Abstract
Background:
Wider clinical applications of 9p status in clear cell renal cell carcinoma (ccRCC) are limited owing to the lack of validation and consensus for interphase fluorescent in situ hybridisation (I-FISH) scoring technique. The aim of this study was to analytically validate the applicability of I-FISH in assessing 9p deletion in ccRCC and to clinically assess its long-term prognostic impact following surgical excision of ccRCC.
Methods:
Tissue microarrays were constructed from 108 renal cell carcinoma (RCC) tumour paraffin blocks. Interphase fluorescent in situ hybridisation analysis was undertaken based on preset criteria by two independent observers to assess interobserver variability. 9p status in ccRCC tumours was determined and correlated to clinicopathological variables, recurrence-free survival and disease-specific survival.
Results:
There were 80 ccRCCs with valid 9p scoring and a median follow-up of 95 months. Kappa statistic for interobserver variability was 0.71 (good agreement). 9p deletion was detected in 44% of ccRCCs. 9p loss was associated with higher stage, larger tumours, necrosis, microvascular and renal vein invasion, and higher SSIGN (stage, size, grade and necrosis) score. Patients with 9p-deleted ccRCC were at a higher risk of recurrence (P=0.008) and RCC-specific mortality (P=0.001). On multivariate analysis, 9p deletion was an independent predictor of recurrence (hazard ratio 4.323; P=0.021) and RCC-specific mortality (hazard ratio 4.603; P=0.007). The predictive accuracy of SSIGN score improved from 87.7% to 93.1% by integrating 9p status to the model (P=0.001).
Conclusions:
Loss of 9p is associated with aggressive ccRCC and worse prognosis in patients following surgery. Our findings independently confirm the findings of previous reports relying on I-FISH to detect 9p (CDKN2A) deletion.
Similar content being viewed by others
Main
The predictive role of the detection of chromosomal and gene copy number variation (CNV) in renal cell carcinoma (RCC) has been explored in several studies (Gunawan et al, 2001; Klatte et al, 2009). Deletion of the short arm of chromosome 9 (9p) has been confirmed, in many studies, to be one of the most frequent non-random chromosomal CNVs in clear cell RCC (ccRCC) (Strefford et al, 2005; Matsuda et al, 2008; Beroukhim et al, 2009; Dalgliesh et al, 2010). Moreover, accumulating evidence from other cancers indicates frequent 9p deletions to be seen in high-grade invasive and aggressive cancers (Sidransky, 1995; Okami et al, 1997; Ellsworth et al, 2008). Potential genes on chromosome 9p have been implicated in ccRCC progression. The cyclin-dependent kinase inhibitor (CDKN2A) gene is a tumour suppressor gene, located on 9p21, encodes two proteins (p16INK4 and p14ARF) that regulate two critical cell cycle regulatory pathways: the p53 pathway and the Rb pathway (Harris and Levine, 2005). Thus, the inactivation of CDKN2A gene via 9p21 deletion, leading to low expression of these regulatory proteins, may be an important biomarker of cancer aggressiveness (Kawada et al, 2001a; Ikuerowo et al, 2007). Also, the low level of expression of carbonic anhydrase IX (CAIX) gene located on locus 9p13 has been associated with shorter survival and poor response in patients on immunotherapy for metastatic lesions (Bui et al, 2003; Atkins et al, 2005; Sandlund et al, 2007; Patard et al, 2008). Thus, established loss of 9p in RCC could act as a marker of tumour aggressiveness and could be responsible for progression (Cairns et al, 1995; Kinoshita et al, 1995; Schullerus et al, 1997; Junker et al, 2003; Klatte et al, 2009; Dagher et al, 2013).
Recently, two studies, using interphase fluorescence in situ hybridisation (I-FISH) as the main detection technique, have concluded that loss of 9p in ccRCC was an independent prognostic factor besides other known pathological parameters (Brunelli et al, 2008; La Rochelle et al, 2010). Although the findings from these studies were promising, they were both of limited clinical applicability owing to lack of standardisation of I-FISH scoring, lack of reporting on degree of interobserver variability and a modest follow-up period. La Rochelle et al (2010) performed I-FISH analysis on a cohort of 316 patients but the cut-off level for 9p deletion was arbitrary, which could have possibly resulted in underestimation of CNVs.
As I-FISH can be performed on both fresh and archived formalin-fixed paraffin-embedded (FFPE) tumour tissue, it has been used widely on tissue microarrays (TMAs)that allow the assessment of many tumours in one experiment, in comparison to conventional cytogenetics that can only be performed on individual cases from fresh tissue after culture. Also, I-FISH assesses CNVs at a cellular level, in contrast to other techniques, such as array comparative genomic hybridisation and microsatellite analysis based on mass DNA extraction, which can conceal CNVs and intratumour cellular genetic complexity and heterogeneity (Bayani and Squire, 2007).
On the other hand, standardisation of I-FISH scoring techniques and reaching a consensus on how to determine the threshold level are required before the technique can be applied in translational renal oncology (Junker et al, 2013). This is due to the fact that the scoring process can be influenced by fading signals, sectioning of paraffin blocks and quality of tissue preserved, which can result in interobserver disagreement.
Our study aims to assess the prognostic effect of 9p deletion in ccRCC subtype. We hypothesised that patients with 9p-deleted ccRCC tumours detected by I-FISH do not carry a worse prognosis (H0). A set of objectives was identified to test our hypothesis: (a) to analytically validate I-FISH scoring technique for assessing 9p status in renal cancer, (b) to assess the degree of interobserver variability in scoring 9p status, (c) to assess the correlation between 9p status and other established clinicopathological prognostic variables and finally, (d) to assess the impact of 9p loss in ccRCC tumours on recurrence-free survival (RFS) and disease-specific survival (DSS) in a cohort of patients with at least 8 years of follow-up.
Materials and Methods
Identification of cohort
The urology department database was searched for consecutive patients who underwent radical nephrectomy for RCC between January 2001 and December 2005 without any exclusion criteria. The period was chosen to allow a sufficient follow-up period of at least 8 years for each patient. Ethical approval (Ref. 12/ES/0083) was obtained from the Tayside Research and Ethics committee. A single pathologist (SF) with special interest in renal cancer, blinded to clinical outcomes, reviewed all the samples using pathology number identifier for each patient.
Tissue microarrays
Haematoxylin and eosin (H&E) slides from all the archived FFPE blocks were reviewed for representative areas in RCC tumours. Tissue microarrays were constructed as described in previous publication from our institution (Hadad et al, 2009). Hundred and twenty six FFPE blocks from 108 tumours, identified from the pathology department archives, were used to construct four tumour TMAs. Sixteen tumours were represented by two blocks and one tumour was represented by three blocks. These extra blocks served as internal control to assess the effect of tumour heterogeneity without the knowledge of the scoring observer. One TMA of normal renal tissue from same cases was prepared to serve as a negative control.
Briefly the technique is described here. Tissue microarrays were constructed using Beecher arraying instrument (Beecher Instruments Inc., Sun Prairie, WI, USA) with the help of TMA Designer 2 software (ALPHELYS, Plaisir, France). The pathologist identified and marked, with a circle on the H&E slide, areas containing only tumour for coring, and avoiding areas of necrosis or normal tissue, before marking the same tumour region on the corresponding formalin-fixed paraffin block. Tissue cores with a diameter of 0.6 mm were punched from the marked tumour regions on paraffin blocks. Cores were then deposited into a master paraffin block and placed 1.2 mm apart from the neighbouring core on the x and y axes. Sections from the resulting master paraffin block measuring 4 μm in thickness were then transferred onto glass slides to form a TMA. Each TMA contained a maximum of 240 tumour cores plus two spleen control cores for orientation. Each case was represented at least with six tumour cores.
Interphase fluorescent in situ hybridisation
Interphase fluorescent in situ hybridisation analysis was performed using the Vysis locus-specific identifier (LSI) CDKN2A spectrum red (R)/(CEP 9) spectrum green (G) probes (Abbott Molecular, Des Plaines, IL, USA). The CDKN2A probe spans ∼222 kb of the 9p21 region across several genes, including p16 (INK4A), p14 (ARF) and p15 (INK4B). CEP 9 is a centromeric probe that hybridises to α-satellite sequences specific to chromosome 9 (9p11-q11).
Tissue microarray slides were baked for 60 min at 60 °C in a dry incubator and then processed in automated Vysis VP-2000 processor (Abbott Molecular) to deparaffinise the slides. The slides were submersed in Xylene basin for 10 min twice, then alcohol 100% basin for 5 min twice before they were left to dry for 2 min. The slides were left in 0.2 N HCL basin for 20 min before they were rinsed in a water chamber for 3 min. The slides were then washed twice in 2 × SSC basin each for 3 min before they were removed from VP-2000. Next, slides were put through sodium thiocyanate (NaSCN) pretreatment reagent (Abbott Molecular) at 80 °C for 30 min followed by protease solution (0.5 mg ml-1 0.01 N HCl) (Abbott Molecular) at 37 °C for 50 min. Slides were then put through three sequential 2-min washes with 2 × SSC and then dehydrated for 2 min each in graded ethanol solutions (70%, 85% and 100%). Once dried, slides were placed in 70% formamide/2 × SSC at 70 °C in a water bath for 5 min, removed, put through a further graded ethanol series (70%, 85% and 100%) for 1 min each and then allowed to dry.
For each slide, a probe mix was made up by combining 4 μl CDKN2A/CEP 9 probe, 21 μl LSI/WCP Vysis hybridisation buffer (Abbott Molecular) and 6 μl dH2O in a 200 μl Eppendorf tube. The probe was mixed and denatured by placing the mix in a 70 °C water bath for 5 min. The mix (31 μl) was then drawn up and pipetted onto a 22 × 50 mm coverslip. The dried slide was then lowered onto the coverslip, ensuring that the probe mix was evenly distributed and that no air bubbles were present. Slides were then sealed around the edges with rubber cement Fixogum (Kreatech Diagnostics, Leica Microsystems, Milton Keynes, UK) and placed overnight (∼12 h) in a humidified hybridisation chamber in a 37 °C incubator.
The following day, cover slips were peeled off. Slides were placed in a hot wash (0.4 × SSC/0.3% NP-40 (Vysis, Abbott Molecular)) for 2 min at 70 °C in a water bath and then in a room temperature wash (2 × SSC/0.1% NP-40) for a further 2 min. Once dried, slides were counterstained with 25 μl DAPI Antifade (Abbott Molecular), mounted onto a fresh coverslip and edges sealed with nail varnish. Slides were stored in covered slide trays at 4 °C before and after scoring.
Interphase fluorescent in situ hybridisation interpretation
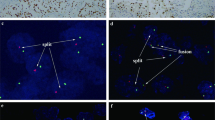
Slides were viewed under an Olympus B × 60 fluorescent microscope (Olympus Corporation, Tokyo, Japan) with filters for red spectrum, green spectrum and DAPI nuclear counterstain. Scoring was undertaken by two independent observers (IEM and JF) blinded to clinical or pathological outcomes. Each observer counted the number of red and green signals in each nucleus using preset criteria. A minimum of 30 intact non-overlapping cells per core, a minimum of three cores per block and a minimum of 100–200 nuclei with good signals per case were required for valid scoring. If these criteria were not met, the scoring was considered invalid. The interpretation of nuclei probe patterns was as follows: 2G2R was interpreted as normal, abnormal patterns included: 2G1R as loss of heterozygosity (LOH), 2G0R as homozygous deletion and 1G1R as monosomy of chromosome 9. Other nuclei patterns were labelled as ‘unclassified’.
The threshold for 9p deletion was determined by calculating the mean percentage (%) of abnormal nuclei from cores on the TMA of normal renal tissue plus four times the standard deviation as previously described in the literature (Cossu-Rocca et al, 2006; Brunelli et al, 2008). This translated to the following cut-off values for 9p deletion: 18% for LOH and homozygous loss of 9p (2G1R/2G0R), 37% for monosomy (1G1R), and 42% for all abnormal patterns. A tumour was scored 9p deleted if the number of cores with 9p deletion was higher than those with normal 9p status. When the number of cores with and without 9p deletion was equal, we relied on the mean percentage of 9p-deleted nuclei in all cores to decide on deletion. Each observer independently determined the deletion status on the basis of the above criteria (Figure 1). In tumours represented with cores from more than one block, they were considered deleted if the cores from one of the blocks were scored deleted. The level of interobserver agreement was assessed and any scoring disagreement was settled by consensus.
Clinicopathological data
The clinicopathological and follow-up data up to December 2013 for each patient were retrieved retrospectively using a unique 10-digit Community Health Index number and recorded on a predesigned electronic database sheet by an independent researcher (JB), blinded to chromosome 9p status. The following Tayside Health Board electronic resources were searched to retrieve and were cross-validated for clinical and follow-up information:
Hospital records: namely, clinical portal, ICE and In-House Surgical Information System Web and Technology (in-site). These databases represent single, secure, electronic, patient-record sources.
Multidisciplinary board meeting records: all renal cancer cases were discussed in weekly uro-oncology meeting during the study period. The records for each patient were available as an MS Word file under the heading ‘kidney’.
Data including patients’ gender, age at diagnosis and pre-operative staging were retrieved. Pathology slides review and reports were used to identify tumour stage based on 2009 TNM staging (Sobin et al, 2009), tumour size, Fuhrman grade, tumour necrosis, pelvicalyceal invasion, renal sinus invasion, microvascular invasion and sarcomatoid features. Each case was assigned a Mayo Clinic SSIGN(stage, size, grade and necrosis) score and stratified into five subcategories (SSIGN score subcategories 0–2, 3–4, 5–6, 7–9 and ⩾10, respectively) (Frank et al, 2002). Stage, size, grade and necrosis score has been externally validated in previous studies and established as a reliable prognostic model (Ficarra et al, 2006; Fujii et al, 2008; Ficarra et al, 2009).
Follow-up was calculated from the time of surgery to the last date of assessment or the date of death. Cause of death was determined on the basis of death certificates and correspondence between clinicians and patients’ general practitioners. All patients were followed up on the basis of a standardised departmental protocol based on tumour stage and grade. Death from renal cancer was defined as disease-related mortality. Recurrence was diagnosed if RCC metastasis or renal bed recurrence was detected on cross-sectional imaging and confirmed in a multidisciplinary team meeting record.
Statistical analysis
The number of cores and nuclei scored by each observer was compared. The level of interobserver agreement upon validity of scoring and deletion status per represented block of tumour was assessed using Kappa statistical analysis.
The clinicopathological data were compared based on 9p status. Proportions between categorical variables were compared using Fisher exact and Pearson chi-square tests, as appropriate. The survival time was summarised using median and interquartile range (IQR). Other continuous variables were summarised as means (s.d.), and compared using Student’s t-tests or Wilcoxon rank test, as appropriate. The Kaplan–Meier method was used to estimate RFS and DSS based on 9p status and other variables. The log-rank test was used to compare the survival differences between the groups.
A univariate Cox proportional hazard model was used to assess the correlation between prognostic variables and recurrence, and RCC-specific mortality. Multivariate analysis was performed for DSS and RFS after excluding the insignificant variables on univariate analysis. For DSS, two models were generated: one included all the significant variables except SSIGN score, and the other was fit with SSIGN score and all the other significant variables not incorporated in the SSIGN algorithm. A backward selection manner with the likelihood ratio criterion (for entry and removal: P⩽0.05 and P>0.1, respectively) and rank of elimination was used to identify the most significant variables to be entered in a final model for RFS and two final prognostic models for DSS. The predictive accuracy of prognostic models was assessed by employing Concordance index (C-index). Statistical analysis was performed using IBM SPSS version 21 (IBM Corporation), with all tests being two-tailed and P<0.05 considered statistically significant.
Results
There were 89 (82.4%) patients with ccRCC tumours, 13 (12%) with papillary RCCs (pRCCs), 5 (4.6%) with chromophobe RCCs and 1 (1%) with collecting duct carcinoma. Ten tumours (nine ccRCCs and one pRCC) could not be scored due to poor signal, or core loss, or excess connective tissue, or tissue damage or necrosis.
Interobserver agreement of FISH scoring
Out of 756 tumour cores, observer 1 scored 534 (70%) compared with 517 (68%) cores by observer 2 on the basis of the preset criteria. The mean number of cores per tumour block scored by each observer was comparable (4.6 out of 6 vs 4.45 out of 6; P=0.055). Also, the mean number of nuclei per tumour block scored by each observer was comparable (mean 151.4 vs 147.8; P=0.645). Both observers agreed upon 9p status or invalid scoring in 105 out of 126 (83%) tumour blocks with a kappa statistic for interobserver agreement of 0.71 (good agreement; 95% CI 0.60–0.82; P<0.0001). Deletion was detected and agreed upon in 37 tumour blocks with LOH being the predominant deletion pattern (n=21), followed by monosomy (n=13) and homozygous deletion (n=3). Both observers agreed on a no-deletion status and invalid scoring in 58 and 10 blocks, respectively. Two out of the 17 tumours represented by more than one block showed variation for 9p status between the blocks.
The pooled mean percentage of abnormal nuclei, in the agreed upon 9p-deleted tumour blocks, was 52.5% (range 35–97%) and 54% (37–98%) for observer 1 and observer 2, respectively (P=0.782). Whereas in the non-9p-deleted tumour blocks, the pooled mean percentage of abnormal nuclei was 23% (range 7–42%) and 25% (6–47%) for observer 1 and observer 2, respectively (P=0.4). In the agreed-upon blocks, the detection of 9p deletion increased with the total number of cores scored. 9p deletion was observed in 25% of blocks where three cores were scored compared with almost 50% of blocks where the six representative cores were scored (Figure 2).
Both observers disagreed on scoring 21 tumour blocks. The majority of discordance was due to the invalid scoring by one observer (n=10). In the rest of discordant cases (n=11), there was a significant difference in the percentage of abnormal nuclei between both observers, which required a slide review to reach a consensus.
We scored 10 whole-tissue sections from tumours on which both observers agreed on 9p status. The five non-deleted tumours showed abnormal nuclei ranging between 11 and 23% (mean 19%) in whole-tissue section compared with 15 and 30% (mean 20.3%) in the representative cores. On the other hand, the five tumours with 9p deletion showed a range of abnormal nuclei between 66 and 93% (mean 78%) on whole sections compared with a range between 53 and 97% (mean 71%) in the corresponding cores. The overall concordance between whole sections and tissue array cores was 100%.
Clinicopathological parameters
There were 98 out of 108 tumours with valid 9p deletion status and complete pathological and follow-up data for analysis. Only patients with ccRCC tumours (n=80), with mean age of 62.96 years (s.d.±11.64) and male to female ratio of 5 : 3, were included in the analysis.
Table 1 summarises the clinicopathological parameters based on chromosome 9p status. 9p deletion was observed in 44% of the tumours (35 out of 80). The mean age of the 9p-deleted group was 65.92 (s.d.±10.7) years compared with 60.4 (s.d.±11.92) years in the non-9p-deleted group (P=0.043).
The mean size of 9p-deleted tumours was 7.7 cm (s.d.±3.42) vs 5.6 cm (s.d.±2.54) for non-9p-deleted ones (P=0.002). Also, 9p loss was significantly associated with higher pathological T-stage (P=0.021), presence of tumour necrosis (P=0.019), microvascular invasion (P=0.032) and renal vein invasion (P=0.046) compared with non-9p-deleted tumours.
Twelve patients (15%) had metastatic disease at diagnosis. Two had regional lymph node metastasis, eight patients had distant metastasis involving typical sites: adrenals, lungs and liver. One patient had metastasis to an unusual site, the bladder wall that was confirmed on resection pathology. Only one patient had both regional lymph node and distant liver metastases. In this cohort, 9p status was not associated with a higher risk of metastasis (P=0.156).
In total, there were 30, 8, 17, 16 and 9 patients with 0–2, 3–4, 5–6, 7–9 and ⩾10 SSIGN scores, respectively. 9p-deleted tumours were associated with higher SSIGN scores (P=0.033).
Survival analysis
The median follow-up for patients with ccRCC was 95 months (IQR 40.5–115.5 months). In total, 40 patients (50%) died at the end of the follow-up period, 20 patients (25%) died from advanced renal cancer, whereas 20 patients died from non-RCC-related causes. Nine cases died from a second advanced malignancy including lung (n=2), breast (n=3), pancreatic (n=1), colorectal (n=2) and melanoma (n=1) with no sign of renal cancer recurrence. One died from sepsis and acute kidney injury on day 8 post nephrectomy. Eleven patients died from other causes including sepsis, cerebrovascular accident, auto-immune hepatitis and severe pancreatitis from gall stones.
The median DSS for tumours with 9p loss was 87 months (IQR 31–112 months) compared with 97 months in non-9p-deleted ones (IQR 53–120 months; P=0.291). At 1, 5 and 10 years of follow-up, 15%, 39% and 55% of patients with 9p loss died from ccRCC, respectively, as compared with 0%, 5% and 11% in patients with normal 9p (P<0.001) (Figure 3a). Also, tumour size ⩾5 cm, pathological T3 and T4 stages, Fuhrman grades 3 and 4, presence of metastasis (N+M+), tumour necrosis, microvascular invasion, renal vein invasion, pelvicalyceal invasion, renal sinus invasion and SSIGN scores were all variables associated significantly with cancer-specific mortality on univariate analysis (Table 2).
There were 19 patients with clinically localised small renal masses (SRMs) (cT1a) in this cohort (<4 cm) with a mean size of 2.9 cm (s.d.±0.66 cm) with a median follow-up of 107 months (IQR 42–128 months). Two out of the five patients (40%) with 9p-deleted SRMs died from advanced renal cancer. Those two SRMs were found to be invading the renal sinus and perinephric fat and were staged as pT3a. On the other hand, none of the patients with normal 9p status SRMs had a ccRCC-specific death (P=0.013).
There were 19 patients with locally advanced non-metastatic ccRCC (pT3N0M0) with a median follow-up of 90 months (IQR 42–105 months). Eight out of 10 patients (80%) with 9p-deleted tumours died from advanced renal cancer compared with 1 out of 9 (11%) with normal 9p status (P=0.009; Figure 3c). In this subgroup of patients, 9p loss was the only significant variable on univariate analysis (hazard ratio 10.071; P=0.026).
Sixteen out of 68 patients (23.5%) with localised ccRCCs at diagnosis developed recurrence with the median time to recurrence being 35.5 months. 9p deletion, pT-stage, microvascular invasion and age were associated with increased risk of recurrence on univariate analysis in our cohort (Table 2). In the non-9p-deleted tumours, only 11% developed recurrences at 5 and 10 years of follow-up, in contrast to 8%, 34% and 55% of patients with 9p-deleted tumours showing recurrences at 1, 5 and 10 years, respectively (P=0.004; Figure 3b). We observed a more aggressive pattern of recurrence in patients with 9p-deleted tumours involving distant organs when first detected; in 11 out of 12 cases disease recurred distantly, whereas 3 out of the 4 patients with non-9p-deleted tumours had the first site of recurrence in the resected renal bed (P=0.026).
In multivariate analysis, 9p deletion was an independent prognostic factor in both models for DSS (Table 3). Patients with 9p-deleted tumours had 4.6- and 4-fold higher risk of mortality from advanced renal cancer compared with the non-deleted ones in DSS models 1 and 2, respectively. The inclusion of 9p status in model 1 beside T-stage and state of metastasis improved the predictive accuracy expressed by C-index from 0.858 to 0.911 (P=0.001). Adding 9p status to the SSIGN score in model 2 increased the predictive accuracy of the SSIGN score expressed by the C-index from 0.877 to 0.931 (P=0.001). In model 3, 9p deletion was an independent predictor of recurrence (hazard ratio 3.863; P=0.021) with pT-stage (Table 3).
Discussion
We performed analytic validation of I-FISH in the assessment of 9p deletion in ccRCC in a cohort of patients with the longest reported period of follow-up. We used the same probe reported in the previous studies, and relied on normal renal tissue as a negative control to determine the threshold level for 9p deletion with a good level of interobserver agreement. The use of a negative control to determine the cut-off threshold for deletion makes the scoring method more reliable, objective and reproducible.
It is known that intratumour heterogeneity and complexity could be underestimated or missed as a result of sampling bias if a small number of biopsy cores are used (Martinez et al, 2013). Also, there is a general realisation that biomarker research using TMA may not be sufficient to adequately represent whole-tissue specimens (Eckel-Passow et al, 2010). The detection rate of 9p deletion in the current study is 44%, which is higher than that reported in the literature—ranging between 14 and 18% in studies employing I-FISH on TMA (Brunelli et al, 2008; La Rochelle et al, 2010).
The higher detection rate of 9p deletion could be partly explained by the difference in cut-off threshold for deletion. It could also be explained by the use of six cores to represent the excised tumours, in the present study, rather than three cores in the previous studies. Our findings show a trend towards higher detection rate of 9p deletion with increasing number of scored cores (Figure 2). We showed a 100% concordance between whole-tissue sections and representative cores, with at least four out of the six cores scored, whether the tumour was 9p deleted or non-deleted.
It is important to note that we also applied the scoring technique suggested by La Rochelle et al (2010) using only the first three cores from each tumour on the TMA. The detection rate of 9p loss was 18% (15 out of 80), which matches the one reported by La Rochelle et al (2010)—14%. There was a significant difference in DSS at 10 years of follow-up between 9p-deleted and non-9p-deleted tumours in the same cohort of patients (20% vs 70%, respectively, P<0.001—Supplementary Figure 1) and 9p status based on La Rochelle’s cut-off threshold remained an independent prognostic variable on multivariate analysis for DSS with T-stage, state of metastasis and sarcomatoid features (hazard ratio 2.956; P=0.025).
Although homozygous deletion, hypermethylation of the promoter region and intragenic mutation have all been reported as mechanisms of CDKN2A inactivation in cell lines (Herman et al, 1995), they all seem to be rare events in primary renal tumours (Cairns et al, 1995; Sanz-Casla et al, 2003). Most of the studies looking at 9p deletion in primary renal tumours, and not in cell lines, found LOH of 9p21 locus (CDKN2A) to be the most commonly reported CNV (Schullerus et al, 1997; Grady et al, 2001; Presti et al, 2002; Brunelli et al, 2008; La Rochelle et al, 2010). In the present study, 91% (32 out of 35) of 9p deletion observed was due to the loss of one copy of the CDKN2A (either LOH or monosomy). Only three tumours showed deletion of both copies (homozygous deletion). It is well known that CNV correlates with gene expression (Knudson’s theory)—loss of one allele allows the inactivation of the gene if the remaining copy is mutated or it could result in lower expression of the tumour suppressor protein. Aberrations, leading to low expression of p16INK4A and p14ARF, have been associated with aggressive ccRCC and poorer prognosis (Kawada et al, 2001b; Ikuerowo et al, 2007).
The principal findings of the present study are that ccRCC with 9p loss is associated with higher stage and larger size tumours, and is more likely to have adverse histopathological features (tumour necrosis, microvascular invasion and renal vein involvement). A statistically nonsignificant trend was observed between 9p-deleted ccRCC tumours and higher Fuhrman grade. Our finding is in agreement with previous reports suggesting an association between 9p deletion and ccRCC progression (Klatte et al, 2009; La Rochelle et al, 2010; Dagher et al, 2013).
The prognostic impact of 9p deletion on ccRCC remains a controversial matter. Several studies assessed the impact of 9p status in ccRCC on survival using different molecular and genetic techniques with median follow-up periods ranging between 31 and 73 months. Two studies showed that 9p deletion was not associated with poor prognosis (Presti et al, 2002; Antonelli et al, 2010). On the other hand, three studies concluded that 9p deletion was associated with worse prognosis on univariate analysis only (Moch et al, 1996; Schraml et al, 2001; Sanjmyatav et al, 2011). Whereas in two studies 9p loss remained an independent predictor of outcome in ccRCC on multivariate analysis. I-FISH was the main technique employed to assess 9p status in those two studies with different scoring techniques and threshold levels for deletion (Brunelli et al, 2008; La Rochelle et al, 2010). In our cohort of ccRCC, with the longest median follow-up of 95 months, survival analysis showed that patients with 9p-deleted tumours had a higher risk of recurrence and cancer-specific mortality. On multivariate analysis, it appears that 9p deletion remains a poor prognosticator for both outcomes, confirming the findings from previous studies based on I-FISH.
Several nomograms to predict the prognosis in RCC, relying on clinical and pathological parameters, have been developed and some have been externally validated. The integration of molecular or genetic biomarker, besides pathological and clinical parameters, has been attempted to improve the prognostication of some nomograms (Kim et al, 2005; Karakiewicz et al, 2007; Klatte et al, 2009). The addition of 9p status to prognostic models for ccRCC improved the predictive accuracy of DSS up to 89% (Klatte et al, 2009; La Rochelle et al, 2010). In the present study, SSIGN score and 9p status were independent predictors of outcome on multivariate analysis, mirroring the findings by Brunelli et al (2008). We have shown that the integration of 9p status to the well-established, externally validated, SSIGN score improved its predictive accuracy of DSS to 93.1%.
We acknowledge that the present study is limited by its retrospective nature. However, we made every effort to avoid bias. The use of preset scoring criteria, assessment of FISH score in a blinded manner by two independent observers, data collection using a validated electronic linkage methodology by third party, characterisation of tumours by a single pathologist with special interest in renal cancer, the representation of tumour by six cores on the TMA, use of normal tissue as a negative control to define threshold of 9p loss and systematic protocol-guided long-term follow-up exceeding those reported in the literature are the strengths of our study.
Lack of reporting of previous studies in a rigorous manner and published articles often showing insufficient information have been highlighted by reporting recommendations for a tumour marker prognostic studies (REMARK guidelines) (McShane et al, 2006) panel following a consensus meeting between US National Cancer Institute and the European Organisation for Research and Treatment of Cancer (NCI-EORTC) experts. We have adhered to these recommendations in reporting the present study (Supplementary Table 1).
The 9p deletion detected by I-FISH in ccRCC, regardless of the cut-off threshold, retained its prognostic value. Therefore, we believe that the potential use of this predictive biomarker in ccRCC should be further explored after agreeing on scoring technique, cut-off threshold for deletion and the number of cores required, keeping in mind intratumour heterogeneity.
We also suggest that a consensus statement is required to standardise I-FISH scoring for chromosomal deletion in renal tissue as this could have an implication on translational research in the area of renal cancer. 9p status could be used to predict behaviour of clinically localised SRMs and help in stratifying patients who may need interventions or surveillance. The change in size of SRMs on serial imaging is currently used as a surrogate marker of tumour progression in most of the centres across the world; however, this may not be the best indicator of aggressiveness. Our findings of worse prognosis in clinically localised SRMs (<4 cm) (Supplementary Figure 2) corroborates with previous reports and suggests that 9p-deleted SRMs could have a different behaviour and hence be a reason for early intervention. Similarly, our observation of poor survival in 9p-deleted locally advanced non-metastatic (pT3N0M0) ccRCC, which was previously reported (Moch et al, 1996; Schraml et al, 2001), may be used to counsel patients for recruitment into adjuvant treatment for locally advanced tumours. This should, ideally, be further assessed in prospective randomised controlled trials.
Conclusion
Loss of 9p is associated with aggressive clear cell carcinoma of kidney and portends a worse prognosis in patients following surgery. Our findings independently confirm the findings of previous reports relying on I-FISH to detect 9p (CDKN2A) deletion.
Change history
23 September 2014
This paper was modified 12 months after initial publication to switch to Creative Commons licence terms, as noted at publication
References
Antonelli A, Arrighi N, Tardanico R, Balzarini P, Zanotelli T, Corti S, Zani D, Cozzoli A, Cunico SC, Simeone C (2010) Prognostic value of cytogenetic analysis in clear cell renal carcinoma: a study on 131 patients with long-term follow-up. Anticancer Res 30: 4705–4709.
Atkins M, Regan M, McDermott D, Mier J, Stanbridge E, Youmans A, Febbo P, Upton M, Lechpammer M, Signoretti S (2005) Carbonic anhydrase IX expression predicts outcome of interleukin 2 therapy for renal cancer. Clin Cancer Res 11: 3714–3721.
Bayani J, Squire JA (2007) Application and interpretation of FISH in biomarker studies. Cancer Lett 249: 97–109.
Beroukhim R, Brunet JP, Di Napoli A, Mertz KD, Seeley A, Pires MM, Linhart D, Worrell RA, Moch H, Rubin MA, Sellers WR, Meyerson M, Linehan WM, Kaelin WG Jr, Signoretti S (2009) Patterns of gene expression and copy-number alterations in von-hippel lindau disease-associated and sporadic clear cell carcinoma of the kidney. Cancer Res 69: 4674–4681.
Brunelli M, Eccher A, Gobbo S, Ficarra V, Novara G, Cossu-Rocca P, Bonetti F, Menestrina F, Cheng L, Eble JN, Martignoni G (2008) Loss of chromosome 9p is an independent prognostic factor in patients with clear cell renal cell carcinoma. Mod Pathol 21: 1–6.
Bui MH, Seligson D, Han KR, Pantuck AJ, Dorey FJ, Huang Y, Horvath S, Leibovich BC, Chopra S, Liao SY, Stanbridge E, Lerman MI, Palotie A, Figlin RA, Belldegrun AS (2003) Carbonic anhydrase IX is an independent predictor of survival in advanced renal clear cell carcinoma: implications for prognosis and therapy. Clin Cancer Res 9: 802–811.
Cairns P, Tokino K, Eby Y, Sidransky D (1995) Localization of tumor suppressor loci on chromosome 9 in primary human renal cell carcinomas. Cancer Res 55: 224–227.
Cossu-Rocca P, Eble JN, Zhang S, Martignoni G, Brunelli M, Cheng L (2006) Acquired cystic disease-associated renal tumors: an immunohistochemical and fluorescence in situ hybridization study. Mod Pathol 19: 780–787.
Dagher J, Dugay F, Verhoest G, Cabillic F, Jaillard S, Henry C, Arlot-Bonnemains Y, Bensalah K, Oger E, Vigneau C, Rioux-Leclercq N, Belaud-Rotureau MA (2013) Histologic prognostic factors associated with chromosomal imbalances in a contemporary series of 89 clear cell renal cell carcinomas. Hum Pathol 44: 2106–2115.
Dalgliesh GL, Furge K, Greenman C, Chen L, Bignell G, Butler A, Davies H, Edkins S, Hardy C, Latimer C, Teague J, Andrews J, Barthorpe S, Beare D, Buck G, Campbell PJ, Forbes S, Jia M, Jones D, Knott H, Kok CY, Lau KW, Leroy C, Lin ML, McBride DJ, Maddison M, Maguire S, McLay K, Menzies A, Mironenko T, Mulderrig L, Mudie L, O'Meara S, Pleasance E, Rajasingham A, Shepherd R, Smith R, Stebbings L, Stephens P, Tang G, Tarpey PS, Turrell K, Dykema KJ, Khoo SK, Petillo D, Wondergem B, Anema J, Kahnoski RJ, Teh BT, Stratton MR, Futreal PA (2010) Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature 463: 360–363.
Eckel-Passow JE, Lohse CM, Sheinin Y, Crispen PL, Krco CJ, Kwon ED (2010) Tissue microarrays: one size does not fit all. Diagn Pathol 5: 48–1596-5-48.
Ellsworth RE, Hooke JA, Love B, Kane JL, Patney HL, Ellsworth DL, Shriver CD (2008) Correlation of levels and patterns of genomic instability with histological grading of invasive breast tumors. Breast Cancer Res Treat 107: 259–265.
Ficarra V, Martignoni G, Lohse C, Novara G, Pea M, Cavalleri S, Artibani W (2006) External validation of the mayo clinic stage, size, grade and necrosis (SSIGN) score to predict cancer specific survival using a European series of conventional renal cell carcinoma. J Urol 175: 1235–1239.
Ficarra V, Novara G, Galfano A, Brunelli M, Cavalleri S, Martignoni G, Artibani W (2009) The ‘Stage, Size, Grade and Necrosis’ score is more accurate than the University of California Los Angeles Integrated Staging System for predicting cancer-specific survival in patients with clear cell renal cell carcinoma. BJU Int 103: 165–170.
Frank I, Blute ML, Cheville JC, Lohse CM, Weaver AL, Zincke H (2002) An outcome prediction model for patients with clear cell renal cell carcinoma treated with radical nephrectomy based on tumor stage, size, grade and necrosis: the SSIGN score. J Urol 168: 2395–2400.
Fujii Y, Saito K, Iimura Y, Sakai Y, Koga F, Kawakami S, Kumagai J, Kihara K (2008) External validation of the Mayo Clinic cancer specific survival score in a Japanese series of clear cell renal cell carcinoma. J Urol 180: 1290–1295, ; discussion 1295-6.
Grady B, Goharderakhshan R, Chang J, Ribeiro-Filho LA, Perinchery G, Franks J, Presti J, Carroll P, Dahiya R (2001) Frequently deleted loci on chromosome 9 may harbor several tumor suppressor genes in human renal cell carcinoma. J Urol 166: 1088–1092.
Gunawan B, Huber W, Holtrup M, von Heydebreck A, Efferth T, Poustka A, Ringert RH, Jakse G, Fuzesi L (2001) Prognostic impacts of cytogenetic findings in clear cell renal cell carcinoma: gain of 5q31-qter predicts a distinct clinical phenotype with favorable prognosis. Cancer Res 61: 7731–7738.
Hadad SM, Baker L, Quinlan PR, Robertson KE, Bray SE, Thomson G, Kellock D, Jordan LB, Purdie CA, Hardie DG, Fleming S, Thompson AM (2009) Histological evaluation of AMPK signalling in primary breast cancer. BMC Cancer 9: 307–2407-9-307.
Harris SL, Levine AJ (2005) The p53 pathway: positive and negative feedback loops. Oncogene 24: 2899–2908.
Herman JG, Merlo A, Mao L, Lapidus RG, Issa JP, Davidson NE, Sidransky D, Baylin SB (1995) Inactivation of the CDKN2/p16/MTS1 gene is frequently associated with aberrant DNA methylation in all common human cancers. Cancer Res 55: 4525–4530.
Ikuerowo SO, Kuczyk MA, von Wasielewski R, Shittu OB, Jonas U, Machtens S, Serth J (2007) p16INK4a expression and clinicopathologic parameters in renal cell carcinoma. Eur Urol 51: 732–737, ; discussion 738.
Junker K, Ficarra V, Kwon ED, Leibovich BC, Thompson RH, Oosterwijk E (2013) Potential role of genetic markers in the management of kidney cancer. Eur Urol 63: 333–340.
Junker K, Weirich G, Amin MB, Moravek P, Hindermann W, Schubert J (2003) Genetic subtyping of renal cell carcinoma by comparative genomic hybridization. Recent Results Cancer Res 162: 169–175.
Karakiewicz PI, Hutterer GC, Trinh QD, Jeldres C, Perrotte P, Gallina A, Tostain J, Patard JJ (2007) C-reactive protein is an informative predictor of renal cell carcinoma-specific mortality: a European study of 313 patients. Cancer 110: 1241–1247.
Kawada Y, Nakamura M, Ishida E, Shimada K, Oosterwijk E, Uemura H, Hirao Y, Chul KS, Konishi N (2001a) Aberrations of the p14(ARF) and p16(INK4a) genes in renal cell carcinomas. Jpn J Cancer Res 92: 1293–1299.
Kawada Y, Nakamura M, Ishida E, Shimada K, Oosterwijk E, Uemura H, Hirao Y, Chul KS, Konishi N (20011b) Aberrations of the p14(ARF) and p16(INK4a) genes in renal cell carcinomas. Jpn J Cancer Res 92: 1293–1299.
Kim HL, Seligson D, Liu X, Janzen N, Bui MH, Yu H, Shi T, Belldegrun AS, Horvath S, Figlin RA (2005) Using tumor markers to predict the survival of patients with metastatic renal cell carcinoma. J Urol 173: 1496–1501.
Kinoshita H, Yamada H, Ogawa O, Kakehi Y, Osaka M, Nakamura E, Mishina M, Habuchi T, Takahashi R, Sugiyama T (1995) Contribution of chromosome 9p21-22 deletion to the progression of human renal cell carcinoma. Jpn J Cancer Res 86: 795–799.
Klatte T, Rao PN, de Martino M, LaRochelle J, Shuch B, Zomorodian N, Said J, Kabbinavar FF, Belldegrun AS, Pantuck AJ (2009) Cytogenetic profile predicts prognosis of patients with clear cell renal cell carcinoma. J Clin Oncol 27: 746–753.
La Rochelle J, Klatte T, Dastane A, Rao N, Seligson D, Said J, Shuch B, Zomorodian N, Kabbinavar F, Belldegrun A, Pantuck AJ (2010) Chromosome 9p deletions identify an aggressive phenotype of clear cell renal cell carcinoma. Cancer 116: 4696–4702.
Martinez P, Birkbak NJ, Gerlinger M, McGranahan N, Burrell RA, Rowan AJ, Joshi T, Fisher R, Larkin J, Szallasi Z, Swanton C (2013) Parallel evolution of tumour subclones mimics diversity between tumours. J Pathol 230: 356–364.
Matsuda D, Khoo SK, Massie A, Iwamura M, Chen J, Petillo D, Wondergem B, Avallone M, Kloostra SJ, Tan MH, Koeman J, Zhang Z, Kahnoski RJ, French Kidney Cancer Study Group, Baba S, Teh BT (2008) Identification of copy number alterations and its association with pathological features in clear cell and papillary RCC. Cancer Lett 272: 260–267.
McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM Statistics Subcommittee of NCI-EORTC Working Group on Cancer Diagnostics (2006) REporting recommendations for tumor MARKer prognostic studies (REMARK). Breast Cancer Res Treat 100: 229–235.
Moch H, Presti JC Jr, Sauter G, Buchholz N, Jordan P, Mihatsch MJ, Waldman FM (1996) Genetic aberrations detected by comparative genomic hybridization are associated with clinical outcome in renal cell carcinoma. Cancer Res 56: 27–30.
Okami K, Cairns P, Westra WH, Linn JF, Ahrendt SA, Wu L, Sidransky D, Jen J (1997) Detailed deletion mapping at chromosome 9p21 in non-small cell lung cancer by microsatellite analysis and fluorescence in situ hybridization. Int J Cancer 74: 588–592.
Patard JJ, Fergelot P, Karakiewicz PI, Klatte T, Trinh QD, Rioux-Leclercq N, Said JW, Belldegrun AS, Pantuck AJ (2008) Low CAIX expression and absence of VHL gene mutation are associated with tumor aggressiveness and poor survival of clear cell renal cell carcinoma. Int J Cancer 123: 395–400.
Presti JC Jr, Wilhelm M, Reuter V, Russo P, Motzer R, Waldman F (2002) Allelic loss on chromosomes 8 and 9 correlates with clinical outcome in locally advanced clear cell carcinoma of the kidney. J Urol 167: 1464–1468.
Sandlund J, Oosterwijk E, Grankvist K, Oosterwijk-Wakka J, Ljungberg B, Rasmuson T (2007) Prognostic impact of carbonic anhydrase IX expression in human renal cell carcinoma. BJU Int 100: 556–560.
Sanjmyatav J, Junker K, Matthes S, Muehr M, Sava D, Sternal M, Wessendorf S, Kreuz M, Gajda M, Wunderlich H, Schwaenen C (2011) Identification of genomic alterations associated with metastasis and cancer specific survival in clear cell renal cell carcinoma. J Urol 186: 2078–2083.
Sanz-Casla MT, Maestro ML, del Barco V, Zanna I, Moreno J, Vidaurreta M, Almansa I, Fernandez C, Blanco J, Maestro C, Resel L (2003) Loss of heterozygosity and methylation of p16 in renal cell carcinoma. Urol Res 31: 159–162.
Schraml P, Struckmann K, Bednar R, Fu W, Gasser T, Wilber K, Kononen J, Sauter G, Mihatsch MJ, Moch H (2001) CDKNA2A mutation analysis, protein expression, and deletion mapping of chromosome 9p in conventional clear-cell renal carcinomas: evidence for a second tumor suppressor gene proximal to CDKN2A. Am J Pathol 158: 593–601.
Schullerus D, Herbers J, Chudek J, Kanamaru H, Kovacs G (1997) Loss of heterozygosity at chromosomes 8p, 9p, and 14q is associated with stage and grade of non-papillary renal cell carcinomas. J Pathol 183: 151–155.
Sidransky D (1995) Molecular genetics of head and neck cancer. Curr Opin Oncol 7: 229–233.
Sobin L, Gospodarowicz MK, Wittekind C (eds) (2009) TNM Classification of Malignant Tumors. Wiley-Blackwell: Oxford, UK.
Strefford JC, Stasevich I, Lane TM, Lu YJ, Oliver T, Young BD (2005) A combination of molecular cytogenetic analyses reveals complex genetic alterations in conventional renal cell carcinoma. Cancer Genet Cytogenet 159: 1–9.
Author information
Authors and Affiliations
Corresponding author
Additional information
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
Supplementary Information accompanies this paper on British Journal of Cancer website
Supplementary information
Rights and permissions
From twelve months after its original publication, this work is licensed under the Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
El-Mokadem, I., Fitzpatrick, J., Bondad, J. et al. Chromosome 9p deletion in clear cell renal cell carcinoma predicts recurrence and survival following surgery. Br J Cancer 111, 1381–1390 (2014). https://doi.org/10.1038/bjc.2014.420
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/bjc.2014.420
Keywords
This article is cited by
-
Association between copy-number alteration of +20q, −14q and −18p and cross-sensitivity to tyrosine kinase inhibitors in clear-cell renal cell carcinoma
Cancer Cell International (2020)
-
A Meta-Analysis Evaluating Clinical Outcomes of Patients with Renal Cell Carcinoma Harboring Chromosome 9P Loss
Molecular Diagnosis & Therapy (2019)
-
Detailed genomic characterization identifies high heterogeneity and histotype-specific genomic profiles in adrenocortical carcinomas
Modern Pathology (2018)
-
Genomics and clinical correlates of renal cell carcinoma
World Journal of Urology (2018)
-
Microfluidics-assisted fluorescence in situ hybridization for advantageous human epidermal growth factor receptor 2 assessment in breast cancer
Laboratory Investigation (2017)