Abstract
Background:
PTEN gene loss occurs frequently in castration-resistant prostate cancer (CRPC) and may drive progression through activation of the PI3K/AKT pathway. Here, we developed a novel CTC-based assay to determine PTEN status and examined the correlation between PTEN status in CTCs and matched tumour tissue samples.
Methods:
PTEN gene status in CTCs was evaluated on an enrichment-free platform (Epic Sciences) by fluorescence in situ hybridisation (FISH). PTEN status in archival and fresh tumour tissue was evaluated by FISH and immunohistochemistry.
Results:
Peripheral blood was collected from 76 patients. Matched archival and fresh cancer tissue was available for 48 patients. PTEN gene status detected in CTCs was concordant with PTEN status in matched fresh tissues and archival tissue in 32 of 38 patients (84%) and 24 of 39 patients (62%), respectively. CTC counts were prognostic (continuous, P=0.001). PTEN loss in CTCs associated with worse survival in univariate analysis (HR 2.05; 95% CI 1.17–3.62; P=0.01) and with high lactate dehydrogenase (LDH) in metastatic CRPC patients.
Conclusions:
Our results illustrate the potential use of CTCs as a non-invasive, real-time liquid biopsy to determine PTEN gene status. The prognostic and predictive value of PTEN in CTCs warrants investigation in CRPC clinical trials of PI3K/AKT-targeted therapies.
Similar content being viewed by others
Main
The PTEN/PI3K/AKT pathway is frequently activated in prostate cancer and appears to be critical for tumour cell survival and proliferation (Sarker et al, 2009). One of the most common molecular events leading to activation of this pathway is loss of the tumour-suppressor phosphate and tensin homolog (PTEN) (Chow and Baker, 2006). PTEN loss is common in prostate cancer (up to 50% of cases) and is associated with poor prognosis (McMenamin et al, 1999; Halvorsen et al, 2003; Verhagen et al, 2006; Yoshimoto et al, 2006; McCall et al, 2008; Reid et al, 2010; Leinonen et al, 2013). The PTEN/PI3K/AKT pathway also has a key role in the regulation of androgen receptor (AR) signalling, and in the development of resistance to hormonal treatment in preclinical prostate cancer models (Lin et al, 2004; Carver et al, 2011; Chen et al, 2013). Strategies that combine novel AR-targeting drugs (i.e., enzalutamide and abiraterone acetate) with PI3K/AKT pathway inhibitors may be more effective than single AR-targeting agents, and could delay or reverse resistance (Carver et al, 2011; Thomas et al, 2013; Toren et al, 2014). The success of clinical studies testing these novel strategies may rely on the molecular characterisation of patients to identify the presence of aberrations in the PI3K/AKT pathway. However, characterisation of castration-resistant prostate cancer (CRPC) disease in clinical studies is challenging, first because of disease heterogeneity; second, because of the high frequency of metastases that are exclusive to the bone and difficult to reach, which may hinder the accrual of patients.
Studies have validated the prognostic significance of circulating tumour cell (CTC) counts in several tumour types, including breast, colorectal and prostate cancer (Cristofanilli et al, 2004; Cohen et al, 2008; de Bono et al, 2008). In addition to the prognostic value, CTCs represent a promising approach to characterise the molecular profile of metastatic CRPC (Attard et al, 2009; Miyamoto et al, 2012; Heitzer et al, 2013). Biomarker characterisation in CTCs could provide insights into the most current state of the disease and an overall picture of intra-tumour heterogeneity. As a non-invasive methodology, CTCs can be evaluated in longitudinal blood draws to monitor in real-time the effect of antitumour therapies. CTCs can also be potentially used as pharmacodynamic endpoints for the clinical development of novel therapies (Yap et al, 2014).
In this study, we report the development and characterisation of a blood-based PTEN FISH assay that will be evaluated in combination trials of abiraterone and PI3K/AKT inhibitors for patients with CRPC (TRIAL GOV NCT01485861). We identified and characterised CTCs using an enrichment-free platform (Epic Sciences). We found that PTEN gene status in CTCs correlated well with PTEN gene status in patient-matched fresh CRPC tissue, and loss of PTEN in CTCs was associated with poorer clinical outcome in univariate analysis in metastatic CRPC.
Materials and methods
Patients and samples
Peripheral blood samples were obtained from 76 patients with metastatic CRPC treated at the Royal Marsden NHS Foundation Trust (London, UK) between March 2012 and February 2013 (Table 1). All patients had received androgen-deprivation therapy with either surgical castration or a luteinising hormone-releasing hormone agonist, and had disease progression according to Prostate Cancer Working Group 2 consensus criteria (Scher et al, 2008).
Prostate cancer tissue was obtained from prostate needle biopsies, transurethral resections of the prostate, or prostatectomies and prostate cancer metastases within bone (bone marrow trephine), lymph node or viscera (needle biopsies). Formalin-fixed and paraffin-embedded tissue blocks were sectioned and reviewed by a pathologist for confirmation of the adequacy of the material. Tumour content was estimated by haematoxylin-eosin staining on 2-μm slices and adjacent slices were used for biomarker analysis.
All patients gave written informed consent and were enrolled on institutional protocol approved by the Royal Marsden NHS Foundation Trust (London, UK) Ethics Review Committees (Reference number: 04/Q0801/60). Demographics and clinical data were retrospectively collected from the hospital electronic record system (EPR).
Circulating tumour cell enumeration
Epic CTC platform
Peripheral blood sample was collected in Cell-free DNA BCT (Streck, Omaha, NE, USA) and shipped immediately to Epic Sciences (San Diego, CA, USA) at ambient temperature. The median blood sample transit time was 32 h (range 29–105 h). Upon receipt, red blood cells were lysed and nucleated cells were dispensed onto glass microscope slides as previously described (Marrinucci et al, 2007; Marrinucci et al, 2009; Mikolajczyk et al, 2011; Marrinucci et al, 2012; Werner et al, 2015) and stored at −80 °C until staining. Up to 16 slides were prepared from each blood sample, with three million nucleated cells deposited onto each slide. The millilitre equivalent of blood plated per slide was calculated based upon the sample’s white blood cell count and the volume of post-RBC lysis cell suspension used.
Circulating tumour cells were identified by immunofluorescence, as described (Marrinucci et al, 2007; Marrinucci et al, 2009; Mikolajczyk et al, 2011; Marrinucci et al, 2012; Werner et al, 2015). Slides were stained for CK (Pan CK 1°Ab (Sigma, St Louis, MO, USA) and CK-19 1°Ab (Dako, Carpinteria, CA, USA) labelled with AlexaFluor555-conjugated anti-mouse IgG1 2°Ab (Life Technologies, Carlsbad, CA, USA)), CD45 (AlexaFluor647-preconjugated (Serotec, Raleigh, NC, USA)), N-terminal AR (1° Ab (Cell Signaling, Danvers, MA, USA) labelled with AlexaFluor488-conjugated anti-rabbit IgG(H+L) 2° Ab (Life Technologies)) and DAPI (Life Technologies). Stained slides were imaged in four channels on a high-speed fluorescent imaging system. Circulating tumour cell candidates were identified by image analysis using an automated algorithm to characterise each of the three million nucleated cells per slide by over 90 different parameters, including protein expression and morphology. Circulating tumour cell candidates were then reviewed by trained technicians blinded to the clinical data. CK+/CD45− cells with intact DAPI+ nuclei exhibiting tumour-associated morphologies were classified as CTCs.
A range of 2–14 slides were tested per patient to evaluate CTC enumeration and biomarker expression levels (protein and DNA markers). Total CTC counts were normalised to CTC per ml by computing the total number of CTCs enumerated over all slides tested, divided by the total volume of blood tested. For this patient cohort, the blood volumes tested ranged from 0.7 to 7.8 ml blood, with a median of 2.7 ml and mean of 3.3 ml (Supplementary Table S6). To compare with CellSearch, Epic CTC enumeration was extrapolated to 7.5 ml by multiplying Epic’s CTC count × 7.5. Given the differences between the volumes evaluated on the Epic and CellSearch platforms as part of their methodologies, a comparison is not possible without extrapolation and therefore carries some bias.
CellSearch System
In parallel to blood collection for CTC analysis on the Epic platform, blood samples were collected in CellSave preservative tubes (Jansen Diagnostics, LLC, NJ, USA) for CTC enumeration on the FDA-approved CellSearch System according to manufacturer’s instructions. The captured images were manually examined by a trained and certified operator. The number of CellSearch CTCs is presented per 7.5 ml of blood.
Molecular characterisation of CTCs on the Epic CTC platform
PTEN FISH analysis on CTCs
Coverslips were removed from slides, and cells were fixed and dehydrated with formaldehyde and ethanol. A Cymogen Dx 2-color probe targeting PTEN and chromosome 10 centromeres (CEP10) was applied across the entire deposition area of each slide and the slides were then coverslipped, denatured and hybridised for 18–24 h at 37 °C. Slides were then washed in saline sodium citrate/Igepal solutions, counterstained with DAPI, and mounted with an anti-fade mounting medium. Circulating tumour cells were relocated through Epic’s software and scored for PTEN FISH. For an internal control, 20 WBCs were evaluated for PTEN on every slide tested.
Each CTC evaluated by FISH was classified according to the number of FISH signals as: PTEN=0 (PTEN=0 and CEP10 ⩾1), PTEN=1 (PTEN=1 and CEP10⩾1), or PTEN⩾2 (PTEN⩾2 and CEP10⩾1). The false positive rates of PTEN loss in WBCs (PTEN=0, PTEN=1, Figure 1C WBCs) and the total number of CTCs were used to establish the criteria to classify the PTEN status of patients. A Homozygous (HO) or Hemizygous (HE) PTEN loss was classified if the 95% confidence interval (CI) on the percentage of PTEN=0 or PTEN=1 in patient samples excluded the 95% CI of PTEN=0 or PTEN=1 in WBCs (Supplementary Table S1). Following these criteria, for example, PTEN=0 was never detected in over 1000 WBCs evaluated, therefore the observation of any number of CTCs exhibiting PTEN=0 and CEP10⩾1 in a patient sample (with at least two evaluable CTCs) was considered significant, and the patient was classified as HO PTEN loss. If additional analysis was required to achieve statistical significance, two additional slides were stained for CTC identification and determination of PTEN status. A second blood sample was requested for PTEN FISH analysis in patients with insufficient number of CTCs in the first blood draw (n=12).
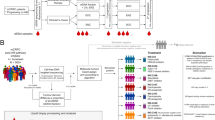
(A, B). Circulating tumour cell capture and FISH analysis in CRPC patients. (A) Flowchart of CTC capture, identification and molecular analysis on the Epic platform (see Methods). (B) Distribution of CTC counts in CRPC patient. Bars represent number of patients with CTC enumeration corresponding to the groups (n=76 patients). Percentages within bars represent the percentage of patients with CTC enumeration corresponding to the groups. Bottom table, % patients that showed successful PTEN evaluation in CTCs. (C, D) Distinct distribution of PTEN and CEP10 genotypes in CTCs and WBCs. (C) Frequencies of CEP10 and PTEN genotypes in CTCs and WBCs. The frequency of every PTEN and CEP10 genotype combination was calculated within each patient and averaged across all patients to generate weighted frequency distributions. x, number of CEP10 signals, y, number of PTEN signals per cell. (D) Representative images of PTEN and CEP10 genotypes in patient CTCs. Upper panel, CTC identification by immunofluorescence. Blue: DAPI, red: CK, green: CD45. Lower panel, PTEN FISH analysis in the CTCs identified above. Green: CEP10 signals, red: PTEN signals, blue: DAPI. Red frame: CTCs with PTEN=0; yellow frame: CTCs with PTEN=1; grey frame: CTCs with PTEN ⩾2.
ERG FISH analysis on CTCs
Two additional slides were assessed for CTC identification and ERG FISH. ERG rearrangements (ERG translocation/split and ERG deletion) were assayed using FISH probes (Cymogen Dx) targeting 3’ERG and 5’ERG. ERG status in CTCs was determined by relative locations of 3’ERG and 5’ERG signals. Cells were classified as ERG-translocation/split if at least one pair of 3’ERG and 5’ERG signals were separated by a distance >2 signal diameters, and ERG deleted if at least one 5’ERG signal was deleted. Cells in which all 3’ERG signals had a corresponding 5’ERG signal within 2 signal diameters were considered ERG-normal. Patient ERG status was classified as ‘ERG rearranged’ if ERG split and/or ERG deletions were present above the cut-offs determined by the classification rules that exclude the 95% CI of ERG rearrangement in WBC (Supplementary Table S2). If additional analysis was required to achieve statistical significance, additional slides were tested for CTC identification and FISH. For internal control, 20 WBCs per slide were evaluated for ERG FISH.
Androgen receptor analysis on CTCs
Androgen receptor was analysed on a subset of patients following staining with the 4-color immunofluorescence assay described previously. Androgen receptor was quantified, using custom image analysis algorithms, as average signal-over-background across the cells of interest. Subcellular localisation of AR was determined by manual visual analysis, classifying each AR+ cell as nuclear (N), cytoplasmic (C) or mixed (NC). Representative images are shown in Supplementary Figure S6. Healthy donor blood samples spiked with AR+ and AR− prostate cancer cell lines (VCaP and PC3, respectively) were analysed in parallel to confirm expected signal intensity and protein localisation.
Tissue FISH analysis
A PTEN-specific probe was developed in-house using a bacterial artificial chromosome as described by Sircar et al (2009), with slight modifications. Labelled DNA from bacterial artificial chromosome RP11-846G17 (Invitrogen, Carlsbad, CA, USA) spanning the PTEN loci at cytoband 10q23.31 (89.6–89.8 Mb) was used to evaluate the PTEN copy number in patient samples. The bacterial artificial chromosome covering the PTEN locus was same as the one used by Sircar et al. A CEP10 centromere probe (Abbott Molecular, Abbott Park, IL, USA) was used as a chromosome 10 control probe. FISH analysis was performed on 4-μM tissue sections, with minor modifications from previously described studies (O'Brien et al, 2008; Modrek et al, 2009).
To establish cut-offs for PTEN analysis, five normal prostate formalin-fixed and paraffin-embedded tissue sections (non-tumour samples from urology clinics) were scored for PTEN, and the % of cells exhibiting PTEN loss was computed. On the basis of the hybridisation in these normal cells, samples were classified as HE PTEN deletion when ⩾30% (mean±3 s.d.) tumour nuclei contained only one PTEN signal and ⩾1 CEP10 signals. Similarly, samples were classified as HO deletion of PTEN if ⩾30% tumour nuclei exhibited ⩾1 CEP10 signal, but lacked any PTEN signal. Minimums of 150 non-overlapping nuclei from areas across the tumour were analysed in each sample.
PTEN immunohistochemistry (IHC)
PTEN protein expression was determined by IHC on 4-uM thick formalin-fixed and paraffin-embedded sections as previously described (Reid et al, 2012; Sandhu et al, 2013; Ferraldeschi et al, 2014). Nuclear and cytoplasmic PTEN staining intensity were semi-quantitatively assessed using the H-score formula: 3 × percentage of strongly staining cells+2 × percentage of moderately staining cells+percentage of weakly staining cells, giving a range of 0 to 300 (Ishibashi et al, 2003). An H-score >10 was considered positive for PTEN protein expression (Ferraldeschi et al, 2014). The IHC sections were evaluated by a pathologist (DNR), who was blinded to the FISH status and clinical information.
Statistical analysis
Positive predictive value, specificity and sensitivity were calculated based on the classification of patients for PTEN status from CTCs, fresh and archival tissue. Patients were included in this comparison only if they had conclusive FISH results in CTCs and tissue. The Kaplan–Meier product-limit method was used to estimate the duration of overall survival from time of blood draw. Patients still alive or lost to follow-up were censored. The survival rates were compared using the log-rank test. Univariate and multivariate analyses of the independent factors for overall survival was performed using the Cox proportional hazard regression model with a 95% CI. Independent sample t-tests and Fisher’s exact tests were used to compare continuous and categorical variables, respectively. All tests were two-sided and a P-value of 0.05 or less was considered statistically significant. Descriptive statistics and survival analyses were performed using IBM SPSS Statistics v22 (IBM, Armonk, NY, USA).
Results
Fluorescence in situ hybridisation analysis reveals changes in ploidy and heterogeneous PTEN and CEP10 genotypes in CTCs
Peripheral blood samples were collected from 76 CRPC patients; of these, 20 were chemotherapy-naïve and 56 were post-docetaxel at the time of study entry. All patients had metastatic disease. Patient characteristics are detailed in Table 1. The PTEN status of CTCs was evaluated using a FISH assay developed on the Epic platform (Figure 1A). Circulating tumour cells were detected in 68 of 76 patients (89%); CTC enumeration was higher than 3 CTCs per ml in 59 of 76 patients (78%, Figure 1B). Assessment of CTC counts in replicate slides demonstrated that enumeration is reproducible on the Epic CTC platform, with very tight counts in duplicate samples with high CTC numbers, and more dispersion in duplicate samples with low CTC numbers (n=89 blood samples, Pearson r=0.99; P<0.0001, Supplementary Figure S1). PTEN FISH was performed on a total of 2631 CTCs and in over 1000 WBCs. Following FISH, 85% of CTCs identified by immunofluorescence were retained on the slides; 74% of retained CTCs showed evaluable FISH signals and were scored for PTEN and the centromeric marker CEP10. In the WBC population, PTEN and CEP10 genotypes distributed narrowly around the diploid wild-type genotype, with 91.7% of WBCs showing two PTEN and two CEP10 signals (Figure 1C WBCs). In contrast, CTCs showed a markedly broader distribution of PTEN and CEP10 genotypes, with only 61% of tumour cells showing the diploid wild-type of two PTEN and two CEP10 signals (Figure 1C CTCs). Twenty-five percent of all CTCs exhibited increased CEP10 copy number (range 3–14 CEP10 signals) compared with 1.38% in WBCs (Figure 1C and D, images ii, v–vii), which is suggestive of the malignant origin of cells defined as CTCs in this assay. PTEN loss was infrequent in WBCs, with no cases of complete PTEN loss observed (PTEN=0) and low frequency of cells with only one copy of PTEN (4.1% WBCs with PTEN=1, Figure 1C WBCs, sum of yellow boxes). Conversely, complete PTEN loss occurred at a frequency of 12% in CTC populations (PTEN=0, Figure 1C CTCs sum of red boxes, and Figure 1D images i and ii). On average, 8% of CTCs exhibited one copy of PTEN (Figure 1C CTCs sum of yellow boxes and Figure 1D, images iii–v), which was often accompanied by increased CEP10 signals, a combination never detected in WBCs (Figure 1C, yellow boxes with CEP10⩾3). Analysis of 400 WBCs from 10 healthy donors revealed PTEN and CEP10 distributions similar to those captured in WBCs from cancer patients, with no complete PTEN loss detected and only 0.75% cells with increased CEP10 signal (CEP10⩾3), validating the use of WBCs as internal controls and further supporting the clear genotypic distinction between CTCs and WBCs (Supplementary Figure S2).
PTEN FISH status in CTCs correlated strongly with PTEN status in fresh tumour tissue
Next, we compared PTEN status in CTCs and patient-matched archival and fresh tumour tissue samples. Of the 76 patients with blood draws, 48 had matched tissue samples (Supplementary Table S3). The median interval between archival tissue collection and blood draw was 5.1 years (interquartile range 2.4–8.9 years) and between fresh tissue collection and CTC blood draw, 0 weeks (interquartile range 0–1 week).
For each patient, PTEN was analysed in all CTCs that were retained after FISH. The number of PTEN signals in CTCs of a patient sample was sometimes heterogeneous, with a mixture of CTCs containing 0, 1 or ⩾2 PTEN signals (Figure 2A, top panel). We defined the PTEN status of a patient using a classification system based on the distinct distributions of PTEN and CEP10 genotypes in CTCs vs WBCs (Figure 1C). The system takes into account the number and percentage of CTCs with PTEN=0, PTEN=1 and PTEN⩾2 to classify the patients’ PTEN status as HO loss, HE loss or No-deletion of PTEN (Methods and Supplementary Table S1). Using this system, all patients with enumeration higher than 10 CTCs per ml, and 74% of patients with 3–10 CTCs per ml were successfully classified (Figure 1B, bottom table). Applying the classification system to the matched cohort, 41 of 48 patients were assigned a PTEN status, and 7 patients had insufficient evaluable CTCs for classification. We found that PTEN loss (HO or HE) occurred in 17 of 41 patients (41%), with 27% HO PTEN loss (11 of 41 patients) and 15% HE PTEN loss (6 of 41 patients, Figure 2A, top panel). Independent evaluation of a patients’ PTEN status in two replicate blood slides demonstrated high reproducibility in patients’ PTEN classification (n=10 patients, Supplementary Figure S1).
PTEN gene status in CTCs and matched archival and fresh tumour tissue specimens. (A) Distribution of PTEN status in patient samples. Upper panel, PTEN status in CTCs; middle panel, PTEN status in fresh tissue; bottom panel, PTEN status in archived tissue. Each bar represents an individual patient, with the proportions of red, yellow and grey colours indicating the percentage of tumour cells with PTEN=0, PTEN=1 and PTEN ⩾2 genotypes. (B) Parameters of concordance in CTCs vs fresh tissue, CTCs vs archival tissue and fresh vs archival tissue.
Analysis of PTEN FISH in tissue was performed on the 41 patients with matched PTEN data in CTCs. Homozygous or HE loss was found in 14 of 38 patients with fresh tissue samples (37%), and in 21 of 39 patients with archival tissue samples (53%, Figure 2A, bottom panel). Comparison of PTEN status in CTCs and tissue revealed concordant PTEN status in 32 of 38 patients with matched CTCs and fresh tissue (84%, Figure 2B left), and 24 of 39 patients with matched CTCs and archival tissue (62%, Figure 2B middle). Representative examples of concordant PTEN status in matched CTCs and fresh tumour tissue samples are shown in Supplementary Figure S3. From 14 patients classified as PTEN Deleted (HO or HE loss) in CTCs, 11 showed PTEN Deleted in fresh tissue (positive predictive value=79%, Figure 2B, left), and from 16 patients who were PTEN Deleted in CTCs, 11 showed PTEN loss in archival tissue (positive predictive value=69%, Figure 2B middle). Sensitivity and specificity values were also higher for CTCs vs fresh tissue than for CTCs vs archived tissue (Figure 2B, left and middle), suggesting that PTEN FISH status in CTCs more accurately reflects PTEN status in fresh tumour tissue than archival tissue.
Comparison of PTEN FISH status in matched archival and fresh tissue revealed discordant PTEN status in 12 of the 36 patients (33%) (Figure 2A arrows, Figure 2B right). In 10 of these 12 cases, PTEN in CTCs was consistent with PTEN in the fresh sample but not in the archival sample; in the other 2 cases, PTEN status in the CTCs correlated to PTEN status in the archival sample (Patients 5086 and 5099). These observations illustrate the intra-patient heterogeneity characteristic of this disease. There were only three cases where CTC data were in poor agreement with tissue data (patients 4002, 4045, 5038). The three discordant patients had sufficient CTC counts for PTEN evaluation, suggesting that the discrepancies were not due to a limited number of CTCs sampled.
Assessment of PTEN by IHC in tissues revealed H-scores ⩽10 in 56% fresh tumour samples (23 of 41), and in 44% archived tissue samples (17 of 39, Supplementary Figure S4). All patients with HO or HE loss in CTCs showed loss of PTEN protein expression in fresh tumour tissue (positive predictive value =100%). Six of the 23 patients (25%) with Non-deleted PTEN in CTCs had loss of PTEN protein expression in fresh tissue. Heterogeneous PTEN protein was observed in archived samples from two patients, but not in fresh samples. PTEN protein status by IHC in fresh tumour tissue was concordant with PTEN protein status in archival tumour tissue in 35 of the remaining 39 patients (90%, Supplementary Table S4). In the discordant four cases, PTEN IHC shifted from H-score>10 in archived tissue to H-score ⩽10 in fresh tissue.
Androgen receptor expression in CTCs does not associate with PTEN or ERG status
Analysis of AR expression in CTCs by immunofluorescence and ERG gene status by FISH was performed in 23 patients who had remaining slides after PTEN evaluation. Patients were classified as ERG rearranged or ERG normal (non-rearranged) following a classification system similar to the one used for PTEN, developed based on the frequency and number of cells that displayed each ERG class in CTCs vs WBCs (Supplementary Table S2). Significant genomic rearrangements of ERG (ERG split and ERG deletion) were detected in CTCs from 10 of the 23 patients (43%). Representative FISH images of ERG rearranged classes (ERG split and ERG deletion) are shown in Figure 3A.
Multi-biomarker evaluation in CTCs (A) Representative images of prostate cancer cells that are ERG-normal, ERG-rearranged by insertion/translocation and ERG-rearranged by deletion. (B) Scatter plots of AR expression within each patient’s CTC population. (Each dot represents one CTC.) Colours represent the tumour-associated or wild-type DNA markers detected in each CTC (Grey=Normal, Orange=Heterozygous PTEN Loss, Red=Homozygous PTEN Loss, Green=ERG Rearrangement by Translocation, Blue=ERG-Rearrangement by Deletion). (C) Summary of biomarker status for each patient (Yellow=AR-Positive, N: nuclear AR localization, C: cytosolic AR localisation, NC: mixed AR localisation, Red=Homozygous PTEN Loss, Orange=Heterozygous PTEN Loss, Teal=ERG-Rearranged (Edel, Esplit, or combination)).
Consistent with observations that PTEN loss is commonly associated with ERG rearrangements (Carver et al, 2009; Taylor et al, 2010), we found 60% PTEN loss in patients classified as ERG rearranged (6 of 10 patients) compared with 38% PTEN loss in patients classified as ERG non-rearranged (5 of 13 patients, Figures 3B and C).
Androgen receptor expression was heterogeneous in CTCs, with intra- and inter-patient variability in AR expression and localisation, and no correlation between AR expression and PTEN status (Figures 3B and C). In the 11 patients classified as PTEN loss, there was no clear segregation of AR expression between CTCs that had loss of PTEN (PTEN=0 or PTEN=1, yellow or red dots) and CTCs that were PTEN normal (PTEN ⩾2, grey dots) (Figures 3B and C). Similarly, AR expression was heterogeneous with respect to ERG status (Figures 3B and C). Only two patients exhibited homogeneous low AR expression with AR levels below the background threshold (patients 4002 and 5089). In these patients, PTEN and ERG were classified as normal in CTCs; patient 4002 was one of the three cases described earlier in which the PTEN status in the CTCs was not concordant with the HO loss observed in the patient tumour tissue.
Correlation of PTEN loss in CTCs with clinical outcome
Enumeration on the CellSearch platform has prognostic value in prostate cancer (de Bono et al, 2008). To evaluate the correlation between enumeration on Epic and CellSearch, we compared CTC counts on the two platforms, assessed on 69 replicate blood samples from 66 of the 76 patients included in this study. CellSearch counts were determined in 7.5 ml blood; Epic counts were extrapolated to 7.5 ml (Supplementary Table S6 and Supplementary Figure S5a). On Epic, the median count was 38 CTCs per 7.5 ml (interquartile range 21–155); on CellSearch, the median count was 18 CTCs per 7.5 ml (interquartile range 4–115). Circulating tumour cell enumeration on Epic correlated with CellSearch (Spearman r=0.75; P<0.0001). The most discordant cases were observed in patients with less than 30 CTC counts per CellSearch. Four out of nine patients with no CTCs detected by CellSearch showed >3 CTCs per 7.5 ml by Epic (Supplementary Figure S5a). In the CTCs of some of the discordant patients, we found ploidy changes and PTEN loss by Epic analysis (patient 5039, with 6 vs 165 CTCs per 7.5 ml, and patient 5059, with 3 vs 57 CTCs per 7.5 ml in CellSearch vs Epic). These features are consistent with malignancy, and demonstrate the tumour origin of the CTCs (Supplementary Figure S5b).
At a median follow-up of 11.4 months, 64 of the 76 patients had died. Circulating tumour cell counts on the Epic platform associated with worse overall survival in univariate (continuous variable; P=0.001) and multivariate analyses that included lactate dehydrogenase levels, presence of visceral metastases, performance status and number of previous systemic lines of therapy (P=0.007, data not shown). Circulating tumour cell enumeration on the CellSearch platform was also significantly associated with survival (data not shown).
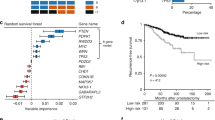
PTEN FISH status in CTCs was available for 60 patients. Patients’ characteristics according to PTEN status in CTCs are detailed in Supplementary Table S5. We have previously shown no difference in outcome between the tumours with HE and those with HO loss (Reid et al, 2010); therefore, our analyses combined HE and HO PTEN loss tumours as one group. Patients with loss of PTEN by FISH in CTCs were younger and had higher lactate dehydrogenase at the time of the blood draw (P=0.02 and P<0.01, respectively). Of note, patients in the PTEN loss group showed a trend to have a higher incidence of visceral metastases compared with patients with no deletion of PTEN. Loss of PTEN in CTCs was significantly associated with a shorter survival in univariate analysis (median survival 7.0 vs 12.1 months; HR 2.05; 95% CI 1.17–3.62; P=0.01) (Figure 4). However, PTEN loss did not retain significance in a multivariate analysis that included Epic CTCs counts, lactate dehydrogenase levels, presence of visceral metastases, performance status and number of previous systemic lines of therapy, in this small cohort (Table 2).
Discussion
In this study, we evaluated the potential utility of CTCs for biomarker characterisation in metastatic CRPC patients. Circulating tumour cell enumeration in the Epic platform correlated well with CTC enumeration by CellSearch. Epic showed increased detection power in samples with low CTC counts by CellSearch. This difference may reflect the distinct CTC identification methodologies of the two platforms (antigen agnostic on the Epic platform, and EpCAM-dependent CTC enrichment on CellSearch). It is possible that the tumour cells detected uniquely on the Epic platform may fail to express EpCAM or have low EpCAM expression, and thus escape the antigen-dependent enrichment method of the CellSearch system (Konigsberg et al, 2011; Mikolajczyk et al, 2011; Gorges et al, 2012). The Epic three-colour CTC enumeration assay has been analytically validated (Werner et al, 2015) and is a CLIA-validated test. The prognostic value of Epic CTC enumeration is being currently investigated in the AKT inhibitor NCT01485861 clinical trial.
This study revealed that CTCs and WBCs are characterised by distinct PTEN and CEP10 genotypes, with increased ploidy and heterogeneous PTEN status in CTCs, but not WBCs. Using a CTC-based classification system developed in this study, we demonstrated a good correlation between the PTEN gene status by FISH determined in CTCs and in patient-matched fresh tumour tissue specimens. Our observations suggest that CTCs can provide an accurate representation of the PTEN gene status in the most current disease and could potentially be used as a non-invasive and real-time solution for biomarker characterisation in CRPC. The perceived intra-patient heterogeneity of PTEN genotypes, with mixtures of cancer cells with PTEN=0, PTEN=1 and PTEN ⩾2 signals, has been previously observed in CTCs from CRPC patients (Attard et al, 2009), and is consistent with the findings of multiple temporal and spatial tumour clones in fresh biopsies and circulating free DNA in CRPC patients (Carreira et al, 2014). Together with the changes we observed in PTEN FISH status from archival to fresh tissue, our results reflect the inherent heterogeneity characteristic of the primary disease where multiple PTEN genotypes might co-exist and selective pressures and additional hits may result in the expansion and evolution of one of these clones during the course of the disease.
The status of PTEN determined by FISH in CTCs also correlated well with the status of PTEN determined by IHC in matched tumour tissue samples, with complete concordance between PTEN loss in CTCs and loss of PTEN expression in fresh tumour tissue. The loss of PTEN expression observed in the tissue of patients that were PTEN Non-Deleted in CTCs could be explained by possible mutations in the PTEN gene locus or epigenetic silencing mechanisms. These results are consistent with previous observations in larger cohort of patients (Ferraldeschi, in press; Verhagen et al, 2006; Han et al, 2009; Lotan et al, 2011; Reid et al, 2012). The shift in PTEN IHC from archival to fresh tissue observed in four patients was consistent with the FISH PTEN data, and may reflect changes in PTEN status with evolution of the disease.
We have previously shown that PTEN protein loss is associated with clinical outcome in metastatic CRPC patients who received abiraterone treatment post-docetaxel (Ferraldeschi et al, 2014). In this exploratory study, Epic CTC count and PTEN loss in CTCs by FISH correlated with a worse prognosis. However, PTEN loss did not retain significance in a multivariate analysis, which could be, at least in part, due to the small sample size and heterogeneous patient population. Our findings encourage the evaluation of PTEN gene status, along with PTEN protein expression in CRPC patients.
In summary, our study demonstrates feasibility of multiple biomarker analysis in CTCs using the enrichment-free Epic platform. The strong concordance between PTEN FISH loss in CTCs and fresh tumour tissue illustrates the potential for using our blood-based PTEN FISH assay to determine PTEN gene status in the patient’s current disease. The shifts in PTEN status between archival and fresh prostate tumour tissue, and the heterogeneity of PTEN gene status in CTCs indicate that CTC analyses could provide important insights on disease heterogeneity, clonal evolution and clone dynamics. The blood-based PTEN FISH assay is being used in ongoing trials of abiraterone/enzalutamide in combination with PI3K/AKT inhibitors for patients suffering from CRPC.
References
Attard G, Swennenhuis JF, Olmos D, Reid AH, Vickers E, A'Hern R, Levink R, Coumans F, Moreira J, Riisnaes R, Oommen NB, Hawche G, Jameson C, Thompson E, Sipkema R, Carden CP, Parker C, Dearnaley D, Kaye SB, Cooper CS, Molina A, Cox ME, Terstappen LW, de Bono JS (2009) Characterization of ERG, AR and PTEN gene status in circulating tumor cells from patients with castration-resistant prostate cancer. Cancer Res 69 (7): 2912–2918.
Carreira S, Romanel A, Goodall J, Grist E, Ferraldeschi R, Miranda S, Prandi D, Lorente D, Frenel JS, Pezaro C, Omlin A, Rodrigues DN, Flohr P, Tunariu N, SdB J, Demichelis F, Attard G (2014) Tumor clone dynamics in lethal prostate cancer. Sci Transl Med 6 (254): 254ra125.
Carver BS, Chapinski C, Wongvipat J, Hieronymus H, Chen Y, Chandarlapaty S, Arora VK, Le C, Koutcher J, Scher H, Scardino PT, Rosen N, Sawyers CL (2011) Reciprocal feedback regulation of PI3K and androgen receptor signaling in PTEN-deficient prostate cancer. Cancer Cell 19 (5): 575–586.
Carver BS, Tran J, Gopalan A, Chen Z, Shaikh S, Carracedo A, Alimonti A, Nardella C, Varmeh S, Scardino PT, Cordon-Cardo C, Gerald W, Pandolfi PP (2009) Aberrant ERG expression cooperates with loss of PTEN to promote cancer progression in the prostate. Nat Genet 41 (5): 619–624.
Chen Y, Chi P, Rockowitz S, Iaquinta PJ, Shamu T, Shukla S, Gao D, Sirota I, Carver BS, Wongvipat J, Scher HI, Zheng D, Sawyers CL (2013) ETS factors reprogram the androgen receptor cistrome and prime prostate tumorigenesis in response to PTEN loss. Nat Med 19 (8): 1023–1029.
Chow LM, Baker SJ (2006) PTEN function in normal and neoplastic growth. Cancer Lett 241 (2): 184–196.
Cohen SJ, Punt CJ, Iannotti N, Saidman BH, Sabbath KD, Gabrail NY, Picus J, Morse M, Mitchell E, Miller MC, Doyle GV, Tissing H, Terstappen LW, Meropol NJ (2008) Relationship of circulating tumor cells to tumor response, progression-free survival, and overall survival in patients with metastatic colorectal cancer. J Clin Oncol 26 (19): 3213–3221.
Cristofanilli M, Budd GT, Ellis MJ, Stopeck A, Matera J, Miller MC, Reuben JM, Doyle GV, Allard WJ, Terstappen LW, Hayes DF (2004) Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med 351 (8): 781–791.
de Bono JS, Scher HI, Montgomery RB, Parker C, Miller MC, Tissing H, Doyle GV, Terstappen LW, Pienta KJ, Raghavan D (2008) Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clin Cancer Res 14 (19): 6302–6309.
Ferraldeschi R, Nava Rodrigues D, Riisnaes R, Miranda S, Figueiredo I, Rescigno P, Ravi P, Pezaro C, Omlin A, Lorente D, Zafeiriou Z, Mateo J, Altavilla A, Sideris S, Bianchini D, Grist E, Thway K, Perez Lopez R, Tunariu N, Parker C, Dearnaley D, Reid A, Attard G, de Bono J (2014) PTEN protein loss and clinical outcome from castration-resistant prostate cancer treated with abiraterone acetate. Eur Urol 67 (4): 795–802.
Gorges TM, Tinhofer I, Drosch M, Rose L, Zollner TM, Krahn T, von Ahsen O (2012) Circulating tumour cells escape from EpCAM-based detection due to epithelial-to-mesenchymal transition. BMC Cancer 12: 178.
Halvorsen OJ, Haukaas SA, Akslen LA (2003) Combined loss of PTEN and p27 expression is associated with tumor cell proliferation by Ki-67 and increased risk of recurrent disease in localized prostate cancer. Clin Cancer Res 9 (4): 1474–1479.
Han B, Mehra R, Lonigro RJ, Wang L, Suleman K, Menon A, Palanisamy N, Tomlins SA, Chinnaiyan AM, Shah RB (2009) Fluorescence in situ hybridization study shows association of PTEN deletion with ERG rearrangement during prostate cancer progression. Mod Pathol 22 (8): 1083–1093.
Heitzer E, Auer M, Gasch C, Pichler M, Ulz P, Hoffmann EM, Lax S, Waldispuehl-Geigl J, Mauermann O, Lackner C, Hofler G, Eisner F, Sill H, Samonigg H, Pantel K, Riethdorf S, Bauernhofer T, Geigl JB, Speicher MR (2013) Complex tumor genomes inferred from single circulating tumor cells by array-CGH and next-generation sequencing. Cancer Res 73 (10): 2965–2975.
Ishibashi H, Suzuki T, Suzuki S, Moriya T, Kaneko C, Takizawa T, Sunamori M, Handa M, Kondo T, Sasano H (2003) Sex steroid hormone receptors in human thymoma. The Journal of clinical endocrinology and metabolism 88 (5): 2309–2317.
Konigsberg R, Obermayr E, Bises G, Pfeiler G, Gneist M, Wrba F, de Santis M, Zeillinger R, Hudec M, Dittrich C (2011) Detection of EpCAM positive and negative circulating tumor cells in metastatic breast cancer patients. Acta Oncol (Madr) 50 (5): 700–710.
Leinonen KA, Saramaki OR, Furusato B, Kimura T, Takahashi H, Egawa S, Suzuki H, Keiger K, Ho Hahm S, Isaacs WB, Tolonen TT, Stenman UH, Tammela TL, Nykter M, Bova GS, Visakorpi T (2013) Loss of PTEN is associated with aggressive behavior in ERG-positive prostate cancer. Cancer Epidemiol Biomarkers Prev 22 (12): 2333–2344.
Lin HK, Hu YC, Lee DK, Chang C (2004) Regulation of androgen receptor signaling by PTEN (phosphatase and tensin homolog deleted on chromosome 10) tumor suppressor through distinct mechanisms in prostate cancer cells. Mol Endocrinol 18 (10): 2409–2423.
Lotan TL, Gurel B, Sutcliffe S, Esopi D, Liu W, Xu J, Hicks JL, Park BH, Humphreys E, Partin AW, Han M, Netto GJ, Isaacs WB, De Marzo AM (2011) PTEN protein loss by immunostaining: analytic validation and prognostic indicator for a high risk surgical cohort of prostate cancer patients. Clin Cancer Res 17 (20): 6563–6573.
Marrinucci D, Bethel K, Bruce RH, Curry DN, Hsieh B, Humphrey M, Krivacic RT, Kroener J, Kroener L, Ladanyi A, Lazarus NH, Nieva J, Kuhn P (2007) Case study of the morphologic variation of circulating tumor cells. Hum Pathol 38 (3): 514–519.
Marrinucci D, Bethel K, Kolatkar A, Luttgen MS, Malchiodi M, Baehring F, Voigt K, Lazar D, Nieva J, Bazhenova L, Ko AH, Korn WM, Schram E, Coward M, Yang X, Metzner T, Lamy R, Honnatti M, Yoshioka C, Kunken J, Petrova Y, Sok D, Nelson D, Kuhn P (2012) Fluid biopsy in patients with metastatic prostate, pancreatic and breast cancers. Phys Biol 9 (1): 016003.
Marrinucci D, Bethel K, Luttgen M, Bruce RH, Nieva J, Kuhn P (2009) Circulating tumor cells from well-differentiated lung adenocarcinoma retain cytomorphologic features of primary tumor type. Arch Pathol Lab Med 133 (9): 1468–1471.
McCall P, Witton CJ, Grimsley S, Nielsen KV, Edwards J (2008) Is PTEN loss associated with clinical outcome measures in human prostate cancer? Br J Cancer 99 (8): 1296–1301.
McMenamin ME, Soung P, Perera S, Kaplan I, Loda M, Sellers WR (1999) Loss of PTEN expression in paraffin-embedded primary prostate cancer correlates with high Gleason score and advanced stage. Cancer Res 59 (17): 4291–4296.
Mikolajczyk SD, Millar LS, Tsinberg P, Coutts SM, Zomorrodi M, Pham T, Bischoff FZ, Pircher TJ (2011) Detection of EpCAM-negative and cytokeratin-negative circulating tumor cells in peripheral blood. J Oncol 2011: 252361.
Miyamoto DT, Lee RJ, Stott SL, Ting DT, Wittner BS, Ulman M, Smas ME, Lord JB, Brannigan BW, Trautwein J, Bander NH, Wu CL, Sequist LV, Smith MR, Ramaswamy S, Toner M, Maheswaran S, Haber DA (2012) Androgen receptor signaling in circulating tumor cells as a marker of hormonally responsive prostate cancer. Cancer Discov 2 (11): 995–1003.
Modrek B, Ge L, Pandita A, Lin E, Mohan S, Yue P, Guerrero S, Lin WM, Pham T, Modrusan Z, Seshagiri S, Stern HM, Waring P, Garraway LA, Chant J, Stokoe D, Cavet G (2009) Oncogenic activating mutations are associated with local copy gain. Mol Cancer Res 7 (8): 1244–1252.
O'Brien C, Cavet G, Pandita A, Hu X, Haydu L, Mohan S, Toy K, Rivers CS, Modrusan Z, Amler LC, Lackner MR (2008) Functional genomics identifies ABCC3 as a mediator of taxane resistance in HER2-amplified breast cancer. Cancer Res 68 (13): 5380–5389.
Reid AH, Attard G, Ambroisine L, Fisher G, Kovacs G, Brewer D, Clark J, Flohr P, Edwards S, Berney DM, Foster CS, Fletcher A, Gerald WL, Moller H, Reuter VE, Scardino PT, Cuzick J, de Bono JS, Cooper CS Transatlantic Prostate Group (2010) Molecular characterisation of ERG, ETV1 and PTEN gene loci identifies patients at low and high risk of death from prostate cancer. Br J Cancer 102 (4): 678–684.
Sandhu SK, Schelman WR, Wilding G, Moreno V, Baird RD, Miranda S, Hylands L, Riisnaes R, Forster M, Omlin A, Kreischer N, Thway K, Gevensleben H, Sun L, Loughney J, Chatterjee M, Toniatti C, Carpenter CL, Iannone R, Kaye SB, de Bono JS, Wenham RM (2013) The poly(ADP-ribose) polymerase inhibitor niraparib (MK4827) in BRCA mutation carriers and patients with sporadic cancer: a phase 1 dose-escalation trial. Lancet Oncol 14 (9): 882–892.
Reid AH, Attard G, Brewer D, Miranda S, Riisnaes R, Clark J, Hylands L, Merson S, Vergis R, Jameson C, Hoyer S, Sorenson KD, Borre M, Jones C, de Bono JS, Cooper CS (2012) Novel, gross chromosomal alterations involving PTEN cooperate with allelic loss in prostate cancer. Mod Pathol 25 (6): 902–910.
Sarker D, Reid AH, Yap TA, de Bono JS (2009) Targeting the PI3K/AKT pathway for the treatment of prostate cancer. Clin Cancer Res 15 (15): 4799–4805.
Scher HI, Halabi S, Tannock I, Morris M, Sternberg CN, Carducci MA, Eisenberger MA, Higano C, Bubley GJ, Dreicer R, Petrylak D, Kantoff P, Basch E, Kelly WK, Figg WD, Small EJ, Beer TM, Wilding G, Martin A, Hussain M Prostate Cancer Clinical Trials Working Group (2008) Design and end points of clinical trials for patients with progressive prostate cancer and castrate levels of testosterone: recommendations of the Prostate Cancer Clinical Trials Working Group. J Clin Oncol 26 (7): 1148–1159.
Sircar K, Yoshimoto M, Monzon FA, Koumakpayi IH, Katz RL, Khanna A, Alvarez K, Chen G, Darnel AD, Aprikian AG, Saad F, Bismar TA, Squire JA (2009) PTEN genomic deletion is associated with p-Akt and AR signalling in poorer outcome, hormone refractory prostate cancer. J Pathol 218 (4): 505–513.
Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, Arora VK, Kaushik P, Cerami E, Reva B, Antipin Y, Mitsiades N, Landers T, Dolgalev I, Major JE, Wilson M, Socci ND, Lash AE, Heguy A, Eastham JA, Scher HI, Reuter VE, Scardino PT, Sander C, Sawyers CL, Gerald WL (2010) Integrative genomic profiling of human prostate cancer. Cancer Cell 18 (1): 11–22.
Thomas C, Lamoureux F, Crafter C, Davies BR, Beraldi E, Fazli L, Kim S, Thaper D, Gleave ME, Zoubeidi A (2013) Synergistic targeting of PI3K/AKT pathway and androgen receptor axis significantly delays castration-resistant prostate cancer progression in vivo. Mol Cancer Ther 12 (11): 2342–2355.
Toren P, Kim S, Cordonnier T, Crafter C, Davies BR, Fazli L, Gleave ME, Zoubeidi A (2014) Combination AZD5363 with enzalutamide significantly delays enzalutamide-resistant prostate cancer in preclinical models. Eur Urol 67 (6): 986–990.
Verhagen PC, van Duijn PW, Hermans KG, Looijenga LH, van Gurp RJ, Stoop H, van der Kwast TH, Trapman J (2006) The PTEN gene in locally progressive prostate cancer is preferentially inactivated by bi-allelic gene deletion. J Pathol 208 (5): 699–707.
Werner SL, Graf RP, Landers M, Valenta DT, Schroeder M, Greene SN, Bales N, Dittamore R, Marrinucci D (2015) Analytical validation and capabilities of the Epic CTC platform: enrichment-free circulating tumour cell detection and characterization. J Circ Biomark 4: 3.
Yap TA, Lorente D, Omlin A, Olmos D, de Bono JS (2014) Circulating tumor cells: a multifunctional biomarker. Clin Cancer Res 20 (10): 2553–2568.
Yoshimoto M, Cutz JC, Nuin PA, Joshua AM, Bayani J, Evans AJ, Zielenska M, Squire JA (2006) Interphase FISH analysis of PTEN in histologic sections shows genomic deletions in 68% of primary prostate cancer and 23% of high-grade prostatic intra-epithelial neoplasias. Cancer Genet Cytogenet 169 (2): 128–137.
Acknowledgements
George Seed (ICR), Mikel Kitchen, Florence Lee, Amanda Anderson (Epic Sciences).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on British Journal of Cancer website
Rights and permissions
This work is licensed under the Creative Commons Attribution-Non-Commercial-Share Alike 4.0 International License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/4.0/
About this article
Cite this article
Punnoose, E., Ferraldeschi, R., Szafer-Glusman, E. et al. PTEN loss in circulating tumour cells correlates with PTEN loss in fresh tumour tissue from castration-resistant prostate cancer patients. Br J Cancer 113, 1225–1233 (2015). https://doi.org/10.1038/bjc.2015.332
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/bjc.2015.332
Keywords
This article is cited by
-
Applications of liquid biopsy in the Pharmacological Audit Trail for anticancer drug development
Nature Reviews Clinical Oncology (2021)
-
Merging new-age biomarkers and nanodiagnostics for precision prostate cancer management
Nature Reviews Urology (2019)
-
Clinical implications of PTEN loss in prostate cancer
Nature Reviews Urology (2018)
-
Development and Application of Liquid Biopsies in Metastatic Prostate Cancer
Current Oncology Reports (2018)
-
Circulating tumour cells as biomarkers of prostate, bladder, and kidney cancer
Nature Reviews Urology (2017)