Abstract
Background:
Extracapsular spread (ECS) in cervical lymph nodes is the single-most prognostic clinical variable in oral squamous cell carcinoma (OSCC), but diagnosis is possible only after histopathological examination. A promising biomarker in the primary tumour, alpha smooth muscle actin (SMA) has been shown to be highly prognostic, however, validated biomarkers to predict ECS prior to primary treatment are not yet available.
Methods:
In 102 OSCC cases, conventional imaging was compared with pTNM staging. SERPINE1, identified from expression microarray of primary tumours as a potential biomarker for ECS, was validated through mRNA expression, and by immunohistochemistry (IHC) on a tissue microarray from the same cohort. Similarly, expression of SMA was also compared with its association with ECS and survival. Expression was analysed separately in the tumour centre and advancing front; and prognostic capability determined using Kaplan–Meier survival analysis.
Results:
Immunohistochemistry indicated that both SERPINE1 and SMA expression at the tumour-advancing front were significantly associated with ECS (P<0.001). ECS was associated with expression of either or both proteins in all cases. SMA+/SERPINE1+ expression in combination was highly significantly associated with poor survival (P<0.001). MRI showed poor sensitivity for detection of nodal metastasis (56%) and ECS (7%). Both separately, and in combination, SERPINE1 and SMA were superior to MRI for the detection of ECS (sensitivity: SERPINE1: 95%; SMA: 82%; combination: 81%).
Conclusion:
A combination of SMA and SERPINE1 IHC offer potential as prognostic biomarkers in OSCC. Our findings suggest that biomarkers at the invasive front are likely to be necessary in prediction of ECS or in therapeutic stratification.
Similar content being viewed by others
Main
Head and neck squamous cell carcinoma (HNSCC) is the sixth commonest cancer worldwide and recent studies have shown an increasing incidence (Argiris et al, 2008). Oral squamous cell carcinoma (OSCC) is the commonest form of HNSCC and currently treatment is relatively uniform, comprising primary surgery, often with the addition of post-operative radiotherapy (PORT) (or chemoradiotherapy) for high-risk cases (Shaw et al, 2011). Unlike oropharyngeal SCC (OPSCC) (Schache et al, 2011), OSCC is only rarely mediated by human papillomavirus (HPV) (Lopes et al, 2011) and also known to be highly molecularly heterogenous, hence the development of clinically validated biomarkers has been slow (Leemans et al, 2011).
Extracapsular spread (ECS) in metastatic cervical lymph nodes is the single-most prognostic clinical variable for recurrence and death in OSCC (Alvi and Johnson, 1996; Myers et al, 2001; Greenberg et al, 2003; Shaw et al, 2010). In our recent large cohort, the overall 5-year survival for patients with ECS was 23%, compared with 60% for patients without ECS(Shaw et al, 2010). Poor outcomes are concentrated in those patients with ECS for whom trials of intensification or novel therapies might be justified. On the other hand, for cases that demonstrate lower biological aggression or a reduced tendency for metastasis, it may be possible to de-escalate therapy (Barry et al, 2013). Although the clinical significance of ECS is well established, its diagnosis currently requires histological examination of lymph nodes and therefore is made only following definitive primary surgery involving neck dissection. There is little in the published literature exploiting ECS as a clinical variable for biological investigation, and likewise, there is limited investigation of its molecular determinants (Zhou et al, 2006). Molecular biomarkers identifying high risk of ECS, or poor prognosis, prior to definitive therapy could direct novel therapies or treatment intensification.
An important observation related to the presence of ECS (ECS+) is that tumour recurrences occur most frequently at the primary site, although regional and distant failures do also occur more frequently than in non-ECS (ECS−) cases (Shaw et al, 2010). This suggests that the biological determinants of ECS will likely be evident in the primary tumour and may even be identified in diagnostic biopsy specimens(Califano et al, 1996). Indeed, molecular fingerprints for metastatic tendencies have been found in primary sites of cancers other than OSCC (Ramaswamy et al, 2003).
Recent studies have also shown that the presence of activated fibroblasts, otherwise known as myofibroblasts, in the tumour-associated stroma are of high prognostic value in OSCC (Kellermann et al, 2007; Vered et al, 2010; Marsh et al, 2011). Myofibroblasts are usually demonstrated by the presence of alpha smooth muscle actin (SMA) expression, and their presence has also been shown to predict for disease recurrence across a number of tumour types (Surowiak et al, 2007; Tsujino et al, 2007). Given the prognostic association of both ECS and SMA, significant overlap of cases demonstrating both features would be expected.
Here, using a well-annotated cohort of OSCC patients, we aim to validate candidate biomarkers within primary tumour samples, using quantitative reverse transcription-PCR (qRT–PCR) and protein expression with immunohistochemistry (IHC). Further, we aim to compare the diagnostic accuracy of conventional imaging with these biomarkers in the diagnosis of ECS, and also their prognostic accuracy.
Patients and methods
Clinical cohort and imaging
Ethical approval was obtained (South Sefton EC 47.01 and REC No 10/H1002/53) and 102 patients were included that were treated at the Aintree University Hospital between June 2003 and June 2010. Inclusion criteria for selection were cases with a new histologically confirmed diagnosis of OSCC and treatment with primary surgery. A discovery cohort of 55 cases from June 2003 to November 2008 (pN staging: 17 pN0; 11pN+ECS−; 27pN+ECS+) and a validation cohort of 47 cases from December 2008 to June 2010 (pN staging: 21 pN0; 11pN+ECS−; 15pN+ECS+) were identified. All cases had both snap-frozen and formalin-fixed paraffin-embedded (FFPE) tumour/normal tissue available and were distinct from previously published cohorts from the same regional unit (Field et al, 1995; Shaw et al, 2006; Rogers et al, 2009; Shaw et al, 2010; Lwin et al, 2012; Shaw et al, 2013).
The demographic features, pathological staging and outcomes of the cohort were collected, with nodal and ECS status were determined using standardised protocols (Woolgar and Triantafyllou, 2009). The data for routine imaging using MRI (magnetic resonance imaging) scans was included, as this represents the current standard of care for pre-treatment staging and diagnosis of ECS. Magnetic resonance imaging scans were re-assessed, blinded to pathological neck staging and outcomes, by a single radiologist (RH). Standard radiological criteria for nodal status were used, with ECS suspected when there was high signal change or oedema surrounding the nodes or the fat suppressed on STIR (short T1 inversion recovery) images, when the margins of the nodal masses were indistinct, or when the nodal masses invaded the overlying sternocleidomastoid muscle (van den Brekel et al, 1990; Lwin et al, 2012).
RNA extraction
Tumour tissue was collected at the time of surgical resection from a superficial, non-necrotic, central aspect of the lesions and was immediately stored at −80 °C. RNA was extracted from 3-mm3 tissue using the AllPrep DNA/RNA/protein mini kit (Qiagen, Valencia, CA, USA) with an on-column RNase-free DNase (Qiagen) treatment to avoid DNA carryover in subsequent RNA preparation. We have previously quantified tumour and stromal proportions in comparable HNSCC specimens, demonstrating tumour proportions >50% in all samples and 80% in two-thirds of samples; microdissection was not undertaken in this series (Schache et al, 2011). Total RNA was quantified spectrophotometrically with the Nanodrop 1000 Spectrophotometer (Wilmington, DA, USA). RNA quality and integrity were determined using the Agilent 2100 Bioanalyser (Agilent Technologies, Waldbron, Germany). RNA inclusion criteria for the single-gene assays were >1.8 260/230 ratio, >1.8 260/280 ratio and an RNA integrity number >6 (Hoffman et al, 2004). Total RNA (500 ng) was used for cDNA synthesis using the SuperScript III First Strand Synthesis System (Invitrogen, Carlsbad, CA, USA) and used as follows.
HPV testing
As some of the tumours encroached on oral cavity/oropharynx site borders, it was deemed prudent to determine HPV status in all samples to clarify the underlying molecular aetiology of the tumour. In brief, real-time PCR reactions of HPV16 E7 were normalised with β-actin and compared with a standard curve generated from a serial dilution of the HPV16-positive cell line SiHa (ATCC-LGC-HTB-35, Manassas, VA, USA) (Schache et al, 2011).
Gene selection and validation
Microarray analysis of the discovery cohort had revealed an eight-gene signature for the presence of ECS (unpublished data, not shown), of which the most promising single genes, HEXIM1 and SERPINE1 were selected for validation on the basis of highest area under the curve on receiver operating curve (ROC) analysis. Quantitative RT–PCR was carried out in duplicate using a multiplexed assay with the target probe FAM-labelled (Hs01126604_m1 and Hs00538918_s1) and a VIC-labelled endogenous control (GAPDH assay, Paisley, UK: Hs02758991_g1) on an Applied Biosystems (Paisley, UK) 7500 FAST thermal cycler. This facilitated internal normalisation and determination of relative quantitation was done using the comparative DdCt method (2−ΔΔCt) as described previously (Livak and Schmittgen, 2001). Relative efficiency was determined using a standard curve generated from a serial dilution of total cDNA (derived from cell lines), which was also used as an internal calibrator between plates.
Tissue microarray (TMA) construction
TMAs were constructed using cores selected from FFPE blocks of primary OSCC tissue, using a manual tissue arrayer (MTA-1, Beecher Instruments, Sun Prairie, WI, USA) as previously described (Parsons and Grabsch, 2009). Areas of the primary tumour centre, tumour-advancing front (tumour–stroma interface) and adjacent tumour-free mucosa were identified and marked on archival haematoxylin and eosin-stained (H&E) sections by an oral and maxillofacial pathologist (AT). Triplicate cores, with a 4-mm depth, were obtained from the marked areas and transferred to recipient array blocks in a randomised distribution, with each replicate located on a different array block. Diameter cores (0.6 mm) were obtained from the tumour centre and tumour-free mucosa; 1-mm diameter cores were obtained from the tumour-advancing front. H&E sections of the TMAs were prepared and examined to confirm the accuracy of tissue sampling.
Immunohistochemistry
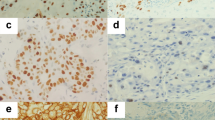
Immunohistochemistry was carried out by standard methods on a DAKO Autostainer (Dako, Ely, UK), using its proprietary kit, the DAKO Envision FLEX/HRP Detection System. In brief, a high-temperature antigen retrieval method was employed using the DAKO PT-Link system (Dako) on 4-μm sections of the TMAs and final dilutions of 1 : 600 of SMA antibody (Clone 1A4, Dako), 1 : 300 of SERPINE1 antibody (3785, American Diagnostica, Stamford, CT, USA) and 1 : 800 anti-HEXIM ChIP grade (ab25388, Abcam, Cambridge, UK). Primary antibody was omitted from negative controls.
Immunohistochemistry-stained TMA blocks were analysed (AT blinded) and validated (JD) using a semi-quantitative method for scoring; SMA expression that reflected the number/density of myofibroblasts at the tumour centre and advancing front conveyed as intensity of immunoreactivity; and graded as high (florid), intermediate or low (sparse or absent). SERPINE1 and HEXIM1 scoring was similarly graded for intensity of immunoreactivity at the tumour centre with a separate score at the tumour–stroma interface for both tumour cells and stromal cells. Concordance was >95% with remaining cases re-analysed and a consensus score agreed.
Data analysis
Statistical analysis was performed using the Software Package for Statistical Analysis, (SPSS) Version 20 (IBM, Armonk, NY, USA). Kaplan–Meier (KM) survival analysis was carried out with a log-rank (Mantel–Cox) test for comparison between curves for overall survival (OS), which was defined by death from any cause. Survival was determined by attendance at the most recent follow-up appointment indicating alive or registered death from case-note review. Gene-expression values between the ECS− and ECS+ groups were compared by non-parametric testing (Wilcoxon test) for determination of significance. The classification power of the individual gene expression was assessed using a ROC curve analysis with calculation of the area under curve. Significance of differences between IHC scores with pathological staining and ECS were determined using χ2-analysis.
Results
The clinical and pathological characteristics of the cohort are shown in Table 1. Tongue and floor of mouth were the commonest sites with the majority of tumours pT2 or pT4. The presence of ECS was highly significant for OS (estimated 5-year survival: pN+ECS 10%; pN+ECS− 70%; pN0 62% P<0.001) with median follow-up 19 months (range 1–87 months). There was a male preponderance in the pN0 cases and a female preponderance in the pN+ECS+ cases (P<0.007). Other than this unexpected gender association, this relationship between ECS and survival and other features were consistent with our previous larger clinical cohort (6).
All patients received primary surgery aiming for pathological resection margins of 5 mm and with dissection of appropriate cervical lymph node levels. The distribution of adjuvant treatment received by the cohort was PORT (50–66 Gy) in 63 cases (62%), post-operative concomitant cisplatin (POCRT) (75 mg m−2 in 3 cycles) chemotherapy in 7 cases (7%), no adjuvant treatment in 31 cases (31%) and data not available in 1 case (1%). Within the pN0 group, 24 (63%) did not receive adjuvant therapy, whereas 14 (37%) had PORT for close or involved margins. Within the pN+ECS-ve group, 4 (21%) did not receive adjuvant therapy, whereas 15(79%) had PORT. Within the ECS group, 3 (7%) had no adjuvant, 34 (77%) had PORT and 7 (16%) POCRT. The reason for some patients not receiving POCRT were advanced age (>70 years), significant contraindicating co-morbidity or that their date of treatment preceded the published evidence supporting POCRT. In addition, for a group of five patients prolonged surgical and medical complications, or patient preference resulted in them not receiving the prescribed adjuvant therapy.
After quality-control assessment, 89 out of 102 RNA samples were suitable for qRT–PCR (Table 1), MRI scans were available for 83 cases and all 102 cases were utilised for TMA construction and subsequent IHC. Four out of 89 cases were identified as HPV positive, all of which were used in the single-gene analysis; only one of these cases was potentially overlapping the oropharynx. As none of the four HPV-positive cases were associated with ECS, they were included in the cohort for biomarker studies because the most important concern regarding known prognostic bias was excluded.
Technical validation of the microarray findings by qRT–PCR showed a moderately positive correlation between the two methods for SERPINE1 expression (r=0.56, P=0.77), but a poorer correlation for HEXIM1 (r=0.22, P=0.15). Area under curve analysis of the ROC curves based on ECS status using single-gene analysis for SERPINE1 was 0.68 and HEXIM1 was 0.67 (Supplementary Table 1). Immunohistochemistry showed that HEXIM1 was ubiquitously expressed with widespread intranuclear high intensity in most cores and therefore non-discriminatory for any pathological features and is not discussed further.
Low-intensity SMA expression was always associated with a focal distribution, whereas intermediate or high-intensity SMA expression was associated with multifocal or diffuse distribution. Significant associations were observed between SMA expression in myofibroblasts at the tumour-advancing front (Figure 1A) and the nodal status (pN0 or pN+) (P<0.001), N-stage (P=0.007) and ECS status (P<0.001) of the tumour (Figure 2). However, differences in SMA expression at the tumour centre did not significantly correlate with any of these variables (P=0.09, P=0.52 and P=0.4, respectively).
SERPINE1 expression in the tumour cells at the tumour-advancing front (Figure 1B) was positively correlated with nodal status, N-stage and ECS (each P<0.001) (Figure 3), but not T-stage. SERPINE 1 expression in stromal cells at the tumour-advancing front did not show these associations. No significant differences in SERPINE1 expression were observed in the tumour centre in relation to T-stage and ECS, but correlations with nodal status and N-stage were highly significant (P=0.002 and P=0.007, respectively).
Analysis of the distribution of SERPINE1 and SMA expression among the whole cohort in relation to ECS status indicate that low positivity for SMA and SERPINE1 effectively excludes ECS-positive tumours (Figure 4).
Neither singular SERPINE1 (P=0.06) nor SMA expression (P=0.042) at the tumour-advancing front matched the prognostic ability of ECS (P<0.001) to predict OS (Shaw et al, 2010) (Figure 5A and B). It became evident that high and intermediate expressions cluster in their prognostic association, so they were considered jointly for further analyses. The combination where both proteins are positively expressed is highly prognostic (P<0.001) (Figure 5C) and highly informative for this cohort, being seen in 32 out of 42 (76%) of ECS cases.
Table 2 indicates that MRI offers very poor sensitivity, although it shows good specificity for overall nodal status and presence of ECS. Either SERPINE1 or SMA expression at the advancing front offer significantly better sensitivity at the cost of specificity for both nodal and ECS status compared with MRI, whereas high or intermediate expression of both SERPINE1 and SMA was sensitive for the diagnosis of ECS, but again lacked specificity. In combination, however, negative SMA and SERPINE1 expressions were able to exclude the presence of ECS (100% specificity) (Table 2).
Discussion
The presence of ECS in OSCC is of high prognostic value and yet existing techniques are inadequate for its detection prior to surgery. Identification of an appropriate biomarker would aid clinical decision-making regarding escalation of therapy as well as selection for clinical trials with novel therapies. The present findings suggest that a combination of SMA and SERPINE1 protein expression in the primary tumour offers promise in this and also shows a significant association with survival. They also confirm that preoperative assessment of patients with MRI has very poor sensitivity for the detection of ECS (Shaw et al, 2010; Liao et al, 2012; Lwin et al, 2012). The advantages of our study are that detailed clinical and outcome data were obtained, together with MRI data re-assessed by a single H&N radiologist with a special interest, so a direct comparison of prognostic variables with prediction of survival could be analysed.
Despite the relatively modest number of cases examined, this is the largest validation study using ECS as a clinical correlate. Tumour heterogeneity is an important feature of HNSCC and is reflected in both clinical and molecular characteristics of the disease (Severino et al, 2008). For example, OSCC has a much greater propensity for spread with poor outcomes compared with OPSCC (Franceschi et al, 1993; Dobrossy, 2005; Timar et al, 2005; Yu et al, 2008). This site specificity is also reflected by the presence of HPV, which is known to have a distinct gene expression profile (Ragin et al, 2006; Slebos et al, 2006; Schlecht et al, 2007; Shaw et al, 2010; van Hooff et al, 2012b). We established the HPV status of the cohort using a gold standard test and identified that the presence of HPV was unlikely to be a confounding variable in our analysis.
Gene-expression studies have previously shown the ability to predict disease progression and poor survival outcomes in OSCC and are often demonstrated to outperform existing clinical methods (Chung et al, 2004; Toruner et al, 2004; O'Donnell et al, 2005; Roepman et al, 2005; Chen et al, 2008; Kondoh et al, 2008; Rickman et al, 2008; Kang et al, 2009; Mendez et al, 2009). Some studies have also compared expression signatures from primary tissue paired with matched metastatic tissue and showed similarities in the patterns of expression observed (Roepman et al, 2006; Mendez et al, 2007; Colella et al, 2008; Liu et al, 2008). This supports our hypothesis that molecular determinants of ECS are present in the primary tumour. In a recent meta-analysis of over 60 studies of differential gene-expression profiling in HNSCC, one study of a limited number of cases predicted ECS by gene-expression analysis from primary site tissue (Zhou et al, 2006; Yu et al, 2008). It is also evident from many studies that there is a high risk for false discovery, and careful validation is mandated before establishing clinical biomarkers (Mroz and Rocco, 2012; van Hooff et al, 2012a).
Our sampling of tissue for gene-expression analysis was superficial, which may be criticised by excluding the more informative tumour-advancing front. The majority of the lesions examined were, however, large pT2 or pT4 tumour, and there was a low likelihood of the tumour-advancing front being included in the samples, even if a deep biopsy were possible. This is a common pathological experience and is reinforced from our previous studies indicating that the stromal component was limited in our research tissues, suggesting the absence of the advancing front (3). The results of the qRT–PCR analysis thereby would be expected to be predominantly of the epithelial compartment. Gene-expression analysis of stroma-rich samples may be more informative in the identification of biomarkers associated with late stages of disease such as metastasis, but methods of obtaining them need to be developed.
SERPINE1, the gene from our ECS signature, was validated with qRT–PCR and IHC on a TMA. In general, most microarray studies with single-gene validation demonstrate a correlation with microarray techniques (Rickman et al, 2008). Mendez et al (2009) showed good correlation between qRT–PCR data and microarray findings for SERPINE1, with a correlation coefficient of 0.74. Our finding of a moderately positive correlation is somewhat surprising, given the greater dynamic range of single-gene assays. This may be due to false discovery associated with microarray analysis or low fold-changes of the selected genes, as some microarray validation studies have shown that consistency was not achievable for genes showing a <four-fold difference in expression (Rajeevan et al, 2001; Chuaqui et al, 2002).
There are several gene-expression studies that have identified SERPINE1 as an important gene associated with metastasis and this has been supported by IHC analysis in OSCC, but those studies did not use a combined approach on the same tumours (Yasuda et al, 1997; Schmalbach et al, 2004; Roepman et al, 2005; Liu et al, 2008; Mendez et al, 2009). To corroborate and extend our RNA expression analyses, we used IHC to determine differences in expression at the tumour centre and the tumour-advancing front, and to determine which site has the greatest discrimination in relation to pathological variables (Chuaqui et al, 2002). We have shown that the intensity of SERPINE1 expression in tumour cells at the tumour-advancing front correlated with N-stage, the presence of nodal metastasis and ECS, while expression in tumour cells at the tumour centre and stromal cells at the tumour-advancing front was not significant. This is in keeping with the biological role of SERPINE1 in invasion and metastasis (Klein et al, 2012).
SERPINE1, also known as plasminogen activator inhibitor-1, is a regulator of the urokinase and tissue-type plasminogen activators. These serine proteases in turn activate the pro-enzyme plasminogen to plasmin that promotes invasion by degradation of the extracellular matrix, as well as activation of matrix metalloproteinases (Baker et al, 2007). The plasminogen activator/plasmin system can also promote invasion by downregulation of cell–cell adhesion molecules such as E-cadherin (Hayashido et al, 2005). The plasminogen activation system in the metastatic cascade also influences proliferation, migration, angiogenesis and extravasation, and it has been implicated in several tumour types including colorectal and breast cancer (Gandolfo et al, 1996; Duffy, 2004; Dano et al, 2005).
We compared these findings with a documented feature associated with poor outcomes in OSCC, SMA expression in stromal cells. The intense SMA expression at the tumour-advancing front, was significantly associated with the N-stage and presence of nodal metastasis and its patterns are in keeping with previous studies (Kellermann et al, 2007; Surowiak et al, 2007; Marsh et al, 2011; Thode et al, 2011). The distribution of SMA immunoreactivity in the tumour stroma varies widely between different tumours and between different areas in the same tumour, and reflects heterogeneity (Vered et al, 2010). However, there is a high level of concordance (up to 98%) reported for triplicate TMA cores (as used in the present study) being representative of the full sections (Hoos and Cordon-Cardo, 2001; Gulmann et al, 2003; Parsons and Grabsch, 2009). Relatively large cores were selected at the tumour-advancing front (1 mm) to ensure that adequate amounts of both tumour and adjacent stroma were present for scoring. Interestingly, high and intermediate levels of SMA expression were also seen at the tumour centre, although SMA expression would be expected to be the greatest at the advancing front where the tumour lies in proximity to stromal cells. This may reflect areas of myofibroblastic reaction/tumour-associated stroma at chronologically earlier sites, which have subsequently become incorporated within the growing tumour.
Both separately and in combination, SMA and SERPINE1 protein expression were superior to MRI for the detection of ECS, which was associated with expression of either, or both, proteins in all cases. Similarly, a combined approach using both SMA and SERPINE1 expression at the advancing front showed greater ability to predict survival than expression of the individual proteins. The most recent and largest study of prognostic stromal features in OSCC reported a greater prognostic significance for SMA expression in the stroma of OSCC than the presence of ECS (Marsh et al, 2011). Our study, however, suggests that ECS is of greater prognostic value, but that SMA plus SERPINE1 expression in combination was highly significantly associated with adverse outcomes.
The prognostic and diagnostic accuracy of SMA and SERPINE1 expression at the invasive front corresponds with observations that histological and molecular determinants at the invasive front are of greater prognostic value than the tumour centre (Bryne et al, 1992; Piffko et al, 1998; Kellermann et al, 2007; Vered et al, 2010; Kato et al, 2011). Our findings indicate that there are differences in the expression of both proteins at the tumour centre and the advancing front. Accordingly, realisation of their prognostic value would only be feasible if the advancing front is included in the preoperative diagnostic incisional biopsy. This, as discussed above, may prove difficult or impossible to achieve, particularly in large established tumours. Nevertheless, as SERPINE1 and SMA expression were more significantly associated with ECS and nodal status at the tumour-advancing front than the tumour centre, insights into the biological perspectives of ECS would be better explored with approaches focusing on the tumour–stroma interface. Future work validating these data in a larger series and in a prospective cohort with deep biopsy may determine the clinical applicability of this approach in future biomarker discovery and disease stratification in OSCC. The biological importance of SERPINE1 in cellular invasion together with the presence of myofibroblasts can also be further studied in three-dimensional co-culture organotypic models that utilise both epithelial and mesenchymal cells in which these genes can be upregulated or downregulated.
The growing interest in the tumour microenvironment and the effects of myofibroblasts in invasion and metastasis open up new avenues for investigation in both biological understanding and potential therapy in the metastatic cascade. Relationships between the advancing front and nodal metastasis/ECS reiterate the importance of this frontier for detection of appropriate biomarkers. We have demonstrated an informative approach by combining microarray findings that identified a biological determinant for ECS (SERPINE1) with a promising immunohistochemical marker of adverse outcome (SMA) to identify patients with poor survival in OSCC.
Change history
25 November 2014
This paper was modified 12 months after initial publication to switch to Creative Commons licence terms, as noted at publication
References
Alvi A, Johnson JT (1996) Extracapsular spread in the clinically negative neck (N0): implications and outcome. Otolaryngol Head Neck Surg 114 (1): 65–70.
Argiris A, Karamouzis MV, Raben D, Ferris RL (2008) Head and neck cancer. Lancet 371 (9625): 1695–1709.
Baker EA, Leaper DJ, Hayter JP, Dickenson AJ (2007) Plasminogen activator system in oral squamous cell carcinoma. Br J Oral Maxillofac Surg 45 (8): 623–627.
Barry CP, Katre C, Papa E, Brown JS, Shaw RJ, Bekiroglu F, Lowe D, Rogers SN (2013) De-escalation of surgery for early oral cancer—is it oncologically safe? Br J Oral Maxillofac Surg 51 (1): 30–36.
Bryne M, Koppang HS, Lilleng R, Kjaerheim A (1992) Malignancy grading of the deep invasive margins of oral squamous cell carcinomas has high prognostic value. J Pathol 166 (4): 375–381.
Califano J, van der Riet P, Westra W, Nawroz H, Clayman G, Piantadosi S, Corio R, Lee D, Greenberg B, Koch W, Sidransky D (1996) Genetic progression model for head and neck cancer: implications for field cancerization. Cancer Res 56 (11): 2488–2492.
Chen C, Mendez E, Houck J, Fan W, Lohavanichbutr P, Doody D, Yueh B, Futran ND, Upton M, Farwell DG, Schwartz SM, Zhao LP (2008) Gene expression profiling identifies genes predictive of oral squamous cell carcinoma. Cancer Epidemiol Biomarkers Prev 17 (8): 2152–2162.
Chuaqui RF, Bonner RF, Best CJ, Gillespie JW, Flaig MJ, Hewitt SM, Phillips JL, Krizman DB, Tangrea MA, Ahram M, Linehan WM, Knezevic V, Emmert-Buck MR (2002) Post-analysis follow-up and validation of microarray experiments. Nat Genet 32 (Suppl): 509–514.
Chung CH, Parker JS, Karaca G, Wu J, Funkhouser WK, Moore D, Butterfoss D, Xiang D, Zanation A, Yin X, Shockley WW, Weissler MC, Dressler LG, Shores CG, Yarbrough WG, Perou CM (2004) Molecular classification of head and neck squamous cell carcinomas using patterns of gene expression. Cancer Cell 5 (5): 489–500.
Colella S, Richards KL, Bachinski LL, Baggerly KA, Tsavachidis S, Lang JC, Schuller DE, Krahe R (2008) Molecular signatures of metastasis in head and neck cancer. Head Neck 30 (10): 1273–1283.
Dano K, Behrendt N, Hoyer-Hansen G, Johnsen M, Lund LR, Ploug M, Romer J (2005) Plasminogen activation and cancer. Thromb Haemost 93 (4): 676–681.
Dobrossy L (2005) Epidemiology of head and neck cancer: magnitude of the problem. Cancer Metastasis Rev 24 (1): 9–17.
Duffy MJ (2004) The urokinase plasminogen activator system: role in malignancy. Curr Pharm Des 10 (1): 39–49.
Field JK, Kiaris H, Risk JM, Tsiriyotis C, Adamson R, Zoumpourlis V, Rowley H, Taylor K, Whittaker J, Howard P (1995) Allelotype of squamous cell carcinoma of the head and neck: fractional allele loss correlates with survival. Br J Cancer 72 (5): 1180–1188.
Franceschi D, Gupta R, Spiro RH, Shah JP (1993) Improved survival in the treatment of squamous carcinoma of the oral tongue. Am J Surg 166 (4): 360–365.
Gandolfo GM, Conti L, Vercillo M (1996) Fibrinolysis components as prognostic markers in breast cancer and colorectal carcinoma. Anticancer Res 16 (4B): 2155–2159.
Greenberg JS, Fowler R, Gomez J, Mo V, Roberts D, El Naggar AK, Myers JN (2003) Extent of extracapsular spread: a critical prognosticator in oral tongue cancer. Cancer 97 (6): 1464–1470.
Gulmann C, Butler D, Kay E, Grace A, Leader M (2003) Biopsy of a biopsy: validation of immunoprofiling in gastric cancer biopsy tissue microarrays. Histopathology 42 (1): 70–76.
Hayashido Y, Hamana T, Yoshioka Y, Kitano H, Koizumi K, Okamoto T (2005) Plasminogen activator/plasmin system suppresses cell-cell adhesion of oral squamous cell carcinoma cells via proteolysis of E-cadherin. Int J Oncol 27 (3): 693–698.
Hoffman EP, Awad T, Palma J, Webster T, Hubbell E, Warrington JA, Spira A, Wright G, Buckley J, Triche T, Davis R, Tibshirani R (2004) Expression profiling—best practices for data generation and interpretation in clinical trials. Nat Rev Genet 5 (3): 229–237.
Hoos A, Cordon-Cardo C (2001) Tissue microarray profiling of cancer specimens and cell lines: opportunities and limitations. Lab Invest 81 (10): 1331–1338.
Kang CJ, Chen YJ, Liao CT, Wang HM, Chang JT, Lin CY, Lee LY, Wang TH, Yen TC, Shen CR, Chen IH, Chiu CC, Cheng AJ (2009) Transcriptome profiling and network pathway analysis of genes associated with invasive phenotype in oral cancer. Cancer Lett 284 (2): 131–140.
Kato K, Kawashiri S, Yoshizawa K, Kitahara H, Okamune A, Sugiura S, Noguchi N, Yamamoto E (2011) Expression form of p53 and PCNA at the invasive front in oral squamous cell carcinoma: correlation with clinicopathological features and prognosis. J Oral Pathol Med 40 (9): 693–698.
Kellermann MG, Sobral LM, da Silva SD, Zecchin KG, Graner E, Lopes MA, Nishimoto I, Kowalski LP, Coletta RD (2007) Myofibroblasts in the stroma of oral squamous cell carcinoma are associated with poor prognosis. Histopathology 51 (6): 849–853.
Klein RM, Bernstein D, Higgins SP, Higgins CE, Higgins PJ (2012) SERPINE1 expression discriminates site-specific metastasis in human melanoma. Exp Dermatol 21 (7): 551–554.
Kondoh N, Ishikawa T, Ohkura S, Arai M, Hada A, Yamazaki Y, Kitagawa Y, Shindoh M, Takahashi M, Ando T, Sato Y, Izumo T, Hitomi K, Yamamoto M (2008) Gene expression signatures that classify the mode of invasion of primary oral squamous cell carcinomas. Mol Carcinog 47 (10): 744–756.
Leemans CR, Braakhuis BJ, Brakenhoff RH (2011) The molecular biology of head and neck cancer. Nat Rev Cancer 11 (1): 9–22.
Liao LJ, Lo WC, Hsu WL, Wang CT, Lai MS (2012) Detection of cervical lymph node metastasis in head and neck cancer patients with clinically N0 neck-a meta-analysis comparing different imaging modalities. BMC Cancer 12: 236.
Liu CJ, Liu TY, Kuo LT, Cheng HW, Chu TH, Chang KW, Lin SC (2008) Differential gene expression signature between primary and metastatic head and neck squamous cell carcinoma. J Pathol 214 (4): 489–497.
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25 (4): 402–408.
Lopes V, Murray P, Williams H, Woodman C, Watkinson J, Robinson M (2011) Squamous cell carcinoma of the oral cavity rarely harbours oncogenic human papillomavirus. Oral Oncol 47 (8): 698–701.
Lwin CT, Hanlon R, Lowe D, Brown JS, Woolgar JA, Triantafyllou A, Rogers SN, Bekiroglu F, Lewis-Jones H, Wieshmann H, Shaw RJ (2012) Accuracy of MRI in prediction of tumour thickness and nodal stage in oral squamous cell carcinoma. Oral Oncology 48 (2): 149–154.
Marsh D, Suchak K, Moutasim KA, Vallath S, Hopper C, Jerjes W, Upile T, Kalavrezos N, Violette SM, Weinreb PH, Chester KA, Chana JS, Marshall JF, Hart IR, Hackshaw AK, Piper K, Thomas GJ (2011) Stromal features are predictive of disease mortality in oral cancer patients. J Pathol 223 (4): 470–481.
Mendez E, Fan W, Choi P, Agoff SN, Whipple M, Farwell DG, Futran ND, Weymuller EA Jr., Zhao LP, Chen C (2007) Tumor-specific genetic expression profile of metastatic oral squamous cell carcinoma. Head Neck 29 (9): 803–814.
Mendez E, Houck JR, Doody DR, Fan W, Lohavanichbutr P, Rue TC, Yueh B, Futran ND, Upton MP, Farwell DG, Heagerty PJ, Zhao LP, Schwartz SM, Chen C (2009) A genetic expression profile associated with oral cancer identifies a group of patients at high risk of poor survival. Clin Cancer Res 15 (4): 1353–1361.
Mroz EA, Rocco JW (2012) Gene expression analysis as a tool in early-stage oral cancer management. J Clin Oncol 30 (33): 4053–4055.
Myers JN, Greenberg JS, Mo V, Roberts D (2001) Extracapsular spread. A significant predictor of treatment failure in patients with squamous cell carcinoma of the tongue. Cancer 92 (12): 3030–3036.
O'Donnell RK, Kupferman M, Wei SJ, Singhal S, Weber R, O'Malley B, Cheng Y, Putt M, Feldman M, Ziober B, Muschel RJ (2005) Gene expression signature predicts lymphatic metastasis in squamous cell carcinoma of the oral cavity. Oncogene 24 (7): 1244–1251.
Parsons M, Grabsch H (2009) How to make tissue microarrays. Diagn Histopathol 15 (3): 142–150.
Piffko J, Bankfalvi A, Tory K, Fuzesi L, Bryne M, Ofner D, Kusch F, Joos U, Schmid KW (1998) Molecular assessment of p53 abnormalities at the invasive front of oral squamous cell carcinomas. Head Neck 20 (1): 8–15.
Ragin CC, Taioli E, Weissfeld JL, White JS, Rossie KM, Modugno F, Gollin SM (2006) 11q13 amplification status and human papillomavirus in relation to p16 expression defines two distinct etiologies of head and neck tumours. Br J Cancer 95 (10): 1432–1438.
Rajeevan MS, Vernon SD, Taysavang N, Unger ER (2001) Validation of array-based gene expression profiles by real-time (kinetic) RT-PCR. J Mol Diagn 3 (1): 26–31.
Ramaswamy S, Ross KN, Lander ES, Golub TR (2003) A molecular signature of metastasis in primary solid tumors. Nat Genet 33 (1): 49–54.
Rickman DS, Millon R, De Reynies A, Thomas E, Wasylyk C, Muller D, Abecassis J, Wasylyk B (2008) Prediction of future metastasis and molecular characterization of head and neck squamous-cell carcinoma based on transcriptome and genome analysis by microarrays. Oncogene 27 (51): 6607–6622.
Roepman P, de Jager A, Groot Koerkamp MJ, Kummer JA, Slootweg PJ, Holstege FC (2006) Maintenance of head and neck tumor gene expression profiles upon lymph node metastasis. Cancer Res 66 (23): 11110–11114.
Roepman P, Wessels LF, Kettelarij N, Kemmeren P, Miles AJ, Lijnzaad P, Tilanus MG, Koole R, Hordijk GJ, van der Vliet PC, Reinders MJ, Slootweg PJ, Holstege FC (2005) An expression profile for diagnosis of lymph node metastases from primary head and neck squamous cell carcinomas. Nat Genet 37 (2): 182–186.
Rogers SN, Brown JS, Woolgar JA, Lowe D, Magennis P, Shaw RJ, Sutton D, Errington D, Vaughan D (2009) Survival following primary surgery for oral cancer. Oral Oncol 45 (3): 201–211.
Schache AG, Liloglou T, Risk JM, Filia A, Jones TM, Sheard J, Woolgar JA, Helliwell TR, Triantafyllou A, Robinson M, Sloan P, Harvey-Woodworth C, Sisson D, Shaw RJ (2011) Evaluation of human papilloma virus diagnostic testing in oropharyngeal squamous cell carcinoma: sensitivity, specificity, and prognostic discrimination. Clin Cancer Res 17 (19): 6262–6271.
Schlecht NF, Burk RD, Adrien L, Dunne A, Kawachi N, Sarta C, Chen Q, Brandwein-Gensler M, Prystowsky MB, Childs G, Smith RV, Belbin TJ (2007) Gene expression profiles in HPV-infected head and neck cancer. J Pathol 213 (3): 283–293.
Schmalbach CE, Chepeha DB, Giordano TJ, Rubin MA, Teknos TN, Bradford CR, Wolf GT, Kuick R, Misek DE, Trask DK, Hanash S (2004) Molecular profiling and the identification of genes associated with metastatic oral cavity/pharynx squamous cell carcinoma. Arch Otolaryngol Head Neck Surg 130 (3): 295–302.
Severino P, Alvares AM, Michaluart P Jr., Okamoto OK, Nunes FD, Moreira-Filho CA, Tajara EH (2008) Global gene expression profiling of oral cavity cancers suggests molecular heterogeneity within anatomic subsites. BMC Res Notes 1: 113.
Shaw RJ, Hobkirk AJ, Nikolaidis G, Woolgar JA, Triantafyllou A, Brown JS, Liloglou T, Risk JM (2013) Molecular staging of surgical margins in oral squamous cell carcinoma using promoter methylation of p16(INK4A), cytoglobin, E-cadherin, and TMEFF2. Ann Surg Oncol 20 (8): 2796–2802.
Shaw RJ, Liloglou T, Rogers SN, Brown JS, Vaughan ED, Lowe D, Field JK, Risk JM (2006) Promoter methylation of P16, RARbeta, E-cadherin, cyclin A1 and cytoglobin in oral cancer: quantitative evaluation using pyrosequencing. Br J Cancer 94 (4): 561–568.
Shaw RJ, Lowe D, Woolgar JA, Brown JS, Vaughan ED, Evans C, Lewis-Jones H, Hanlon R, Hall GL, Rogers SN (2010) Extracapsular spread in oral squamous cell carcinoma. Head Neck 32 (6): 714–722.
Shaw RJ, Pace-Balzan A, Butterworth C (2011) Contemporary clinical management of oral squamous cell carcinoma. Periodontol 2000 57 (1): 89–101.
Slebos RJ, Yi Y, Ely K, Carter J, Evjen A, Zhang X, Shyr Y, Murphy BM, Cmelak AJ, Burkey BB, Netterville JL, Levy S, Yarbrough WG, Chung CH (2006) Gene expression differences associated with human papillomavirus status in head and neck squamous cell carcinoma. Clin Cancer Res 12 (3 Pt 1): 701–709.
Surowiak P, Murawa D, Materna V, Maciejczyk A, Pudelko M, Ciesla S, Breborowicz J, Murawa P, Zabel M, Dietel M, Lage H (2007) Occurence of stromal myofibroblasts in the invasive ductal breast cancer tissue is an unfavourable prognostic factor. Anticancer Res 27 (4C): 2917–2924.
Thode C, Jorgensen TG, Dabelsteen E, Mackenzie I, Dabelsteen S (2011) Significance of myofibroblasts in oral squamous cell carcinoma. J Oral Pathol Med 40 (3): 201–207.
Timar J, Csuka O, Remenar E, Repassy G, Kasler M (2005) Progression of head and neck squamous cell cancer. Cancer Metastasis Rev 24 (1): 107–127.
Toruner GA, Ulger C, Alkan M, Galante AT, Rinaggio J, Wilk R, Tian B, Soteropoulos P, Hameed MR, Schwalb MN, Dermody JJ (2004) Association between gene expression profile and tumor invasion in oral squamous cell carcinoma. Cancer Genet Cytogenet 154 (1): 27–35.
Tsujino T, Seshimo I, Yamamoto H, Ngan CY, Ezumi K, Takemasa I, Ikeda M, Sekimoto M, Matsuura N, Monden M (2007) Stromal myofibroblasts predict disease recurrence for colorectal cancer. Clin Cancer Res 13 (7): 2082–2090.
van den Brekel MW, Castelijns JA, Stel HV, Valk J, Croll GA, Golding RP, Luth WJ, Meyer CJ, Snow GB (1990) Detection and characterization of metastatic cervical adenopathy by MR imaging: comparison of different MR techniques. J Comput Assist Tomogr 14 (4): 581–589.
van Hooff SR, Leusink FK, Roepman P, Baatenburg de Jong RJ, Speel EJ, van den Brekel MW, van Velthuysen ML, van Diest PJ, van Es RJ, Merkx MA, Kummer JA, Leemans CR, Schuuring E, Langendijk JA, Lacko M, De Herdt MJ, Jansen JC, Brakenhoff RH, Slootweg PJ, Takes RP, Holstege FC (2012a) Validation of a gene expression signature for assessment of lymph node metastasis in oral squamous cell carcinoma. J Clin Oncol 30 (33): 4104–4110.
van Hooff SR, Leusink FK, Roepman P, Baatenburg de Jong RJ, Speel EJ, van den Brekel MW, van Velthuysen ML, van Diest PJ, van Es RJ, Merkx MA, Kummer JA, Leemans CR, Schuuring E, Langendijk JA, Lacko M, De Herdt MJ, Jansen JC, Brakenhoff RH, Slootweg PJ, Takes RP, Holstege FC (2012b) Validation of a Gene Expression Signature for Assessment of Lymph Node Metastasis in Oral Squamous Cell Carcinoma. J Clin Oncol 30 (33): 4104–4110.
Vered M, Dobriyan A, Dayan D, Yahalom R, Talmi YP, Bedrin L, Barshack I, Taicher S (2010) Tumor-host histopathologic variables, stromal myofibroblasts and risk score, are significantly associated with recurrent disease in tongue cancer. Cancer Sci 101 (1): 274–280.
Woolgar JW, Triantafyllou A (2009) Pitfalls and procedures in the histopathological diagnosis of oral and oropharyngeal squamous cell carcinoma and a review of the role of pathology in prognosis: University of Liverpool. Oral Oncol 45 (4-5): 361–385.
Yasuda T, Sakata Y, Kitamura K, Morita M, Ishida T (1997) Localization of plasminogen activators and their inhibitor in squamous cell carcinomas of the head and neck. Head Neck 19 (7): 611–616.
Yu YH, Kuo HK, Chang KW (2008) The evolving transcriptome of head and neck squamous cell carcinoma: a systematic review. PLoS One 3 (9): e3215.
Zhou X, Temam S, Oh M, Pungpravat N, Huang BL, Mao L, Wong DT (2006) Global expression-based classification of lymph node metastasis and extracapsular spread of oral tongue squamous cell carcinoma. Neoplasia 8 (11): 925–932.
Acknowledgements
J Dhanda received funding to conduct this research from the Royal College of Surgeons of England Research Department, Saving Faces—The Facial Surgery Research Foundation, The Faculty of Dental Surgery—The Royal College of Surgeons of England, The British Association of Oral & Maxillofacial Surgeons. DR Sibson and B Lloyd received funding to conduct this research from the Clatterbridge Cancer Research Trust (now North West Cancer Research).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
Supplementary Information accompanies this paper on British Journal of Cancer website
Supplementary information
Rights and permissions
From twelve months after its original publication, this work is licensed under the Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Dhanda, J., Triantafyllou, A., Liloglou, T. et al. SERPINE1 and SMA expression at the invasive front predict extracapsular spread and survival in oral squamous cell carcinoma. Br J Cancer 111, 2114–2121 (2014). https://doi.org/10.1038/bjc.2014.500
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/bjc.2014.500
Keywords
This article is cited by
-
MMP14 expression levels accurately predict the presence of extranodal extensions in oral squamous cell carcinoma: a retrospective cohort study
BMC Cancer (2023)
-
Vessel size as a marker of survival in estrogen receptor positive breast cancer
Breast Cancer Research and Treatment (2023)
-
Novel pathological predictive factors for extranodal extension in oral squamous cell carcinoma: a retrospective cohort study based on tumor budding, desmoplastic reaction, tumor-infiltrating lymphocytes, and depth of invasion
BMC Cancer (2022)
-
Prioritization of genes involved in endothelial cell apoptosis by their implication in lymphedema using an analysis of associative gene networks with ANDSystem
BMC Medical Genomics (2019)
-
Keratin 14-high subpopulation mediates lung cancer metastasis potentially through Gkn1 upregulation
Oncogene (2019)