Abstract
Metastatic breast cancer is one of the leading causes of cancer-related mortality among women in the Western world. To date most research efforts have focused on the molecular analysis of the primary tumour to dissect the genotypes of the disease. However, accumulating evidence supports a molecular evolution of breast cancer during its life cycle, with metastatic lesions acquiring new molecular aberrations. Recognising this critical gap of knowledge, the Breast International Group is launching AURORA, a large, multinational, collaborative metastatic breast cancer molecular screening programme. Approximately 1300 patients with metastatic breast cancer who have received no more than one line of systemic treatment for advanced disease will, after giving informed consent, donate archived primary tumour tissue, as well as will donate tissue collected prospectively from the biopsy of metastatic lesions and blood. Both tumour tissue types, together with a blood sample, will then be subjected to next generation sequencing for a panel of cancer-related genes. The patients will be treated at the discretion of their treating physicians per standard local practice, and they will be followed for clinical outcome for 10 years. Alternatively, depending on the molecular profiles found, patients will be directed to innovative clinical trials assessing molecularly targeted agents. Samples of outlier patients considered as ‘exceptional responders’ or as ‘rapid progressors’ based on the clinical follow-up will be subjected to deeper molecular characterisation in order to identify new prognostic and predictive biomarkers. AURORA, through its innovative design, will shed light onto some of the unknown areas of metastatic breast cancer, helping to improve the clinical outcome of breast cancer patients.
Similar content being viewed by others
Main
We have recently entered an exciting new era of molecular oncology, with powerful high-throughput molecular profiling tools coupled with sophisticated bioinformatics pipelines enabling a detailed molecular characterisation of breast cancer. Repeatedly observed findings from studies applying these techniques suggest extensive genetic diversity of this common disease, with relatively few cancer-related genes showing recurrent mutations at high frequency (Banerji et al, 2012; Ellis et al, 2012; Koboldt et al, 2012; Shah et al, 2012; Stephens et al, 2012). At the same time, extensive preclinical research has led to the development of molecularly targeted anticancer agents that hold the promise for dramatically improving the clinical outcome of breast cancer patients. These agents undergo initial clinical assessment mostly in the metastatic setting (Saini et al, 2013; Zardavas et al, 2013a). Accumulating scientific evidence indicates that such agents will be effective in breast cancer patients carrying the targeted molecular aberrations. Indeed, at the preclinical level detailed genetic characterisation of extensive collections of human cancer cell lines coupled with their pharmacological profiles identifies genetic predictors of response (Barretina et al, 2012; Garnett et al, 2012). Furthermore, emerging evidence from clinical trials assessing targeted agents provides increasing proof that the genetic landscape of any given tumour will dictate its sensitivity or resistance profile to these anticancer agents (Tsimberidou et al, 2012).
A rapidly growing body of evidence suggests that both inter- and intra-tumour genetic heterogeneity of breast cancer can be a severe impediment to the successful clinical development of innovative anticancer treatments. Breast cancer is increasingly fragmented into smaller molecular subpopulations and a successful coupling of patients with the corresponding targeted treatment based on the genotype of their disease will be essential. Recent evidence supports the concept that breast cancer behaves as an evolving entity following the principles of clonal evolution, with metastases acquiring new molecular aberrations as compared to their matched primary tumours (Ding et al, 2010; Navin et al, 2011). This molecular evolution can be exacerbated by the selective pressures imposed by treatment during the life cycle of the disease, thus altering its sensitivity and resistance profile to anticancer treatments (Duncan et al, 2012).
The importance of molecular re-characterisation of metastatic breast cancer has been prospectively assessed (de Dueñas et al, 2014) and it has been recently acknowledged in the clinical recommendations of major cancer societies. The European Society for Medical Oncology (ESMO) clinical practice guidelines for metastatic breast cancer suggest that histopathological confirmation should be pursued especially in the case of an isolated metastatic lesion (Cardoso et al, 2011). Additionally, the National Comprehensive Cancer Network (NCCN) guidelines suggest that biopsy should be performed for oestrogen receptor (ER), progesterone receptor (PgR) and human epidermal growth factor receptor (HER2) assessment in a recurrent lesion, if the status of these markers in the primary tumour was either negative or unknown (NCCN Guidelines for Treatment of Breast Cancer: http://www.nccn.org/professionals/physician_gls/pdf/breast.pdf).
Despite this ever-growing recognition of the clinical significance of the molecular characterisation of metastatic lesions in breast cancer (Shah et al, 2009; Ding et al, 2010; Navin et al, 2011), most of the molecular profiling studies reported so far have assessed primary breast cancer tumours, thus failing to inform us about the molecular evolution and the landscape of molecular changes underpinning metastatic disease (Banerji et al, 2012; Curtis et al, 2012; Ellis et al, 2012; Koboldt et al, 2012; Shah et al, 2012; Stephens et al, 2012). However, cancer cells are able to molecularly evolve, adapting to selective pressures posed by treatment, as has been shown both in the preclinical and clinical setting (Duncan et al, 2012; Balko et al, 2014). This ability to adapt results in longitudinal tumour heterogeneity, with cancer behaving like an evolving ecosystem. In terms of the firmly established triplet of biomarkers used in the clinical management of breast cancer patients, namely ER/PgR/HER2, multiple lines of evidence indicate relevant rates of discordance between primary tumour and subsequent metastatic disease (Cardoso et al, 2001; Liedtke et al, 2009; Amir et al, 2012; Lindström et al, 2012). In terms of more recently recognised genomic aberrations observed in breast cancer, even for the most common ones such as PIK3CA mutations, only a few studies have interrogated their prevalence in metastatic disease in comparison to what is observed in primary tumours (Dupont Jensen et al, 2010; Gonzalez-Angulo et al, 2011; Kalinsky et al, 2011; Fumagalli et al, 2013). In order to address these important clinical gaps, the Breast International Group (BIG), a non-profit organisation for academic research groups from around the world, is launching a very ambitious programme called AURORA – Aiming to Understand the Molecular Aberrations in Metastatic Breast Cancer. This is a large longitudinal cohort study with downstream biologically driven clinical trials focusing on patients with metastatic breast cancer, which will be conducted using the extensive collaborative network of BIG.
Study design – objectives
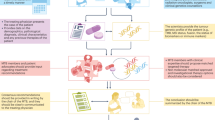
AURORA is a multinational, collaborative metastatic breast cancer molecular profiling programme. Initially, 1300 patients with metastatic breast cancer, either newly diagnosed with metastatic burden or treated with no more than one line of systemic treatment in the metastatic setting, will participate through a large number of leading European hospitals (Figure 1). Currently, countries anticipated to participate in the AURORA molecular screening programme are: Austria, Belgium, Finland, France, Germany, Iceland, Italy, Luxemburg, the Netherlands, Poland, Portugal, Spain, Sweden, Switzerland and the United Kingdom. At a later time, the programme may be extended to enrol patients from other parts of the world.
The AURORA study design. Abbreviations: DCT=downstream clinical trial; MBC=metastatic breast cancer; RNASeq=RNA sequencing; TGS=targeted gene sequencing; WES=whole exome sequencing.
Patients who consent to enter AURORA will provide the following biological material:
-
Tissue from at least one metastatic lesion (if possible two or more core biopsies), either collected prospectively or from archived samples up to 6 months old (formalin-fixed paraffin-embedded (FFPE) and frozen tissue)
-
Archived FFPE tissue from the primary tumour
-
Whole blood samples
-
Plasma samples
-
Serum samples
The tumour tissues provided, together with a blood sample, will then be subjected to next generation sequencing of an extensive targeted panel of cancer-related genes in real time, as well as for RNA sequencing (RNAseq) performed at a central laboratory in batches. This is a commercially available panel of 411 genes (Supplementary Figure 1), which have been shown to be altered in cancer, be potential drivers of oncogenesis and/or potentially modulating the response, or lack of response, to several anticancer drugs on the market or currently under development. A specialized IT infrastructure has been developed to enable reporting of the results to clinicians. The central laboratory has documented quality assurance and quality control procedures as well as validated assay procedures in place for the matrix that will be used in the programme, with BIG conducting regular audits. Patients enrolled in AURORA will be treated as per their treating physician’s choice, following local standard practice. In addition, patients will be given the opportunity to enter clinical trials developed by BIG or other by organisations based on the genotype of their disease. In both cases, patients enrolled in AURORA will be followed with systematic collection of clinical data and plasma and serum samples every 6 months to determine response (locally assessed) and clinical outcome end points for a period of 10 years.
The objectives of the AURORA programme are as follows:
-
To improve our understanding of metastatic breast cancer, delineate its molecular evolution life cycle and explore intra-tumour heterogeneity through high coverage next generation sequencing of a large panel of cancer-related genes. In particular, the parallel sequencing of both primary tumours and their matched metastatic lesions will be informative about the longitudinal intra-tumour heterogeneity, since the genomic landscape of metastatic breast cancer will be compared to that of its primary precursor.
-
To identify potential predictive biomarkers for response and resistance to commonly applied anticancer agents.
-
To provide evidence supporting the feasibility of a global molecular screening platform for metastatic breast cancer.
-
To identify patients with candidate driving mutations in their tumours that can be matched to biomarker-driven clinical trials.
-
To build new therapeutic hypotheses based on findings generated by targeted gene sequencing.
-
To determine patients’ prognosis based on the nature and number of genomic aberrations found in both tumour metastatic biopsies and archived primary tumour tissue.
-
To correlate these genomic aberrations found in patients with the efficacy end points (response rate, progression-free survival and overall survival).
Innovative clinical trials
Downstream to the AURORA programme, BIG is launching innovative phase II clinical trials to assess emerging targeted agents in metastatic breast cancer, either as monotherapy or in combination regimens. Two different scenarios can be identified regarding these trials:
-
Genotype-driven clinical trials, where patients will be matched to the trials according to the genotype of their disease as defined by the results of the targeted gene sequencing. These trials will be developed on the basis of a priori biologic assumptions about specific molecular aberrations dictating sensitivities to targeted therapeutics. In the hypothetical scenario, where two different targetable aberrations are identified, conferring eligibility to more than one clinical trials, the decision will be made by the treating physician, based on the rest of the disease characteristics, patient’s status and comorbidities, patient’s preferences and expected benefits, molecular characteristics, namely allelic frequency of the reported alterations.
-
Clinically driven clinical trials (or non-genotype-driven), where the eligibility of patients to participate will depend on their clinicopathological characteristics and treatment history.
In both scenarios, within any given trial, independent approval processes will be followed with patients being required to sign a separate informed consent form than the one granting access to the AURORA programme. For each patient participating in such a trial, the AURORA molecular screening programme will generate a molecular profile. This information, coupled with the clinical outcome of the experimental treatments, will help identify biomarkers for response and/or resistance to the agents under clinical development. Moreover, a strong translational research component will be developed in a ‘tailored’ manner within the separate trials to maximise the clinically relevant knowledge produced. The extensive biological material being collected will be subjected to further molecular analyses, including exploration for plasma-based mutations and detection, and molecular profiling of circulating tumour cells. These further analyses will be carried out not as part of the AURORA protocol, but in the context of subsequent translational research studies. Thus, AURORA will serve as a molecular screening filter, guiding patients with metastatic breast cancer to innovative clinical trials and accelerating the clinical development of promising targeted agents within the appropriate molecular niches of the disease.
Deeper molecular characterisation
Patients enrolled in the AURORA programme will be treated at the discretion of their treating physicians, either per standard local practice or within downstream clinical trials assessing innovative anticancer agents, as already described. In both cases, treatment history, disease status and clinical outcome data will be prospectively collected every 6 months in an electronic clinical report form that has been developed for the programme. For ‘outlier’ patients, that is, those showing exceptional response or rapid progression, additional in-depth molecular characterisation will be performed on the collected biological samples. The members of the AURORA Steering Committee will identify these outlier patients; however, the following definitions will be used: ‘rapid progressors’ will be initially identified as patients on first- or second-line treatment progressing within the first 3 months since its initiation. ‘Exceptional responders’ will be initially identified as patients showing (nearly) complete response for a duration exceeding 1 year. In particular, paired primary and metastatic lesions as well as blood samples will be subjected to whole exome sequencing to complement the targeted gene sequencing and RNAseq. Only ‘exceptional responders’ enrolled in the downstream clinical trials will be eligible for this in-depth molecular characterisation. The combination of these high-throughput molecular assessment methods should allow us to study the phenotypic changes and related signalling pathways associated with the identified mutational patterns and signatures.
Importantly, detailed dissection of the molecular characteristics of the biological samples of patients experiencing either a striking response to any given anticancer treatment or a particularly aggressive course of the disease will potentiate our ability to identify biomarkers of sensitivity or resistance to the applied therapeutics. Such retrospective molecular analyses of tumour samples derived from patients exposed to targeted therapeutics have already led to the identification of previously occult predictive biomarkers in other cancer types. A recent example is the identification of TSC1 (tuberous sclerosis complex 1) mutations as predictors of sensitivity to everolimus for patients with metastatic urothelial cancer, since this molecular aberration was associated with significant improvement of time-to-recurrence under everolimus treatment for patients bearing these mutations as compared to their wild-type counterparts (P=0.001; Iyer et al, 2012). This methodology is now referred to as the ‘phenotype to genotype’ example.
Expected benefits and challenges
AURORA represents a large-scale, international effort aiming to improve the clinical management of metastatic breast cancer patients. The magnitude of this project entails multiple organisational, logistical and technical challenges. To address these issues, before launching AURORA, a dedicated pilot study was performed involving 30 patients from several European hospitals who provided biopsies of metastatic lesions and blood samples. After central assessment for standard biomarkers (i.e., ER/PgR/HER2), the metastatic lesions and blood samples were subjected to next generation sequencing, with the results of these analyses becoming available within a maximum of 15 working days after the receipt of samples at central laboratories. Of note, a specialised web-based IT platform was developed to manage the different activities and support the information flow required: patient registration; acquisition of data from participating hospitals and central laboratories; sample bio-tracking; reporting of molecular and pathology results to clinicians; and importantly, the automated matching of these results to the eligibility criteria of clinical trials. This platform captures baseline information about the patient’s clinical status, the primary tumour and the biopsied metastatic lesion, as well as the patient’s preferences, expressed in the informed consent form, regarding future re-contacting and the reporting of germline sequence variants. It also allows members of the Molecular Advisory Board to access the patients’ molecular data and to add annotations pertaining to these data, such as additional information on the clinical relevance of molecular aberrations. All this information, pathology, sequencing data and annotations, is then presented to the investigator in a secure, integrated and intuitive manner.
We expect to generate multiple layers of clinically relevant knowledge from breast cancer patients through the AURORA programme. First, the molecular characterisation of the primary tumour combined with (neo)-adjuvant treatment history and recurrence information hold the promise for hypothesis-generating findings in terms of predictive and prognostic biomarkers in early-stage disease. Second, such information coupled with the history of the disease after metastatic relapse and in particular the deeper molecular characterisation of the clinical ‘outliers’ can lead to similar findings in the metastatic setting. Third, the downstream-targeted clinical trials will speed-up the successful clinical development of molecularly targeted agents in the metastatic setting. Fourth, the coupling of molecular analyses of the primary tumour and a subsequent metastatic lesion will provide insight into the temporal intra-tumour heterogeneity of breast cancer. Fifth, AURORA will generate a unique clinically and molecularly annotated database of 1300 patients with metastatic breast cancer, with an abundance of biological material stored for future research with state-of-the-art high-throughput technologies. Last, AURORA should be acknowledged as a valuable educational opportunity to familiarise treating physicians with genomics-based oncology. Through its extensive collaborative network, AURORA will expose clinicians from a large number of hospitals across Europe initially to reports containing genomic information originating from their patients. Furthermore, an open channel of communication will become available for clinicians from the participating centres with an AURORA-dedicated multidisciplinary molecular advisory board that will support the clinical interpretation of the genomic profiling results.
We expect for such a complex initiative to encounter different challenges. First, the logistic complexities underlying AURORA are such a challenge, with the small lead-in pilot study that preceded it, having offered valuable guidance. Second, the selection of the molecular profiling platform to be selected is another important issue, since there is an increasing repertoire of different options (whole genome, whole exome or sequencing of targeted panels of genes, with different platforms available for each one of them). The selection of multiplexed testing techniques over multiple individual molecular aberrations’ assessment offers the compelling advantage of interrogating multiple alterations from one sample. The final selection is based on factors related to sensitivity and specificity, tissue requirements and turn-around times to name a few. Third, the assessment of the clinical relevance of the molecular information generated by such an initiative is another challenge. In AURORA, the formation of a dedicated molecular advisory board, consisting of experts from different disciplines such as (molecular) oncology, molecular pathology, genetics and bioinformatics, is a valuable asset of the programme to this end. Fourth, ethical challenges are expected to arise for any initiative interrogating the genetic landscapes of cancer diseases, in particular in regard to the evaluation of the germline alterations that can hold significance for family members of the patients subjected to sequencing. In our programme we allow patients entering the AURORA programme to opt out from the assessment of germline variants. Finally, as with any other academic research initiative funding is a major challenge, since healthcare providers do not cover the costs of the intensive molecular profiling foreseen for AURORA.
Conclusion
AURORA is an academia-driven initiative that aims to boost genomic and clinical knowledge generated from metastatic breast cancer patients. Detailed molecular profiling of breast cancer is deepening our understanding of the molecular biology underpinning this common disease, promising to lead to personalised cancer medicine (Zardavas et al, 2013b). Currently, there are multiple initiatives worldwide aiming to provide a detailed molecular characterisation of several types of cancer (Table 1). The goal of such initiatives is to assess the feasibility and clinical utility of genomics-based medicine, eventually establishing it in oncology. First experiences of such initiatives have been already reported, indicating feasibility of personalisation of medicine for patients with metastatic breast cancer (André et al, 2014).
AURORA is an effort dedicated to metastatic breast cancer, also aiming to interrogate the intra-tumour heterogeneity of the disease through the coupled genomic analyses of both primary tumour and metastatic lesions. The clinical development of promising targeted anticancer compounds is reshaping the design of future clinical trials. Indeed, the ‘one size fits all’ clinical trial design paradigm is being gradually abandoned in favour of genotype-driven clinical trials, with the realistic assumption that targeted compounds will show overt activity against tumours bearing the targeted molecular aberrations (Table 2; Sleijfer et al, 2013). AURORA promises to boost the successful clinical development of targeted agents through its downstream clinical trials. Overall, the AURORA programme, through its innovative design, will shed light onto the unknown areas of metastatic breast cancer, bringing clinicians several steps closer to the implementation of precision medicine for their patients.
References
Amir E, Miller N, Geddie W, Freedman O, Kassam F, Simmons C, Oldfield M, Dranitsaris G, Tomlinson G, Laupacis A, Tannock IF, Clemons M (2012) Prospective study evaluating the impact of tissue confirmation of metastatic disease in patients with breast cancer. J Clin Oncol 30: 587–592.
André F, Bachelot T, Commo F, Campone M, Arnedos M, Dieras V, Lacroix-Triki M, Lacroix L, Cohen P, Gentien D, Adélaide J, Dalenc F, Goncalves A, Levy C, Ferrero J-M, Bonneterre J, Lefeuvre C, Jimenez M, Filleron T, Bonnefoi H (2014) Comparative genomic hybridisation array and DNA sequencing to direct treatment of metastatic breast cancer: a multicentre, prospective trial (SAFIR01/UNICANCER). Lancet Oncol 15: 267–274.
Balko JM, Giltnane JM, Wang K, Schwarz LJ, Young CD, Cook RS, Owens P, Sanders ME, Kuba MG, Sánchez V, Kurupi R, Moore PD, Pinto JA, Doimi FD, Gómez H, Horiuchi D, Goga A, Lehmann BD, Bauer JA, Pietenpol JA, Ross JS, Palmer GA, Yelensky R, Cronin M, Miller VA, Stephens PJ, Arteaga CL (2014) Molecular profiling of the residual disease of triple-negative breast cancers after neoadjuvant chemotherapy identifies actionable therapeutic targets. Cancer Discov 4: 232–245.
Banerji S, Cibulskis K, Rangel-Escareno C, Brown KK, Carter SL, Frederick AM, Lawrence MS, Sivachenko AY, Sougnez C, Zou L, Cortes ML, Fernandez-Lopez JC, Peng S, Ardlie KG, Auclair D, Bautista-Piña V, Duke F, Francis J, Jung J, Maffuz-Aziz A, Onofrio RC, Parkin M, Pho NH, Quintanar-Jurado V, Ramos AH, Rebollar-Vega R, Rodriguez-Cuevas S, Romero-Cordoba SL, Schumacher SE, Stransky N, Thompson KM, Uribe-Figueroa L, Baselga J, Beroukhim R, Polyak K, Sgroi DC, Richardson AL, Jimenez-Sanchez G, Lander ES, Gabriel SB, Garraway LA, Golub TR, Melendez-Zajgla J, Toker A, Getz G, Hidalgo-Miranda A, Meyerson M (2012) Sequence analysis of mutations and translocations across breast cancer subtypes. Nature 486: 405–409.
Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehár J, Kryukov GV, Sonkin D, Reddy A, Liu M, Murray L, Berger MF, Monahan JE, Morais P, Meltzer J, Korejwa A, Jané-Valbuena J, Mapa FA, Thibault J, Bric-Furlong E, Raman P, Shipway A, Engels IH, Cheng J, Yu GK, Yu J, Aspesi P Jr, de Silva M, Jagtap K, Jones MD, Wang L, Hatton C, Palescandolo E, Gupta S, Mahan S, Sougnez C, Onofrio RC, Liefeld T, MacConaill L, Winckler W, Reich M, Li N, Mesirov JP, Gabriel SB, Getz G, Ardlie K, Chan V, Myer VE, Weber BL, Porter J, Warmuth M, Finan P, Harris JL, Meyerson M, Golub TR, Morrissey MP, Sellers WR, Schlegel R, Garraway LA (2012) The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483: 603–607.
Cardoso F, Di Leo A, Larsimont D, Gancberg D, Rouas G, Dolci S, Ferreira F, Paesmans M, Piccart M (2001) Evaluation of HER2, p53, bcl-2, topoisomerase II-alpha, heat shock proteins 27 and 70 in primary breast cancer and metastatic ipsilateral axillary lymph nodes. Ann Oncol 12: 615–620.
Cardoso F, Fallowfield L, Costa A, Castiglione M, Senkus E On behalf of the ESMO Guidelines Working Group (2011) Locally recurrent or metastatic breast cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 22: vi25–vi30.
Curtis C, Shah SP, Chin S-F, Turashvili G, Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y, Gräf S, Ha G, Haffari G, Bashashati A, Russell R, McKinney S METABRIC Group Langerød A, Green A, Provenzano E, Wishart G, Pinder S, Watson P, Markowetz F, Murphy L, Ellis I, Purushotham A, Børresen-Dale A-L, Brenton JD, Tavaré S, Caldas C, Aparicio S (2012) The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 486: 346–352.
de Dueñas EM, Hernández AL, Zotano AG, Carrión RMP, López-Muñiz JIC, Novoa SA, Rodríguez AL, Fidalgo JAP, Lozano JF, Gasión OB, Carrascal EC, Capilla AH, López-Barajas IB, Mateu MM, de Ceballos Reyna MHL, Ferrando AO, Jañez NM, Ballerini VC, Torres AA, Catalán G, Sáenz JAG, Menjón S, González-Angulo AM (2014) Prospective evaluation of the conversion rate in the receptor status between primary breast cancer and metastasis: results from the GEICAM 2009-03 ConvertHER study. Breast Cancer Res Treat 143: 507–515.
Ding L, Ellis MJ, Li S, Larson DE, Chen K, Wallis JW, Harris CC, McLellan MD, Fulton RS, Fulton LL, Abbott RM, Hoog J, Dooling DJ, Koboldt DC, Schmidt H, Kalicki J, Zhang Q, Chen L, Lin L, Wendl MC, McMichael JF, Magrini VJ, Cook L, McGrath SD, Vickery TL, Appelbaum E, Deschryver K, Davies S, Guintoli T, Lin L, Crowder R, Tao Y, Snider JE, Smith SM, Dukes AF, Sanderson GE, Pohl CS, Delehaunty KD, Fronick CC, Pape KA, Reed JS, Robinson JS, Hodges JS, Schierding W, Dees ND, Shen D, Locke DP, Wiechert ME, Eldred JM, Peck JB, Oberkfell BJ, Lolofie JT, Du F, Hawkins AE, O’Laughlin MD, Bernard KE, Cunningham M, Elliott G, Mason MD, Thompson DM Jr, Ivanovich JL, Goodfellow PJ, Perou CM, Weinstock GM, Aft R, Watson M, Ley TJ, Wilson RK, Mardis ER (2010) Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature 464: 999–1005.
Duncan JS, Whittle MC, Nakamura K, Abell AN, Midland AA, Zawistowski JS, Johnson NL, Granger DA, Jordan NV, Darr DB, Usary J, Kuan P-F, Smalley DM, Major B, He X, Hoadley KA, Zhou B, Sharpless NE, Perou CM, Kim WY, Gomez SM, Chen X, Jin J, Frye SV, Earp HS, Graves LM, Johnson GL (2012) Dynamic reprogramming of the kinome in response to targeted MEK inhibition in triple-negative breast cancer. Cell 149: 307–321.
Dupont Jensen J, Laenkholm A-V, Knoop A, Ewertz M, Bandaru R, Liu W, Hackl W, Barrett JC, Gardner H (2010) PIK3CA mutations may be discordant between primary and corresponding metastatic disease in breast cancer. Clin Cancer Res 17: 667–677.
Ellis MJ, Ding L, Shen D, Luo J, Suman VJ, Wallis JW, Van Tine BA, Hoog J, Goiffon RJ, Goldstein TC, Ng S, Lin L, Crowder R, Snider J, Ballman K, Weber J, Chen K, Koboldt DC, Kandoth C, Schierding WS, McMichael JF, Miller CA, Lu C, Harris CC, McLellan MD, Wendl MC, DeSchryver K, Allred DC, Esserman L, Unzeitig G, Margenthaler J, Babiera GV, Marcom PK, Guenther JM, Leitch M, Hunt K, Olson J, Tao Y, Maher CA, Fulton LL, Fulton RS, Harrison M, Oberkfell B, Du F, Demeter R, Vickery TL, Elhammali A, Piwnica-Worms H, McDonald S, Watson M, Dooling DJ, Ota D, Chang L-W, Bose R, Ley TJ, Piwnica-Worms D, Stuart JM, Wilson RK, Mardis ER (2012) Whole-genome analysis informs breast cancer response to aromatase inhibition. Nature 486 (7403): 353–360.
Fumagalli D, Salgado R, Criscitiello C, Pugliano L, Laios I, Wilson T, Larsimont D, Piccart M, Michiels S, Lackner MR, Sotiriou C, Loi S (2013) Use of mutational profiling of metastatic ER+/HER2- breast cancers and the coexistence of KRAS, MET, BRAF, and FGFR3 with PIK3CA mutations. J Clin Oncol 31: 2013 (suppl; abstr 11003).
Garnett MJ, Edelman EJ, Heidorn SJ, Greenman CD, Dastur A, Lau KW, Greninger P, Thompson IR, Luo X, Soares J, Liu Q, Iorio F, Surdez D, Chen L, Milano RJ, Bignell GR, Tam AT, Davies H, Stevenson JA, Barthorpe S, Lutz SR, Kogera F, Lawrence K, McLaren-Douglas A, Mitropoulos X, Mironenko T, Thi H, Richardson L, Zhou W, Jewitt F, Zhang T, O’Brien P, Boisvert JL, Price S, Hur W, Yang W, Deng X, Butler A, Choi HG, Chang JW, Baselga J, Stamenkovic I, Engelman JA, Sharma SV, Delattre O, Saez-Rodriguez J, Gray NS, Settleman J, Futreal PA, Haber DA, Stratton MR, Ramaswamy S, McDermott U, Benes CH (2012) Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature 483: 570–575.
Gonzalez-Angulo AM, Ferrer-Lozano J, Stemke-Hale K, Sahin A, Liu S, Barrera JA, Burgues O, Lluch AM, Chen H, Hortobagyi GN, Mills GB, Meric-Bernstam F (2011) PI3K Pathway Mutations and PTEN Levels in Primary and Metastatic Breast Cancer. Mol Cancer Ther 10: 1093–1101.
Iyer G, Hanrahan AJ, Milowsky MI, Al-Ahmadie H, Scott SN, Janakiraman M, Pirun M, Sander C, Socci ND, Ostrovnaya I, Viale A, Heguy A, Peng L, Chan TA, Bochner B, Bajorin DF, Berger MF, Taylor BS, Solit DB (2012) Genome sequencing identifies a basis for everolimus sensitivity. Science 338: 221.
Kalinsky K, Heguy A, Bhanot UK, Patil S, Moynahan ME (2011) PIK3CA mutations rarely demonstrate genotypic intratumoral heterogeneity and are selected for in breast cancer progression. Breast Cancer Res Treat 129: 635–643.
Koboldt DC, Fulton RS, McLellan MD, Schmidt H, Kalicki-Veizer J, McMichael JF, Fulton LL, Dooling DJ, Ding L, Mardis ER, Wilson RK, Ally A, Balasundaram M, Butterfield YSN, Carlsen R, Carter C, Chu A, Chuah E, Chun H-JE, Coope RJN, Dhalla N, Guin R, Hirst C, Hirst M, Holt RA, Lee D, Li HI, Mayo M, Moore RA, Mungall AJ, Pleasance E, Gordon Robertson A, Schein JE, Shafiei A, Sipahimalani P, Slobodan JR, Stoll D, Tam A, Thiessen N, Varhol RJ, Wye N, Zeng T, Zhao Y, Birol I, Jones SJM, Marra MA, Cherniack AD, Saksena G, Onofrio RC, Pho NH, Carter SL, Schumacher SE, Tabak B, Hernandez B, Gentry J, Nguyen H, Crenshaw A, Ardlie K, Beroukhim R, Winckler W, Getz G, Gabriel SB, Meyerson M, Chin L, Park PJ, Kucherlapati R, Hoadley KA, Todd Auman J, Fan C, Turman YJ, Shi Y, Li L, Topal MD, He X, Chao H-H, Prat A, Silva GO, Iglesia MD, Zhao W, Usary J, Berg JS, Adams M, Booker J, Wu J, Gulabani A, Bodenheimer T, Hoyle AP, Simons JV, Soloway MG, Mose LE, Jefferys SR, Balu S, Parker JS, Neil Hayes D, Perou CM, Malik S, Mahurkar S, Shen H, Weisenberger DJ, Triche T Jr, Lai PH, Bootwalla MS, Maglinte DT, Berman BP, Van Den Berg DJ, Baylin SB, Laird PW, Creighton CJ, Donehower LA, Getz G, Noble M, Voet D, Saksena G, Gehlenborg N, DiCara D, Zhang J, Zhang H, Wu C-J, Yingchun Liu S, Lawrence MS, Zou L, Sivachenko A, Lin P, Stojanov P, Jing R, Cho J, Sinha R, Park RW, Nazaire M-D, Robinson J, Thorvaldsdottir H, Mesirov J, Park PJ, Chin L, Reynolds S, Kreisberg RB, Bernard B, Bressler R, Erkkila T, Lin J, Thorsson V, Zhang W, Shmulevich I, Ciriello G, Weinhold N, Schultz N, Gao J, Cerami E, Gross B, Jacobsen A, Sinha R, Arman Aksoy B, Antipin Y, Reva B, Shen R, Taylor BS, Ladanyi M, Sander C, Anur P, Spellman PT, Lu Y, Liu W, Verhaak RRG, Mills GB, Akbani R, Zhang N, Broom BM, Casasent TD, Wakefield C, Unruh AK, Baggerly K, Coombes K, Weinstein JN, Haussler D, Benz CC, Stuart JM, Benz SC, Zhu J, Szeto CC, Scott GK, Yau C, Paull EO, Carlin D, Wong C, Sokolov A, Thusberg J, Mooney S, Ng S, Goldstein TC, Ellrott K, Grifford M, Wilks C, Ma S, Craft B, Yan C, Hu Y, Meerzaman D, Gastier-Foster JM, Bowen J, Ramirez NC, Black AD, Pyatt RE, White P, Zmuda EJ, Frick J, Lichtenberg TM, Brookens R, George MM, Gerken MA, Harper HA, Leraas KM, Wise LJ, Tabler TR, McAllister C, Barr T, Hart-Kothari M, Tarvin K, Saller C, Sandusky G, Mitchell C, Iacocca MV, Brown J, Rabeno B, Czerwinski C, Petrelli N, Dolzhansky O, Abramov M, Voronina O, Potapova O, Marks JR, Suchorska WM, Murawa D, Kycler W, Ibbs M, Korski K, Spychała A, Murawa P, Brzeziński JJ, Perz H, Łaźniak R, Teresiak M, Tatka H, Leporowska E, Bogusz-Czerniewicz M, Malicki J, Mackiewicz A, Wiznerowicz M, Van Le X, Kohl B, Viet Tien N, Thorp R, Van Bang N, Sussman H, Duc Phu B, Hajek R, Phi Hung N, Viet The Phuong T, Quyet Thang H, Zaki Khan K, Penny R, Mallery D, Curley E, Shelton C, Yena P, Ingle JN, Couch FJ, Lingle WL, King TA, Maria Gonzalez-Angulo A, Mills GB, Dyer MD, Liu S, Meng X, Patangan M, Waldman F, Stöppler H, Kimryn Rathmell W, Thorne L, Huang M, Boice L, Hill A, Morrison C, Gaudioso C, Bshara W, Daily K, Egea SC, Pegram MD, Gomez-Fernandez C, Dhir R, Bhargava R, Brufsky A, Shriver CD, Hooke JA, Leigh Campbell J, Mural RJ, Hu H, Somiari S, Larson C, Deyarmin B, Kvecher L, Kovatich AJ, Ellis MJ, King TA, Hu H, Couch FJ, Mural RJ, Stricker T, White K, Olopade O, Ingle JN, Luo C, Chen Y, Marks JR, Waldman F, Wiznerowicz M, Bose R, Chang L-W, Beck AH, Maria Gonzalez-Angulo A, Pihl T, Jensen M, Sfeir R, Kahn A, Chu A, Kothiyal P, Wang Z, Snyder E, Pontius J, Ayala B, Backus M, Walton J, Baboud J, Berton D, Nicholls M, Srinivasan D, Raman R, Girshik S, Kigonya P, Alonso S, Sanbhadti R, Barletta S, Pot D, Sheth M, Demchok JA, Mills Shaw KR, Yang L, Eley G, Ferguson ML, Tarnuzzer RW, Zhang J, Dillon LAL, Buetow K, Fielding P, Ozenberger BA, Guyer MS, Sofia HJ, Palchik JD (2012) Comprehensive molecular portraits of human breast tumours. Nature 490: 61–70.
Liedtke C, Broglio K, Moulder S, Hsu L, Kau S-W, Symmans WF, Albarracin C, Meric-Bernstam F, Woodward W, Theriault RL, Kiesel L, Hortobagyi GN, Pusztai L, Gonzalez-Angulo AM (2009) Prognostic impact of discordance between triple-receptor measurements in primary and recurrent breast cancer. Ann Oncol 20: 1953–1958.
Lindström LS, Karlsson E, Wilking UM, Johansson U, Hartman J, Lidbrink EK, Hatschek T, Skoog L, Bergh J (2012) Clinically used breast cancer markers such as estrogen receptor, progesterone receptor, and human epidermal growth factor receptor 2 are unstable throughout tumor progression. J Clin Oncol 30: 2601–2608.
Navin N, Kendall J, Troge J, Andrews P, Rodgers L, McIndoo J, Cook K, Stepansky A, Levy D, Esposito D, Muthuswamy L, Krasnitz A, McCombie WR, Hicks J, Wigler M (2011) Tumour evolution inferred by single-cell sequencing. Nature 472: 90–94.
Saini KS, Loi S, de Azambuja E, Metzger-Filho O, Saini ML, Ignatiadis M, Dancey JE, Piccart-Gebhart MJ (2013) Targeting the PI3K/AKT/mTOR and Raf/MEK/ERK pathways in the treatment of breast cancer. Cancer Treat Rev 39: 935–946.
Shah SP, Morin RD, Khattra J, Prentice L, Pugh T, Burleigh A, Delaney A, Gelmon K, Guliany R, Senz J, Steidl C, Holt RA, Jones S, Sun M, Leung G, Moore R, Severson T, Taylor GA, Teschendorff AE, Tse K, Turashvili G, Varhol R, Warren RL, Watson P, Zhao Y, Caldas C, Huntsman D, Hirst M, Marra MA, Aparicio S (2009) Mutational evolution in a lobular breast tumour profiled at single nucleotide resolution. Nature 461: 809–813.
Shah SP, Roth A, Goya R, Oloumi A, Ha G, Zhao Y, Turashvili G, Ding J, Tse K, Haffari G, Bashashati A, Prentice LM, Khattra J, Burleigh A, Yap D, Bernard V, McPherson A, Shumansky K, Crisan A, Giuliany R, Heravi-Moussavi A, Rosner J, Lai D, Birol I, Varhol R, Tam A, Dhalla N, Zeng T, Ma K, Chan SK, Griffith M, Moradian A, Cheng S-WG, Morin GB, Watson P, Gelmon K, Chia S, Chin S-F, Curtis C, Rueda OM, Pharoah PD, Damaraju S, Mackey J, Hoon K, Harkins T, Tadigotla V, Sigaroudinia M, Gascard P, Tlsty T, Costello JF, Meyer IM, Eaves CJ, Wasserman WW, Jones S, Huntsman D, Hirst M, Caldas C, Marra MA, Aparicio S (2012) The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature 486 (7403): 395–399.
Sleijfer S, Bogaerts J, Siu LL (2013) Designing transformative clinical trials in the cancer genome era. J Clin Oncol 31: 1834–1841.
Stephens PJ, Tarpey PS, Davies H, Van Loo P, Greenman C, Wedge DC, Zainal SN, Martin S, Varela I, Bignell GR, Yates LR, Papaemmanuil E, Beare D, Butler A, Cheverton A, Gamble J, Hinton J, Jia M, Jayakumar A, Jones D, Latimer C, Lau KW, McLaren S, McBride DJ, Menzies A, Mudie L, Raine K, Rad R, Spencer Chapman M, Teague J, Easton D, Langerød A OSBREAC Lee MTM, Shen C-Y, Tee BTK, Huimin BW, Broeks A, Vargas AC, Turashvili G, Martens J, Fatima A, Miron P, Chin S-F, Thomas G, Boyault S, Mariani O, Lakhani SR, van de Vijver M, van’t Veer L, Foekens J, Desmedt C, Sotiriou C, Tutt A, Caldas C, Reis-Filho JS, Aparicio SAJR, Salomon AV, Børresen-Dale A-L, Richardson A, Campbell PJ, Futreal PA, Stratton MR, Karesen R, Schlichting E, Naume B, Sauer T, Ottestad L (2012) The landscape of cancer genes and mutational processes in breast cancer. Nature 486 (7403): 400–404.
Tsimberidou A-M, Iskander NG, Hong DS, Wheler JJ, Falchook GS, Fu S, Piha-Paul S, Naing A, Janku F, Luthra R, Ye Y, Wen S, Berry D, Kurzrock R (2012) Personalized medicine in a phase I clinical trials program: the MD Anderson Cancer Center initiative. Clin Cancer Res 18: 6373–6383.
Zardavas D, Baselga J, Piccart M (2013a) Emerging targeted agents in metastatic breast cancer. Nat Rev Clin Oncol 10: 191–210.
Zardavas D, Pugliano L, Piccart M (2013b) Personalized therapy for breast cancer: a dream or a reality? Future Oncol 9: 1105–1119.
Acknowledgements
We would like to express our sincere gratitude to the Breast Cancer Research Foundation, the Luxemburg Cancer Foundation, the Belgian Loterie Nationale, NIF Trust and BIG Against Breast Cancer for enabling the AURORA programme through their generosity.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies this paper on British Journal of Cancer website
Supplementary information
PowerPoint slides
Rights and permissions
This work is licensed under the Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Zardavas, D., Maetens, M., Irrthum, A. et al. The AURORA initiative for metastatic breast cancer. Br J Cancer 111, 1881–1887 (2014). https://doi.org/10.1038/bjc.2014.341
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/bjc.2014.341
Keywords
This article is cited by
-
International research to address the challenges of metastatic breast cancer: the AURORA Program (BIG 14-01)
npj Breast Cancer (2023)
-
AURKAIP1 actuates tumor progression through stabilizing DDX5 in triple negative breast cancer
Cell Death & Disease (2023)
-
Precision oncology for breast cancer through clinical trials
Clinical & Experimental Metastasis (2022)
-
Prevalence of mutations in BRCA and homologous recombination repair genes and real-world standard of care of Asian patients with HER2-negative metastatic breast cancer starting first-line systemic cytotoxic chemotherapy: subgroup analysis of the global BREAKOUT study
Breast Cancer (2022)
-
Association of BRCA1, ERCC1, RAP80, PKM2, RRM1, RRM2, TS, TSP1, and TXR1 mRNA expression levels between primary tumors and infiltrated regional lymph nodes in patients with resectable non-small cell lung cancer
The Pharmacogenomics Journal (2019)