higashi Y, Darling D, Singh M, Shroyer KR, Broaddus RR, Horwitz KB, Richer JK (2006) The transcription factor ZEB1 is aberrantly expressed in aggressive uterine cancers. Cancer Res 66 (7): 3893–3902.
Stockinger A, Eger A, Wolf J, Beug H, Foisner R (2001) E-cadherin regulates cell growth by modulating proliferation-dependent beta-catenin transcriptional activity. J Cell Biol 154 (6): 1185–1196.
Thiery JP, Acloque H, Huang RY, Nieto MA (2009) Epithelial-mesenchymal transitions in development and disease. Cell 139 (5): 871–890.
van Roy F, Berx G (2008) The cell-cell adhesion molecule E-cadherin. Cell Mol Life Sci 65 (23): 3756–3788.
Wang S, Liu Z, Wang L, Zhang X (2009) NF-kappaB signaling pathway, inflammation and colorectal cancer. Cell Mol Immunol 6 (5): 327–334.
Wei J, Xu G, Wu M, Zhang Y, Li Q, Liu P, Zhu T, Song A, Zhao L, Han Z, Chen G, Wang S, Meng L, Zhou J, Lu Y, Wang S, Ma D (2008) Overexpression of vimentin contributes to prostate cancer invasion and metastasis via src regulation. Anticancer Res 28 (1a): 327–334.
Witta SE, Gemmill RM, Hirsch FR, Coldren CD, Hedman K, Ravdel L, Helfrich B, Dziadziuszko R, Chan DC, Sugita M, Chan Z, Baron A, Franklin W, Drabkin HA, Girard L, Gazdar AF, Minna JD, Bunn PA Jr (2006) Restoring E-cadherin expression increases sensitivity to epidermal growth factor receptor inhibitors in lung cancer cell lines. Cancer Res 66 (2): 944–950.
Wyler L, Napoli CU, Ingold B, Sulser T, Heikenwalder M, Schraml P, Moch H (2014) Brain metastasis in renal cancer patients: metastatic pattern, tumour-associated macrophages and chemokine/chemoreceptor expression. Br J Cancer 110 (3): 686–694.
Yamazaki T, Yang XO, Chung Y, Fukunaga A, Nurieva R, Pappu B, Martin-Orozco N, Kang HS, Ma L, Panopoulos AD, Craig S, Watowich SS, Jetten AM, Tian Q, Dong C (2008) CCR6 regulates the migration of inflammatory and regulatory T cells. J Immunol 181 (12): 8391–8401.
Yokoyama K, Kamata N, Hayashi E, Hoteiya T, Ueda N, Fujimoto R, Nagayama M (2001) Reverse correlation of E-cadherin and snail expression in oral squamous cell carcinoma cells in vitro. Oral Oncol 37 (1): 65–71.
Zhao Y, Yan Q, Long X, Chen X, Wang Y (2008) Vimentin affects the mobility and invasiveness of prostate cancer cells. Cell Biochem Funct 26 (5): 571–577.
Zhu QS, Rosenblatt K, Huang KL, Lahat G, Brobey R, Bolshakov S, Nguyen T, Ding Z, Belousov R, Bill K, Luo X, Lazar A, Dicker A, Mills GB, Hung MC, Lev D (2011) Vimentin is a novel AKT1 target mediating motility and invasion. Oncogene 30 (4): 457–470.
Acknowledgements
The content of this manuscript benefited from many fruitful conversations with members of the Morehouse School of Medicine, Atlanta, GA, USA. This study was supported in part by the funds (R21 CA169716, SC1 CA180212, U54 CA118638 and UO1 CA179701) from NCI and Morehouse School of Medicine flow cytometry core (G12 MD007602). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author contributions
NK conducted experiments, analysed data and drafted the manuscript. HM assisted in flow cytometry analysis and manuscript preparation. UK validated the immunohistochemistry results and edited the manuscript. CEC, III and DJB provided their clinical inputs in manuscript preparation and edited the manuscript. JWL, Jr provided his expertise in manuscript preparation. SS conceptualised the work, developed study design, supervised the authors throughout the study and manuscript preparation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License.
Rights and permissions
From twelve months after its original publication, this work is licensed under the Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/4.0/
About this article
Cite this article
Kapur, N., Mir, H., Clark III, C. et al. CCR6 expression in colon cancer is associated with advanced disease and supports epithelial-to-mesenchymal transition. Br J Cancer 114, 1343–1351 (2016). https://doi.org/10.1038/bjc.2016.113
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/bjc.2016.113
Keywords
This article is cited by
-
Chemokine Receptor Antagonists: Role in Oncology
Current Oncology Reports (2021)
-
MBD1 promotes the malignant behavior of gallbladder cancer cells and induces chemotherapeutic resistance to gemcitabine
Cancer Cell International (2019)
-
Altered T Cell Migratory Capacity in the Progression from Barrett Oesophagus to Oesophageal Adenocarcinoma
Cancer Microenvironment (2019)
-
Bone marrow mesenchymal stromal cells attenuate silica-induced pulmonary fibrosis potentially by attenuating Wnt/β-catenin signaling in rats
Stem Cell Research & Therapy (2018)