Abstract
Background:
Colorectal cancers arise from benign adenomas, although not all adenomas progress to cancer and there are marked interpatient differences in disease progression. We have previously associated KRAS mutations with disease progression and reduced survival in colorectal cancer patients.
Methods:
We used TaqMan low-density array (TLDA) qRT–PCR analysis to identify miRNAs differentially expressed in normal colorectal mucosa, adenomas and cancers and in isogeneic KRAS WT and mutant HCT116 cells, and used a variety of phenotypic assays to assess the influence of miRNA expression on KRAS activity, chemosensitivity, proliferation and invasion.
Results:
MicroRNA-224 was differentially expressed in dysplastic colorectal disease and in isogeneic KRAS WT and mutant HCT116 cells. Antagomir-mediated miR-224 silencing in HCT116 KRAS WT cells phenocopied KRAS mutation, increased KRAS activity and ERK and AKT phosphorylation. 5-FU chemosensitivity was significantly increased in miR-224 knockdown cells, and in NIH3T3 cells expressing KRAS and BRAF mutant proteins. Bioinformatics analysis of predicted miR-224 target genes predicted altered cell proliferation, invasion and epithelial–mesenchymal transition (EMT) phenotypes that were experimentally confirmed in miR-224 knockdown cells.
Conclusions:
We describe a novel mechanism of KRAS regulation, and highlight the clinical utility of colorectal cancer-specific miRNAs as disease progression or clinical response biomarkers.
Similar content being viewed by others
Main
Colorectal cancer (ICD-10 C18-C20) is the fourth most common cancer in the United Kingdom and a leading cause of cancer death (Cancer Research UK, 2014). Colorectal cancers initially present as benign polyps (adenomas), the most aggressive of which become malignant, promoting metastasis to lymph node and liver secondary sites. Identification and removal of premalignant adenomas is therefore a clinical priority, and the focus of NHS (National Health Service, 2014) and Scottish Bowel Screening programmes (Scottish Bowel Screening Programme, 2014). Not all adenomas progress to cancer, however, and we therefore need to better understand the molecular events that influence disease progression, and to develop more specific screening tests for early detection and the identification of polyps at greater risk of progression.
Progression from colorectal adenoma to cancer is driven by the acquisition of loss of function mutations in tumour suppressor genes such as APC and TP53, and activating mutations in oncogenes including Kirsten RAS (KRAS). KRAS mutations, and phenotypically similar mutations in BRAF, promote cell growth by prolonged activation of RAS/MAPK-regulated signalling cascades (Barbacid, 1990). We have described significantly increased KRAS mutation burden in advanced Dukes’ C cancers and shown that mutation burden is negatively associated with survival (Smith et al, 2002; Conlin et al, 2005).
Colorectal cancer treatment routinely combines surgical resection with adjuvant or neoadjuvant treatment with 5-fluorouracil (5-FU) or its prodrug capecitabine (Scottish Intercollegiate Guidelines Network, 2011). As with rates of disease progression, response to treatment varies widely, and sensitive, patient-specific biomarkers for treatment selection and assessment of prognosis are urgently required. KRAS and BRAF mutation status are clinically relevant response biomarkers in patients treated with cetuximab and related drugs, monoclonal antibodies targeted to the epidermal growth factor receptor, a key node in the RAS/MAPK signalling pathway (Lièvre et al, 2006; Amado et al, 2008; Bokemeyer et al, 2008). As biomarker assessment is often limited by the availability of serial tissue samples, recent research has focussed on identification of serum-based biomarkers including microRNAs (miRNAs), small (18–25 nucleotides) noncoding RNAs that function as negative regulators of gene expression, repressing protein translation by binding to target messenger mRNAs (Bartel, 2004). MicroRNAs are frequently differentially expressed in cancer, and have been suggested to influence cancer progression, following negative regulation of both tumour suppressor genes and proto-oncogenes. Previous studies have highlighted significant differences in miRNA expression in colorectal cancer relative to matched normal tissue, where specific miRNAs have consistently increased (e.g., miR-21, miR-135b, miR-183, miR-31, miR-201) or decreased (e.g., miR-145, miR-378) expression (Bandrés et al, 2006; Monzo et al, 2008; Schetter et al, 2008; Chen et al, 2009; Motoyama et al, 2009; Earle et al, 2010; Wang et al, 2010; Chang et al, 2011; Faltejskova et al, 2012; Fu et al, 2012; Hamfjord et al, 2012; Mosakhani et al, 2012; Reid et al, 2012). In contrast, miRNA expression in colorectal adenomas is less well described, although recent studies have highlighted differential expression of several miRNAs, including miR-135b (Wu et al, 2014) and miR-17 (Kanaan et al, 2013).
Individual miRNAs can target multiple mRNAs because of imperfect complementarity in 3′-UTR seed sequences (Lewis et al, 2005). Robust bioinformatics approaches are therefore required to predict miRNA target genes, combined with phenotypic validation of predicted miRNA/mRNA interactions. To address these issues in colorectal cancer development, we have performed a quantitative, systematic comparison of miRNA expression in colorectal cancers, matched healthy tissue and colorectal adenomas with low-grade and high-grade dysplasia to identify miRNAs that promote, or that may have utility as biomarkers of colorectal cancer progression, with a particular focus on novel regulators of the RAS/MAPK signalling pathway.
Materials and methods
Ethical approval
All studies involving human tissues were approved by the Tayside Tissue Bank Research Ethics Committee, a devolved sub-committee of the Tayside Committee on Medical Research Ethics.
RNA extraction and quantitation
RNA was extracted from cell lines using Qiagen RNeasy mini kits (Qiagen, Manchester, UK) following the manufacturer’s protocol for mammalian cells, with on-column DNase digestion (Qiagen RNase free DNase kit). Tissue samples (∼20 mg) were disrupted by homogenisation using a rotor–stator homogeniser (Polytron, Thermo Fisher Scientific, Waltham, MA, USA) before RNA extraction, following the manufacturer’s protocol for animal tissue. RNA was additionally extracted from three 20 μm FFPE tissue sections (combined) using RecoverAll Total Nucleic Acid isolation kits (Life Technologies, Paisley, UK).
RNA yield and integrity was initially quantitated from A260 and A280 absorbance readings obtained from a Nanodrop 1000 spectrophotometer (Thermo Fisher Scientific, Loughborough, UK) – all samples selected for further analysis had an A260:A280 ratio between 1.7 and 2.0. RNA integrity was further assessed in human tissue samples using an Agilent Bioanalyser 2100 and RNA 6000 Nano LabChip Kit (Agilent Technologies, Wokingham, UK); samples selected for further analysis had RIN (RNA Integrity Number) values >7.
TaqMan quantitative real-time PCR analysis
MiRNA expression was initially investigated using TaqMan low-density miRNA A card arrays (377 miRNAs, 3 endogenous controls and negative control; Life Technologies; http://tools.lifetechnologies.com/content/sfs/manuals/cms_054742.pdf). Reverse transcription (RT) reactions were performed using TaqMan RT reagent kits and Megaplex RT primers (Life Technologies), following the manufacturer’s protocol. PCR reactions, combining Megaplex RT products with TaqMan Universal PCR Master Mix II (Life Technologies), were added to each Taqman low-density array (TLDA) card port and PCR reactions run in triplicate on the standard Real-Time PCR program on the 7900 TaqMan real-time system (Applied Biosystems, Warrington, UK). For single probe qRT–PCR miRNA analysis, RT reactions were performed using TaqMan MicroRNA Reverse Transcription Kits (Life Technologies) according to the manufacturer’s protocol. Resulting cDNA samples were analysed in triplicate using TaqMan Small RNA Assays, combining miRNA-specific probes (Life Technologies) with TaqMan Universal PCR Master Mix II and run as previously described. Let-7a was used as an endogenous control for all miRNA qRT–PCR analyses.
The mRNA RT reactions were performed, primed with random hexamers, using TaqMan reverse transcription reagent kits (Life Technologies) according to the manufacturer’s protocol. PCR reactions were then performed in triplicate using TaqMan Gene Expression Assays (Life Technologies), as described above, using 18S ribosomal RNA as an endogenous control. Gene expression differences in both single probe and TLDA card experiments were identified using SDS 2.3 software (Applied Biosystems) to determine Ct (cycle threshold) values for each target and endogenous control gene for each sample, as previously described (Smith et al, 2012).
Cell lines
Isogeneic HCT116 cell lines specifically engineered to express the WT or mutant form of KRAS were kindly donated by Dr Bert Vogelstein (John Hopkins University, Baltimore, MD, USA). Isogeneic cell lines were created from the parental HCT116 cell line (heterozygous for KRAS WT and G13D KRAS mutated alleles) by deleting an allele by targeted homologous recombination to create KRAS mutant(KRAS G13D/−) and WT(KRASWT/−) HCT116 colorectal cancer cell lines (Shirasawa et al, 1993; Torrance et al, 2001). Cells were cultured in McCoy’s 5A medium with 10% fetal bovine serum (FBS) and 1% antibiotics (penicillin and streptomycin) and incubated at 37 °C and 5% carbon dioxide (CO2). The mouse NIH3T3 embryonic fibroblast cell line was obtained from Cancer Research UK cell services. NIH3T3 cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% calf serum, at 37 °C in 5% CO2. 2.5 × 105 cells were transiently transfected in six-well plates with 250 ng of plasmids encoding either WT KRAS, WT BRAF, KRAS G12V, KRAS G13D or BRAF V6000E using Lipofectamine (Invitrogen, Paisley, UK), and harvested 48 h after transfection as previously described (Smith et al, 2010). To create stable cell lines, cells were simultaneously transfected with 250 ng pcDNA3 (Invitrogen), containing a selection marker for geneticin (G418). Geneticin-resistant colonies (formed up to 6 weeks following transfection) were picked using cloning cylinders and expanded in selective media, and each stable cell line tested for KRAS or BRAF mRNA and protein expression by qRT–PCR and western blot analysis, respectively.
MiR-224 transfection experiments
HCT116 KRAS WT cells (1 × 105 cells per well) were seeded in six-well plates and incubated for 24 h before lipofectamine-based transient transfections with a final concentration of 30 nM miR-224-specific antagomir or miRNA inhibitor negative control (Life Technologies) in serum-free medium. The extent of miR-224 knockdown was assessed by qRT–PCR analysis 24 h following transfection, as previously described.
RAS GTPase ELISA
The amount of GTP-bound KRAS in cellular extracts was determined using RAS GTPase Chemi ELISAs (Active Motif, La Hulpe, Belgium) according to the manufacturer’s guidelines. Cellular extracts were obtained from cells 24 h following transfection with miR-224 inhibitors or from untransfected cells. Cells were washed with ice-cold PBS and lysed by the addition of complete lysis/binding buffer, and the collection of cell supernatants. Protein concentrations were determined by Bradford assay (Bradford, 1976) and RAS ELISAs performed in triplicate according to the manufacturer’s protocol, with endpoint luminescence assessment.
PathScan intracellular signalling array
Protein lysates were obtained as previously described and diluted to 1 mg ml−1. PathScan Intracellular Signalling Array kits (Cell Signaling Technology, Hitchin, UK), slide-based antibody arrays for the simultaneous detection of 18 signalling molecules including ERK and AKT, were analysed according to the manufacturer’s protocol and scanned using Li-Cor Odyssey (Li-Cor Biosciences, Cambridge, UK; excitation 680 nm and detection 700 nm). Relative fluorescent intensities were quantified using Image Studio Software (Li-Cor Biosciences).
MTT cytotoxicity assays
MTT cytotoxicity assays were performed to compare chemosensitivity of HCT116 KRAS WT and mutant cells, HCT116 KRAS WT cells following miR-224 knockdown and NIH3T3 KRAS and BRAF WT and mutant cells following treatment with 5-FU, oxaliplatin and irinotecan. Cells (3000 cells per well) were seeded in triplicate in 96-well plates and incubated for 24 h before drug treatment, previously optimised in preliminary experiments (5-FU, 1.25–80 μ M; oxaliplatin, 1.25–80 μ M; irinotecan, 1–64 μ M). Cells were incubated with drug (or vehicle control) for 72 h before addition of 100 μl 5 mg ml−1 MTT in phenol red-deficient DMEM. Following a 3-h incubation, the resulting formazan crystals were solubilised in DMSO and absorbance read at 570 nm. Mean absorbance readings for each treatment were calculated as percentages of vehicle controls (designated 100%), plotted against [log10 drug] and IC50 values calculated.
Cell growth assays
Cells were seeded in 6-well plates (1 × 105 cells per well) and incubated for 24 h before trypsinisation at 16, 24, 32 and 48 h time points, counted using a haemocytometer and cell counts plotted against time. Cell doubling time was also compared in HCT116 KRAS WT and mutant cells and KRAS WT miR-224 knockdown cells using CellTrace Violet Cell Proliferation Kits (Life Technologies) according to the manufacturer’s guidelines. Cells were seeded (1 × 106 cells per reaction) in triplicate, labelled with 5 μ M CellTrace dye and harvested at 16, 24, 32, 48, 64 and 72 h time points as previously described. Proliferation was quantitated by flow cytometry (Attune Flow cytometer using Attune Cytometric software (Life Technologies)) and ModFit LT software (version 4; Verity Software House, Topsham, ME, USA) used to create a proliferation model and calculate proliferation index (total number of divisions/number of cells that went into division). Proliferation index was plotted against time and doubling time calculated as previously described.
Cell invasion assays
Invasion in HCT116 KRAS WT and mutant cells and KRAS WT miR-224 knockdown cells was compared using InnoCyte invasion assays (Merck Millipore, Watford, UK) according to the manufacturer’s protocol. Cells (1 × 106 per well) were placed in the top chamber in serum-free media, allowed to migrate, fluorescently labelled and relative fluorescence assessed (excitation 485 nm, emission 530 nm).
Bioinformatics and statistics
MicroRNA data from TLDA miRNA cards were analysed using Bioconductor 1.9 (Gentleman et al, 2004) running on R 2.6.0 (R Development Core Team, 2008). Initial miRNA screens were performed using individual TLDA cards, with all subsequent experiments performed in triplicate. Cross-sample normalisation of CT values was performed, where the expression of individual miRNAs in each sample was normalised to corresponding let-7a expression. Statistically significantly differentially expressed miRNAs were identified using an empirical Bayes t-test (Smyth, 2004) and P-values adjusted for multiple testing correction using the Benjamini–Hochberg method (Benjamini and Hochberg, 1995). MicroRNAs with adjusted P-values of <0.05 were used to predict target genes identified by at least two of the miRNA target prediction databases: mirDB (www.mirdb.org/miRDB/), miRANDA (www.microrna.org), miRBase (www.mirbase.org), TargetMiner (www.isical.ac.in) and TargetScan (www.targetscan.org). Pathway and biological process enrichment analyses were performed using Metacore (www.thomsonreuters.com/metacore/), employing a hypergeometric distribution to determine enriched gene sets. Independent t-tests were used to assess statistically significant (P⩽0.05) differences in the means of continuous variables in cell lines, and Mann–Whitney tests were used to compare the medians of continuous variables in patient groups. In all figures, *P⩽0.05, **P⩽0.01, ***P⩽0.0001 and ****P⩽0.0001.
Results
Identification of miRNAs differentially expressed in colorectal adenomas and cancers
To perform an initial unbiased analysis of miRNA expression in colorectal cancer, qRT–PCR analysis using TLDA cards was performed as described in Materials and Methods. Colorectal cancer samples (n=12), previously analysed for KRAS and BRAF mutation status (Weidlich et al, 2011), were profiled for miRNA expression and marked interpatient differences in miRNA expression were identified (Figure 1A and Supplementary Figure 1).
Identification of miRNAs differentially expressed in normal colorectal mucosae, colorectal adenomas and colorectal cancers. qRT–PCR analysis (TaqMan low-density array A cards, n=377 unique miRNA targets) was used, as described in Materials and Methods, in an initial screen to identify miRNAs expressed in normal mucosae (n=3), colorectal adenomas (n=6) and colorectal cancers (n=3), and to assess interindividual differences in miRNA expression. (A) Representative data summarising the expression of the first 55 miRNAs on the TLDA array are illustrated, with additional data summarised in Supplementary Figure 1. MicroRNA expression is illustrated relative to the expression of the invariant miRNA let-7a. MicroRNAs with consistently increased (n=7; B) or decreased (n=6; C) expression, comparing normal mucosae with colorectal adenomas and cancers, are highlighted.
Of the 377 miRNAs analysed, 153 (40.6%) were not expressed, 199 (52.7%) expressed in at least 2 cancers and 108 (28.6%) expressed in all 12 cancers, including 18 miRNAs (miR-193b, miR-323-3p, miR-324-3p, miR-331-5p, miR-339-3p, miR-362-5p, miR-365, miR-374, miR-449b, miR-487a, miR-491-5p, miR-494, miR-501, miR-532-5p, miR-574-3p, miR-671-3p, miR-744 and miR-99b) not previously described in colorectal cancer (Supplementary Information and Supplementary Table 1A).
To identify differentially expressed miRNAs, normal patient-matched colorectal mucosa samples (n=3) and colorectal adenomas (high-grade dysplasia (n=3), low-grade dysplasia (n=3)) were analysed. In all colorectal tissues, 111 miRNAs (29.4%) were expressed (Supplementary Information and Supplementary Table 1B). The expression of seven miRNAs was significantly increased (Supplementary Table 1B) and six miRNAs decreased (Supplementary Table 1C) in low- and high-grade adenomas and cancers compared with normal colorectal mucosa.
Prioritisation of miRNAs associated with KRAS mutation status
To investigate the influence of KRAS mutation status on miRNA expression, we used TLDA qRT–PCR analysis to compare miRNA expression in isogeneic HCT116 colorectal cancer cell lines, expressing either WT or mutant KRAS (Shirasawa et al, 1993; Torrance et al, 2001). We identified 12 differentially expressed miRNAs (Figure 2A), 3 of which (miR-224, miR-29c and miR-139-5p) were also differentially expressed in colorectal cancer. Unlike miR-29c and miR-139-5p, miR-224 expression was increased in both colorectal adenomas and cancers, and was prioritised for more detailed analysis, following replication of our TLDA data using a small TaqMan miR-224 RNA assay to confirm differential miR-224 expression in HCT116 KRAS WT and mutant cells (3.3-fold; P=0.0002; Figure 2B), and in colorectal cancers relative to normal colorectal mucosa (n=12; P=0.005; Figure 2C). To further investigate whether miR-224 expression was influenced by KRAS or BRAF mutation status, we compared miR-224 expression in an extended cancer panel, selected by previously determined mutation status (n=41; 18 KRAS mutant, 9 BRAFV600E mutant and 14 KRAS and BRAF WT cancers; Supplementary Information and Supplementary Table 2). MiR-224 expression was significantly decreased (P=0.012) in BRAF mutant cancers (Figure 2D), although not in KRAS mutant cancers, possibly as a consequence of mixed KRAS genotype; ideally, we would have restricted our analysis to KRAS G13D mutant cancers to facilitate direct comparison with our cell line data, but insufficient cancers with this rare genotype were available for analysis.
Identification of miRNAs differentially expressed in isogeneic KRAS WT and mutant HCT116 colorectal cell lines. qRT–PCR analysis (TaqMan low-density array A cards, n=377 unique miRNA targets) was used, as described in Materials and Methods, to identify miRNAs differentially expressed in isogeneic KRAS WT and mutant HCT116 colorectal cell lines. (A) Heatmap summarising differentially expressed miRNAs, highlighting miR-224 (*), the expression of which was also increased in both colorectal adenomas and colorectal cancers (Figure 1B). qRT–PCR analysis was also used to confirm increased miR-224 expression in (B) KRAS WT HCT116 colorectal cancer cells, (C) colorectal cancers relative to normal mucosae (n=12) and (D) an extended colorectal cancer series (n=41) subdivided according to KRAS and BRAF genotype, using an independent TaqMan small RNA assay, as described in Materials and Methods. MicroRNA-224 expression is illustrated relative to the expression of the invariant miRNA let-7a; all samples were analysed in triplicate, with experimental errors calculated as previously described (Smith et al, 2012).
As pre-mir-224 is transcribed from the q arm of the X chromosome, we hypothesised that expression may vary in male (n=18) and female (n=23) patients, although no significant expression differences were identified (P=0.331). Similarly, miR-224 expression did not vary with cancer stage.
Phenotypic consequences of miR-224 knockdown
To investigate the functional role of miR-224, a miR-224-specific antagomir was transiently transfected into HCT116 KRAS WT cells. MicroRNA-224 and negative control antagomir concentrations were optimised, with maximum knockdown occurring 24 h following transfection with 30 nM miR-224 antagomir (P=1.1 × 10−6; Supplementary Figure 2A). Knockdown (>85% relative to basal expression) was maintained for 96 h following transfection (Supplementary Figure 2A), highlighting an experimental window in which to investigate the phenotypic consequences of reduced miR-224 expression.
As miR-224 expression is significantly higher in KRAS WT cells, we hypothesised that miR-224 knockdown may phenocopy KRAS mutation by increasing KRAS activity. The effect of miR-224 knockdown on the amount of GTP-bound KRAS was therefore investigated using a RAS ELISA. KRAS activity was higher (2.5-fold, P=0.002) in KRAS mutant than in WT HCT116 cells (Figure 3A) and, consistent with our hypothesis, miR-224 knockdown in KRAS WT cells significantly increased RAS activity (1.9-fold, P=0.001). To investigate KRAS specificity, NRAS and HRAS expression was additionally compared in HCT116 KRAS WT and miR-224 knockdown cells, but no significant differences identified (Supplementary Figure 3), although KRAS expression was itself significantly increased following miR-224 knockdown (1.7-fold, P=0.007).
Characterisation of miR-224 knockdown phenotypes. (A) A quantitative RAF-binding RAS ELISA was used, as described in Materials and Methods, to assess the influence of miR-224 knockdown on the amount of GTP-bound RAS in cell extracts from KRAS WT, KRAS WT-miR-224 knockdown and KRAS mutant HCT116 cells. Each sample was assessed in triplicate and data represent mean±s.e.m. of relative luminescence units. (B) PathScan intracellular signalling arrays were used, as described in Materials and Methods, to assess the influence of miR-224 knockdown on the phosphorylation of ERK1/2 and additional substrates summarised in Supplementary Figure 2. Each sample was analysed in duplicate, with relative fluorescence at 680/700 nm quantified using a Li-Cor Odyssey fluorescence imaging system. MTT chemosensitivity assays were used, as described in Materials and Methods, to compare sensitivity to 5-FU in (C) KRAS WT HCT116 cells, cells transfected with a nonspecific negative control antagomir and miR-224 knockdown cells and (D) NIH3T3 mouse fibroblast cells stably expressing equivalent levels of WT KRAS and BRAF or KRAS (G12V and G13D) or BRAF (V600) mutant proteins. All experiments were performed in triplicate and IC50 values for each cell line calculated using GraphPad Prism (La Jolla, CA, USA).
PathScan intracellular signalling array targets were then compared in WT HCT116 and miR-224 knockdown cells. Consistent with increased KRAS activity, ERK phosphorylation was increased in miR-224 knockdown cells (1.6-fold; P=0.006; Figure 3B). Similarly, AKT phosphorylation was significantly higher (tyrosine 308 1.4-fold, P=0.01; serine 473 1.3-fold, P=0.04) in miR-224 knockdown cells, suggesting that miR-224 directly influences the pro-survival RAS/AKT/PI3K pathway. Our analysis identified several additional signalling molecules (STAT1, STAT3, AMPK, HSP27, PRAS40 and p38 (MAPK11)) the expression of which was also significantly increased following miR-224 knockdown, highlighting novel candidate miR-224 target genes (Supplementary Figure 4).
MTT cytotoxicity assays were used to investigate the effect of miR-224 knockdown on sensitivity to 5-FU, oxaliplatin and irinotecan, the drugs used to treat colorectal cancer. MicroRNA-224 knockdown increased the sensitivity of HCT116 KRAS WT cells to 5-FU, with a 1.9-fold decrease in IC50 (P=0.04, Figure 3C) but did not influence sensitivity to oxaliplatin or irinotecan (data not shown). To investigate whether 5-FU chemosensitivity was KRAS dependent, we additionally compared 5-FU sensitivity in novel NIH3T3-derived cell lines, stably transfected with WT KRAS, WT BRAF, various KRAS mutants and BRAFV600E (Figure 3D). Consistent with our miR-224 knockdown data, all mutant KRAS cells (G12V and G13D mutants illustrated as examples) and BRAF mutant (V600E) cells were more sensitive to 5-FU (Figure 3D).
Bioinformatics prediction of miR-224-regulated pathways
Individual miRNAs regulate multiple target genes because of imperfect complementarity with 3′UTR seed sequences. MiRNA target prediction databases use unique algorithms to predict miRNA target genes in silico, ranked by predicted affinity. We used five miRNA target prediction databases (mirDB, miRANDA, miRBase, TargetMiner and TargetScan) to predict targets of miRNAs differentially expressed in HCT116 KRAS WT and mutant cells (Figure 2A), prioritising targets common to at least two databases for further analysis. Enriched pathways and processes significantly associated with each predicted target were identified using the bioinformatics tool Metacore, and several common cancer-related processes and pathways identified, including the epithelial–mesenchymal transition (EMT), cell adhesion, cytoskeleton remodelling and cell proliferation (Supplementary Information and Supplementary Table 3). A similar approach was used to compare HCT116 WT cells before and after miR-224 knockdown (Supplementary Information and Supplementary Table 4).
Experimental validation of bioinformatics predictions
The influence of miR-224 knockdown on cell proliferation was investigated by quantitative cell growth assays and flow cytometry analysis. Consistent with our bioinformatics predictions, miR-224 knockdown significantly increased cell doubling time (23.5 to 26.1 h; P<0.001 and 19.8 to 27.5 h; P=0.01, respectively). The effect of miR-224 knockdown on cell cycle kinetics was investigated by flow cytometry analysis, but no significant differences in cell cycle progression were identified, either in untreated cells or following acute treatment with 5-FU (data not shown).
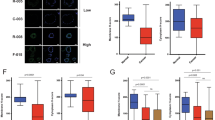
To explore the predicted association between miR-224 and EMT, qRT–PCR analysis was used to compare the expression of candidate EMT genes CDH1 (E-cadherin), CDH2 (N-cadherin) and VIM (Vimentin) in KRAS WT and miR-224 knockdown cells. CDH1 (1.12-fold; P=0.005, Figure 4A) expression was significantly increased in miR-224 knockdown cells, although such subtle variation in gene expression is unlikely to significantly influence the EMT phenotype. In contrast, Vimentin expression was decreased (1.7-fold; P=0.04, Figure 4B), whereas CDH2 was not expressed in HCT116 cells (data not shown).
Confirmation of predicted EMT and altered invasion phenotypes in miR-224 knockdown cells. qRT–PCR analysis was used, as described in Materials and Methods, to compare the expression of (A) E-cadherin (CDH1) and (B) Vimentin in WT KRAS HCT116 and WT KRAS HCT116-miR-224 knockdown cells, with gene expression differences represented relative to the expression of 18S ribosomal RNA. All samples were analysed in triplicate, with experimental errors calculated as previously described (Smith et al, 2012). (C) Innocyte invasion assays were used to model and quantify cell invasion in vitro following Calcein AM labelling, as described in Materials and Methods. Each sample was analysed in triplicate and invasion in WT KRAS, mutant KRAS and WT KRAS HCT116-miR-224 knockdown cells compared with noninvasive cells (no serum control).
InnoCyte invasion assays were then used to show that miR-224 knockdown significantly reduced invasion in HCT116 KRAS WT cells (1.75-fold; P=0.009, Figure 4C). HCT116 KRAS mutant cells were significantly less invasive (1.2-fold; P=0.04) than HCT116 KRAS WT cells, further confirming our hypothesis that miR-224 knockdown in KRAS WT cells phenocopies KRAS mutation. In NIH3T3 cells stably expressing WT or mutant KRAS, however, G13D KRAS mutant cells were significantly more invasive than NIH3T3 KRAS WT cells (4.6-fold; P=0.002).
MiR-224 expression in metastatic colorectal disease
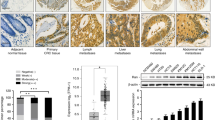
To investigate whether miR-224-dependent invasion differences influenced disease progression, a matched series of primary colorectal cancers and lymph node (n=15) and liver (n=20) metastases from 24 patients were selected for analysis and miR-224 expression compared within each matched pair by qRT–PCR analysis as previously described. MicroRNA-224 expression was significantly decreased in the majority (11 of 15) of lymph node metastasis (Figure 5A) but significantly and more abundantly increased in 13 of 20 liver metastases (Figure 5B). We attempted to further correlate miR-224 expression with patient gender, differentiation, cancer site and KRAS genotype, but found no significant associations, although our patient cohort was too small for meaningful assessment.
Comparison of miR-224 expression in paired primary and metastatic colorectal cancers. qRT–PCR analysis was used, as described in Materials and Methods, to compare miR-224 expression in primary colorectal cancers (n=20) and matched (A) liver and (B) lymph node metastases. MicroRNA-224 expression is illustrated relative to the expression of the invariant miRNA let-7a; all samples were analysed in triplicate, with experimental errors calculated as previously described (Smith et al, 2012). Fold changes in miR-224 expression and associated P-values are summarised for sample pairs where significant differences in gene expression were identified.
Discussion
We have previously described the importance of dysregulated KRAS signalling in the development and progression of colorectal cancer (Smith et al, 2002; Conlin et al, 2005), and now aimed to identify miRNAs associated with disease progression and/or regulation of the RAS/MAPK signalling pathway. We identified several candidate miRNAs, one of which, miR-224, was differentially expressed in colorectal adenomas and cancers, and also in KRAS WT and mutant HCT116-derived colorectal cancer cell lines. MicroRNA-224 is one of the several miRNAs that have previously shown to be differentially expressed in colorectal cancer (Monzo et al, 2008; Arndt et al, 2009; Sarver et al, 2009; Wang et al, 2010; Fu et al, 2012; Liao et al, 2013; Yuan et al, 2013), and increased miR-224 expression in colorectal adenomas has been described by Bartley et al (2011) and Oberg et al (2011), suggesting that it is an early event in colorectal cancer development.
A miR-224/KRAS regulatory interaction has not previously been described, although miR-224 has been shown to repress the tumour suppressor gene RKIP (RAF kinase inhibitory protein) that inhibits metastasis in various cancers by dissociating the interaction between RAF1 and MEK (Huang et al, 2012). Inhibition of miR-224 would therefore be predicted to increase RAS/MAPK signalling, either through activation of KRAS or promotion of RAF1/MEK binding following RKIP activation. Consistent with this hypothesis, we described increased KRAS activity and increased ERK and AKT phosphorylation following miR-224 knockdown in HCT116 colorectal cancer cells, whereas in colorectal cancer patients, RKIP expression has previously been shown to inversely correlate with patient survival and predict metastatic spread (Al-Mulla et al, 2006). Increased RAS activity following miR-224 knockdown is of particular interest, as our bioinformatics analysis identified the guanine nucleotide exchange factor SOS2 (sons of sevenless 2) gene as a candidate miR-224 binding partner, promoting dissociation of KRAS and GDP, and favouring GTP binding (Pierre et al, 2011).
Several additional miRNAs (miR-146b-3p, miR-486-5p, miR-92a, miR-127-3p and miR-378) have previously been shown to be differentially expressed in KRAS WT and mutant colorectal cancers (Ragusa et al, 2010; Mosakhani et al, 2012). Of these, miR-92a, a member of the miR-17-92 cluster, has been most extensively characterised, shown to directly target the anti-apoptotic protein BIM (Tsuchida et al, 2011), and to predict both metastatic spread and survival (Zhou et al, 2013). It will therefore be of particular interest to investigate in future experiments whether the expression of miR-92a, like miR-224, correlates with sensitivity to 5-FU-based chemotherapy in colorectal cancer patients.
Similar to our findings in colorectal cancer, previous studies have described increased miR-224 expression (and increased expression of other genes located on chromosome Xq28) in hepatocellular carcinoma (HCC), as a result of alternative epigenetic regulation of miRNA expression by the histone deacetylases HDAC1 and HDAC3, and the histone acetylase protein EP300 (Wang et al, 2012). Like colorectal cancer, progression of HCC has been associated with increased miR-224 expression, as a consequence of both increased EP300 expression and NF-κB-mediated response to inflammation (Scisciani et al, 2012). We have previously described amplification of the q arm of the X chromosome in both colorectal adenomas (Leslie et al, 2006) and colorectal cancers (Leslie et al, 2003); it will therefore be of particular interest in future studies to investigate whether similar epigenetic and inflammation-mediated mechanisms of miR-224 regulation are important in colorectal cancer.
MicroRNA-224 knockdown in HCT116 KRAS WT cells increased cell doubling time, suggesting that miR-224 influences disease progression by increasing cell proliferation or by inhibiting apoptosis. MicroRNA-224 has recently been shown to promote proliferation through repression of the tumour suppressor gene SMAD4 (Wang et al, 2013; Zhang et al, 2013b), and to influence apoptosis in hepatocellular carcinoma through negative regulation of apoptosis inhibitor protein-5 (API-5; (Wang et al, 2008). Consistent with these findings, we confirmed increased pro-proliferative ERK and STAT3 phosphorylation following miR-224 knockdown, and increased STAT1 phosphorylation that influences tumour suppressor and anti-angiogenesis pathways (Battle et al, 2006). A recent report describing miR-224 targeting of the cyclin-dependent kinase inhibitor CDKN1A (p21cip1; Olaru et al, 2013) is of particular interest, as our own bioinformatics analysis identified the related gene CDKN1B (p27kip1) as a miR-224 target. Consistent with our findings, Liao et al (2013) have recently suggested that miR-224 regulates the G1/S cell cycle transition by downregulating p21cip1 and p27kip1 in colorectal cancers, and may therefore have clinical utility as both a prognostic marker and therapeutic target.
Intriguingly, we have shown that miR-224 knockdown increased 5-FU chemosensitivity, and have suggested that this may result from the creation of a KRAS ‘mutant-like’ phenotype. In support of this hypothesis, we further demonstrated that introduction of mutant KRAS or BRAF to NIH3T3 cells also significantly increased 5-FU chemosensitivity, whereas Klampfer et al (2005) reported protection from 5-FU-induced apoptosis after targeted deletion of a mutant KRAS allele in HCT116 cells, and promotion of cell death following 5-FU challenge in cells transfected with inducible mutant KRAS (V12). However, as KRAS mutation status independently influences disease progression (Smith et al, 2002), it is challenging to assess the unique influence of mutation status on chemotherapy response in colorectal cancer patients.
Our bioinformatics analysis suggested that miR-224 may play a role in regulation of the EMT, and may therefore influence cell invasion and migration. Consistent with this hypothesis, miR-224 knockdown resulted in increased E-cadherin and decreased Vimentin expression. Our bioinformatics data predict miR-224 targeting of additional EMT-associated genes including claudins (CLDN11 and CLDN14) and desmogleins (DGS2), groups of proteins involved in cell adhesion (Lamouille et al, 2014). Our own data in HCT116 colorectal cancer cells and recent similar studies in HepG2 hepatocellular cancer cells suggest that miR-224 promotes cell migration and invasion by activation of AKT signalling and direct targeting of the tumour suppressor gene PPP2R1B (Ma et al, 2012) and rho GTPase-activating proteins ARHGAP9 and ARHGAP21 that deactivate CDC42 (Scisciani et al, 2012). MicroRNA-224 may also influence invasion and metastasis by direct or indirect regulation of matrix metalloproteinases, enzymes secreted by cancer cells that degrade the extracellular matrix and aid dissemination. For example, miR-224 has been shown to regulate both PAK4, a CDC42 effector that plays a role in cytoskeleton remodelling, and the matrix metalloproteinase MMP9 (Li et al, 2010) through direct targeting of the transcription factor HOXD10 (Li et al, 2014). Consistent with our own data, MMP9, like miR-224, was previously shown to be differentially expressed in primary colorectal cancers and matched liver and lymph node metastases (Illemann et al, 2006). Similar phenotypic associations have been proposed for additional miRNAs, including miR-29c and miR-139-5p that, like miR-224, are differentially expressed in colorectal cancer and in KRAS WT and mutant HCT116 cells. MicroRNA-29c, for example, has recently been shown to promote EMT by regulation of β-catenin signalling in colorectal tumours (Zhang et al, 2014a), whereas decreased miR-29c expression has been associated with relapse in colorectal cancer patients (Kuo et al, 2012). In previous TLDA analysis, miR-139-5p was shown to inhibit colorectal cancer cell growth (Schepeler et al, 2012), and has recently been described as a Notch1-dependent tumour suppressor in colorectal cancer cell lines and xenograft models (Zhang et al, 2014b), emphasising the need to extend our miR-224 analysis to additional candidate miRNAs in future studies.
Our data highlight significant differences in miR-224 expression in alternative metastatic sites, where expression is decreased in the majority of lymph node metastases, but increased in liver metastases. These differences may help to rationalise apparent discrepancies in the conclusions of recent studies by Zhang et al (2013a) who confirm increased miR-224 expression in colorectal cancer and suggest that miR-224 may have utility as a relapse biomarker, whereas Yuan et al (2013) alternatively suggest that miR-224 suppresses metastasis in SW480 colorectal cancer cell line-derived xenografts. Although the regulatory role of miR-224 (and additional microRNAs) will almost certainly be influenced by the genetic background of individual colorectal cancers, our data suggest that miRNA expression is influenced by anatomical site, and highlight the potential utility, particularly of stably expressed serum microRNAs, as clinical response biomarkers, although additional quantitative analyses of much larger clinical series will be required to support this conclusion. Although serum levels of miR-224 have not yet been studied as clinical response biomarkers in colorectal cancer, serum miR-224 expression has recently been associated with the development of monogenic diabetes (HNF1A-MODY; (Bonner et al, 2013), shown to be increased in Crohn’s disease (Fujioka et al, 2014) and renal cell carcinoma patients (Cheng et al, 2013) and to predict response to combination (R-CHOP) chemotherapy in diffuse large B-cell lymphoma patients (Song et al, 2014).
Change history
28 April 2015
This paper was modified 12 months after initial publication to switch to Creative Commons licence terms, as noted at publication
References
Al-Mulla F, Hagan S, Behbehani AI, Bitar MS, George SS, Going JJ, García JJC, Scott L, Fyfe N, Murray GI, Kolch W (2006) Raf kinase inhibitor protein expression in a survival analysis of colorectal cancer patients. J Clin Oncol 24: 5672–5679.
Amado RG, Wolf M, Peeters M, Van Cutsem E, Siena S, Freeman DJ, Juan T, Sikorski R, Suggs S, Radinsky R, Patterson SD, Chang DD (2008) Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol 26: 1626–1634.
Arndt GM, Dossey L, Cullen LM, Lai A, Druker R, Eisbacher M, Zhang C, Tran N, Fan H, Retzlaff K, Bittner A, Raponi M (2009) Characterization of global microRNA expression reveals oncogenic potential of miR-145 in metastatic colorectal cancer. BMC Cancer 9: 374.
Bandrés E, Cubedo E, Agirre X, Malumbres R, Zárate R, Ramirez N, Abajo A, Navarro A, Moreno I, Monzó M, García-Foncillas J (2006) Identification by real-time PCR of 13 mature microRNAs differentially expressed in colorectal cancer and non-tumoral tissues. Mol Cancer 5: 29.
Barbacid M (1990) ras oncogenes: their role in neoplasia. Eur J Clin Invest 20: 225–235.
Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281–297.
Bartley AN, Yao H, Barkoh BA, Ivan C, Mishra BM, Rashid A, Calin GA, Luthra R, Hamilton SR (2011) Complex patterns of altered microRNA expression during the adenoma-adenocarcinoma sequence for microsatellite-stable colorectal cancer. Clin Cancer Res 17: 7283–7293.
Battle TE, Lynch RA, Frank DA (2006) Signal transducer and activator of transcription 1 activation in endothelial cells is a negative regulator of angiogenesis. Cancer Res 66: 3649–3657.
Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B Methodol 57: 289–300.
Bokemeyer C, Bondarenko I, Hartmann JT, De Braud FG, Volovat C, Nippgen J, Stroh C, Celik I, Koralewski P (2008) KRAS status and efficacy of first-line treatment of patients with metastatic colorectal cancer (mCRC) with FOLFOX with or without cetuximab: the OPUS experience. ASCO Meet Abstr 26: 4000.
Bonner C, Nyhan KC, Bacon S, Kyithar MP, Schmid J, Concannon CG, Bray IM, Stallings RL, Prehn JHM, Byrne MM (2013) Identification of circulating microRNAs in HNF1A-MODY carriers. Diabetologia 56: 1743–1751.
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254.
Cancer Research UK (2014) Bowel cancer key facts http://www.cancerresearchuk.org/cancer-info/cancerstats/keyfacts/bowel-cancer/#survival accessed: 29 June 2014.
Chang KH, Miller N, Kheirelseid EAH, Lemetre C, Ball GR, Smith MJ, Regan M, McAnena OJ, Kerin MJ (2011) MicroRNA signature analysis in colorectal cancer: identification of expression profiles in stage II tumors associated with aggressive disease. Int J Colorectal Dis 26: 1415–1422.
Chen X, Guo X, Zhang H, Xiang Y, Chen J, Yin Y, Cai X, Wang K, Wang G, Ba Y, Zhu L, Wang J, Yang R, Zhang Y, Ren Z, Zen K, Zhang J, Zhang C-Y (2009) Role of miR-143 targeting KRAS in colorectal tumorigenesis. Oncogene 28: 1385–1392.
Cheng T, Wang L, Li Y, Huang C, Zeng L, Yang J (2013) Differential microRNA expression in renal cell carcinoma. Oncol Lett 6: 769–776.
Conlin A, Smith G, Carey FA, Wolf CR, Steele RJC (2005) The prognostic significance of K-ras, p53, and APC mutations in colorectal carcinoma. Gut 54: 1283–1286.
Earle JSL, Luthra R, Romans A, Abraham R, Ensor J, Yao H, Hamilton SR (2010) Association of microRNA expression with microsatellite instability status in colorectal adenocarcinoma. J Mol Diagn 12: 433–440.
Faltejskova P, Svoboda M, Srutova K, Mlcochova J, Besse A, Nekvindova J, Radova L, Fabian P, Slaba K, Kiss I, Vyzula R, Slaby O (2012) Identification and functional screening of microRNAs highly deregulated in colorectal cancer. J Cell Mol Med 16: 2655–2666.
Fu J, Tang W, Du P, Wang G, Chen W, Li J, Zhu Y, Gao J, Cui L (2012) Identifying microRNA-mRNA regulatory network in colorectal cancer by a combination of expression profile and bioinformatics analysis. BMC Syst Biol 6: 68.
Fujioka S, Nakamichi I, Esaki M, Asano K, Matsumoto T, Kitazono T (2014) Serum microRNA levels in patients with Crohn’s disease during induction therapy by infliximab. J Gastroenterol Hepatol 29: 1207–1214.
Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JY, Zhang J (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5: R80.
Hamfjord J, Stangeland AM, Hughes T, Skrede ML, Tveit KM, Ikdahl T, Kure EH (2012) Differential expression of miRNAs in colorectal cancer: comparison of paired tumor tissue and adjacent normal mucosa using high-throughput sequencing. PloS One 7: e34150.
Huang L, Dai T, Lin X, Zhao X, Chen X, Wang C, Li X, Shen H, Wang X (2012) MicroRNA-224 targets RKIP to control cell invasion and expression of metastasis genes in human breast cancer cells. Biochem Biophys Res Commun 425: 127–133.
Illemann M, Bird N, Majeed A, Sehested M, Laerum OD, Lund LR, Danø K, Nielsen BS (2006) MMP-9 is differentially expressed in primary human colorectal adenocarcinomas and their metastases. Mol Cancer Res 4: 293–302.
Kanaan Z, Roberts H, Eichenberger MR, Billeter A, Ocheretner G, Pan J, Rai SN, Jorden J, Williford A, Galandiuk S (2013) A plasma microRNA panel for detection of colorectal adenomas: a step toward more precise screening for colorectal cancer. Ann Surg 258: 400–408.
Klampfer L, Swaby L-A, Huang J, Sasazuki T, Shirasawa S, Augenlicht L (2005) Oncogenic Ras increases sensitivity of colon cancer cells to 5-FU-induced apoptosis. Oncogene 24: 3932–3941.
Kuo T-Y, Hsi E, Yang I-P, Tsai P-C, Wang J-Y, Juo S-HH (2012) Computational analysis of mRNA expression profiles identifies microRNA-29a/c as predictor of colorectal cancer early recurrence. PloS One 7: e31587.
Lamouille S, Xu J, Derynck R (2014) Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol 15: 178–196.
Leslie A, Pratt NR, Gillespie K, Sales M, Kernohan NM, Smith G, Wolf CR, Carey FA, Steele RJC (2003) Mutations of APC, K-ras, and p53 are associated with specific chromosomal aberrations in colorectal adenocarcinomas. Cancer Res 63: 4656–4661.
Leslie A, Stewart A, Baty DU, Mechan D, McGreavey L, Smith G, Wolf CR, Sales M, Pratt NR, Steele RJC, Carey FA (2006) Chromosomal changes in colorectal adenomas: relationship to gene mutations and potential for clinical utility. Genes Chromosomes Cancer 45: 126–135.
Lewis BP, Burge CB, Bartel DP (2005) Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120: 15–20.
Li Q, Ding C, Chen C, Zhang Z, Xiao H, Xie F, Lei L, Chen Y, Mao B, Jiang M, Li J, Wang D, Wang G (2014) miR-224 promotion of cell migration and invasion by targeting Homeobox D 10 gene in human hepatocellular carcinoma. J Gastroenterol Hepatol 29: 835–842.
Li Q, Wang G, Shan J-L, Yang Z-X, Wang H-Z, Feng J, Zhen J-J, Chen C, Zhang Z-M, Xu W, Luo X-Z, Wang D (2010) MicroRNA-224 is upregulated in HepG2 cells and involved in cellular migration and invasion. J Gastroenterol Hepatol 25: 164–171.
Liao W-T, Li T-T, Wang Z-G, Wang S-Y, He M-R, Ye Y-P, Qi L, Cui Y-M, Wu P, Jiao H-L, Zhang C, Xie Y-J, Wang J-X, Ding Y-Q (2013) microRNA-224 promotes cell proliferation and tumor growth in human colorectal cancer by repressing PHLPP1 and PHLPP2. Clin Cancer Res 19: 4662–4672.
Lièvre A, Bachet J-B, Le Corre D, Boige V, Landi B, Emile J-F, Côté J-F, Tomasic G, Penna C, Ducreux M, Rougier P, Penault-Llorca F, Laurent-Puig P (2006) KRAS mutation status is predictive of response to cetuximab therapy in colorectal cancer. Cancer Res 66: 3992–3995.
Ma D, Tao X, Gao F, Fan C, Wu D (2012) miR-224 functions as an onco-miRNA in hepatocellular carcinoma cells by activating AKT signaling. Oncol Lett 4: 483–488.
Monzo M, Navarro A, Bandres E, Artells R, Moreno I, Gel B, Ibeas R, Moreno J, Martinez F, Diaz T, Martinez A, Balagué O, Garcia-Foncillas J (2008) Overlapping expression of microRNAs in human embryonic colon and colorectal cancer. Cell Res 18: 823–833.
Mosakhani N, Sarhadi VK, Borze I, Karjalainen-Lindsberg M-L, Sundström J, Ristamäki R, Osterlund P, Knuutila S (2012) MicroRNA profiling differentiates colorectal cancer according to KRAS status. Genes Chromosomes Cancer 51: 1–9.
Motoyama K, Inoue H, Takatsuno Y, Tanaka F, Mimori K, Uetake H, Sugihara K, Mori M (2009) Over- and under-expressed microRNAs in human colorectal cancer. Int J Oncol 34: 1069–1075.
National Health Service (2014) NHS Bowel Cancer Screening Programme, 2014 http://www.cancerscreening.nhs.uk/bowel/ accessed: 29 July 2014.
Oberg AL, French AJ, Sarver AL, Subramanian S, Morlan BW, Riska SM, Borralho PM, Cunningham JM, Boardman LA, Wang L, Smyrk TC, Asmann Y, Steer CJ, Thibodeau SN (2011) miRNA expression in colon polyps provides evidence for a multihit model of colon cancer. PloS One 6: e20465.
Olaru AV, Yamanaka S, Vazquez C, Mori Y, Cheng Y, Abraham JM, Bayless TM, Harpaz N, Selaru FM, Meltzer SJ (2013) MicroRNA-224 negatively regulates p21 expression during late neoplastic progression in inflammatory bowel disease. Inflamm Bowel Dis 19: 471–480.
Pierre S, Bats A-S, Coumoul X (2011) Understanding SOS (Son of Sevenless). Biochem Pharmacol 82: 1049–1056.
R Development Core Team (2008) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing: Vienna, Austria.
Ragusa M, Majorana A, Statello L, Maugeri M, Salito L, Barbagallo D, Guglielmino MR, Duro LR, Angelica R, Caltabiano R, Biondi A, Di Vita M, Privitera G, Scalia M, Cappellani A, Vasquez E, Lanzafame S, Basile F, Di Pietro C, Purrello M (2010) Specific alterations of microRNA transcriptome and global network structure in colorectal carcinoma after cetuximab treatment. Mol Cancer Ther 9: 3396–3409.
Reid JF, Sokolova V, Zoni E, Lampis A, Pizzamiglio S, Bertan C, Zanutto S, Perrone F, Camerini T, Gallino G, Verderio P, Leo E, Pilotti S, Gariboldi M, Pierotti MA (2012) miRNA profiling in colorectal cancer highlights miR-1 involvement in MET-dependent proliferation. Mol Cancer Res 10: 504–515.
Sarver AL, French AJ, Borralho PM, Thayanithy V, Oberg AL, Silverstein KAT, Morlan BW, Riska SM, Boardman LA, Cunningham JM, Subramanian S, Wang L, Smyrk TC, Rodrigues CMP, Thibodeau SN, Steer CJ (2009) Human colon cancer profiles show differential microRNA expression depending on mismatch repair status and are characteristic of undifferentiated proliferative states. BMC Cancer 9: 401.
Schepeler T, Holm A, Halvey P, Nordentoft I, Lamy P, Riising EM, Christensen LL, Thorsen K, Liebler DC, Helin K, Ørntoft TF, Andersen CL (2012) Attenuation of the beta-catenin/TCF4 complex in colorectal cancer cells induces several growth-suppressive microRNAs that target cancer promoting genes. Oncogene 31: 2750–2760.
Schetter AJ, Leung SY, Sohn JJ, Zanetti KA, Bowman ED, Yanaihara N, Yuen ST, Chan TL, Kwong DLW, Au GKH, Liu C-G, Calin GA, Croce CM, Harris CC (2008) MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA 299: 425–436.
Scisciani C, Vossio S, Guerrieri F, Schinzari V, De Iaco R, D’Onorio de Meo P, Cervello M, Montalto G, Pollicino T, Raimondo G, Levrero M, Pediconi N (2012) Transcriptional regulation of miR-224 upregulated in human HCCs by NFκB inflammatory pathways. J Hepatol 56: 855–861.
Scottish Bowel Screening Programme (2014) Why is screening important? http://www.bowelscreening.scot.nhs.uk/why-is-screening-important accessed: 29 June 2014.
Scottish Intercollegiate Guidelines Network (2011) Diagnosis and management of colorectal cancer. SIGN: Edinburgh, 2014. Available at http://www.sign.ac.uk.
Shirasawa S, Furuse M, Yokoyama N, Sasazuki T (1993) Altered growth of human colon cancer cell lines disrupted at activated Ki-ras. Science 260: 85–88.
Smith G, Bounds R, Wolf H, Steele RJC, Carey FA, Wolf CR (2010) Activating K-Ras mutations outwith ‘hotspot’ codons in sporadic colorectal tumours - implications for personalised cancer medicine. Br J Cancer 102: 693–703.
Smith G, Carey FA, Beattie J, Wilkie MJV, Lightfoot TJ, Coxhead J, Garner RC, Steele RJC, Wolf CR (2002) Mutations in APC, Kirsten-ras, and p53—alternative genetic pathways to colorectal cancer. Proc Natl Acad Sci USA 99: 9433–9438.
Smith G, Ng MTH, Shepherd L, Herrington CS, Gourley C, Ferguson MJ, Wolf CR (2012) Individuality in FGF1 expression significantly influences platinum resistance and progression-free survival in ovarian cancer. Br J Cancer 107: 1327–1336.
Smyth GK (2004) Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3: Article3.
Song G, Gu L, Li J, Tang Z, Liu H, Chen B, Sun X, He B, Pan Y, Wang S, Cho WC (2014) Serum microRNA expression profiling predict response to R-CHOP treatment in diffuse large B cell lymphoma patients. Ann Hematol 93: 1735–1743.
Torrance CJ, Agrawal V, Vogelstein B, Kinzler KW (2001) Use of isogenic human cancer cells for high-throughput screening and drug discovery. Nat Biotechnol 19: 940–945.
Tsuchida A, Ohno S, Wu W, Borjigin N, Fujita K, Aoki T, Ueda S, Takanashi M, Kuroda M (2011) miR-92 is a key oncogenic component of the miR-17-92 cluster in colon cancer. Cancer Sci 102: 2264–2271.
Wang Y, Lee ATC, Ma JZI, Wang J, Ren J, Yang Y, Tantoso E, Li K-B, Ooi LLPJ, Tan P, Lee CGL (2008) Profiling microRNA expression in hepatocellular carcinoma reveals microRNA-224 up-regulation and apoptosis inhibitor-5 as a microRNA-224-specific target. J Biol Chem 283: 13205–13215.
Wang Y, Ren J, Gao Y, Ma JZI, Toh HC, Chow P, Chung AYF, Ooi LLPJ, Lee CGL (2013) MicroRNA-224 targets SMAD family member 4 to promote cell proliferation and negatively influence patient survival. PloS One 8: e68744.
Wang Y, Toh HC, Chow P, Chung AYF, Meyers DJ, Cole PA, Ooi LLPJ, Lee CGL (2012) MicroRNA-224 is up-regulated in hepatocellular carcinoma through epigenetic mechanisms. FASEB J 26: 3032–3041.
Wang YX, Zhang XY, Zhang BF, Yang CQ, Chen XM, Gao HJ (2010) Initial study of microRNA expression profiles of colonic cancer without lymph node metastasis. J Dig Dis 11: 50–54.
Weidlich S, Walsh K, Crowther D, Burczynski ME, Feuerstein G, Carey FA, Steele RJC, Wolf CR, Miele G, Smith G (2011) Pyrosequencing-based methods reveal marked inter-individual differences in oncogene mutation burden in human colorectal tumours. Br J Cancer 105: 246–254.
Wu CW, Ng SC, Dong Y, Tian L, Ng SSM, Leung WW, Law WT, Yau TO, Chan FKL, Sung JJY, Yu J (2014) Identification of microRNA-135b in stool as a potential noninvasive biomarker for colorectal cancer and adenoma. Clin Cancer Res 20: 2994–3002.
Yuan K, Xie K, Fox J, Zeng H, Gao H, Huang C, Wu M (2013) Decreased levels of miR-224 and the passenger strand of miR-221 increase MBD2, suppressing maspin and promoting colorectal tumor growth and metastasis in mice. Gastroenterology 145: 853–864, e9.
Zhang G-J, Zhou H, Xiao H-X, Li Y, Zhou T (2013a) Up-regulation of miR-224 promotes cancer cell proliferation and invasion and predicts relapse of colorectal cancer. Cancer Cell Int 13: 104.
Zhang JX, Mai SJ, Huang XX, Wang FW, Liao YJ, Lin MC, Kung HF, Zeng YX, Xie D (2014a) MiR-29c mediates epithelial-to-mesenchymal transition in human colorectal carcinoma metastasis via PTP4A and GNA13 regulation of β-catenin signaling. Ann Oncol 25: 2196–2204.
Zhang L, Dong Y, Zhu N, Tsoi H, Zhao Z, Wu CW, Wang K, Zheng S, Ng SS, Chan FK, Sung JJ, Yu J (2014b) microRNA-139-5p exerts tumor suppressor function by targeting NOTCH1 in colorectal cancer. Mol Cancer 13: 124.
Zhang Y, Takahashi S, Tasaka A, Yoshima T, Ochi H, Chayama K (2013b) Involvement of microRNA-224 in cell proliferation, migration, invasion, and anti-apoptosis in hepatocellular carcinoma. J Gastroenterol Hepatol 28: 565–575.
Zhou T, Zhang G, Liu Z, Xia S, Tian H (2013) Overexpression of miR-92a correlates with tumor metastasis and poor prognosis in patients with colorectal cancer. Int J Colorectal Dis 28: 19–24.
Acknowledgements
We acknowledge Dr Gino Miele, Dr Dan Crowther and the Translational Medicine Research Collaboration project team, Professor Bert Vogelstein for the generous gift of HCT116 KRAS WT and mutant cells, Professor Richard Marais for the generous gift of WT and V600E BRAF expression plasmids and Tayside Tissue Bank for help with our human tissue studies. We gratefully acknowledge the award of a Medical Research Council CASE PhD studentship, and additional funding from Cancer Research UK (C4639/A10822), the Translational Medicine Research Collaboration and a Strategic Research Development Grant from the Scottish Funding Council.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License.
Supplementary Information accompanies this paper on British Journal of Cancer website
Supplementary information
Rights and permissions
From twelve months after its original publication, this work is licensed under the Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/4.0/
About this article
Cite this article
Amankwatia, E., Chakravarty, P., Carey, F. et al. MicroRNA-224 is associated with colorectal cancer progression and response to 5-fluorouracil-based chemotherapy by KRAS-dependent and -independent mechanisms. Br J Cancer 112, 1480–1490 (2015). https://doi.org/10.1038/bjc.2015.125
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/bjc.2015.125
Keywords
This article is cited by
-
The diagnostic potential of miR-196a-1 in colorectal cancer
BMC Cancer (2024)
-
MiRNA-mediated EMT and CSCs in cancer chemoresistance
Experimental Hematology & Oncology (2021)
-
Current Evidence on miRNAs as Potential Theranostic Markers for Detecting Chemoresistance in Colorectal Cancer: A Systematic Review and Meta-Analysis of Preclinical and Clinical Studies
Molecular Diagnosis & Therapy (2019)
-
Mechanistic regulation of epithelial-to-mesenchymal transition through RAS signaling pathway and therapeutic implications in human cancer
Journal of Cell Communication and Signaling (2018)
-
MiR-215-5p is a tumor suppressor in colorectal cancer targeting EGFR ligand epiregulin and its transcriptional inducer HOXB9
Oncogenesis (2017)