Abstract
Background:
Mutations in HBx gene are frequently found in HBV-associated hepatocellular carcinoma (HCC). Activation of hypoxia-inducible factor-1α (HIF-1α) contributes to HCC development and progression. Wild-type HBx has been demonstrated to activate HIF-1α, but the effect of HBx mutations on HIF-1α has not been elucidated.
Methods:
HBx mutations were identified by gene sequencing in 101 HCC tissues. Representative HBx mutants were cloned and transfected into HCC cells. Expression and activation of HIF-1α were analysed by western blot and luciferase assays, respectively. The relationship between HBx mutants and HIF-1α expression in HCC tissues was also evaluated.
Results:
The dual mutations K130M/V131I enhanced the functionality of HBx as they upregulated the expression and transcriptional activity of HIF-1α. The C-terminal truncations and deletion mutations, however, weakened the ability of HBx to upregulate HIF-1α. Meanwhile, the C-terminus was further found to be essential for the stability and transactivation of HBx. In the HCC tissues, there was a positive association between the HBx mutants and HIF-1α expression.
Conclusion:
Different mutations of HBx exert differentiated effects on the functionality of HIF-1α, however, the overall activity of HBx mutants appears to increase the expression and transcriptional activity of HIF-1α.
Similar content being viewed by others
Main
Hepatocellular carcinoma (HCC) is the fifth most common cancer worldwide and the third most common cause of cancer mortality (Siegel et al, 2012). Of the risk factors involved in liver carcinogenesis, hepatitis B virus (HBV) chronic infection has been shown to have a major role (Chen et al, 2006). The double-stranded DNA genome of HBV contains four overlapping open-reading frames that encode the surface protein, the core protein, a polymerase and the X protein (HBx) (Neuveut et al, 2010). HBx is a multifunctional protein, which does not bind directly to DNA, but exerts transcriptional activation by its interaction with nuclear transcription factors and modulation of cytoplasmic signal transduction pathways, such as RAS/RAF/MAP signalling (Ng and Lee, 2011). HBx has been demonstrated to accelerate the progress of HCC in many aspects, involving in apoptosis, proliferation, inflammation, angiogenesis, immune responses and multi-drug resistance (Liu et al, 2010; Neuveut et al, 2010; Ng and Lee, 2011). Overall, an increasing body of evidence reveals the crucial role of HBx in HCC development and progress.
Other key factors that account for the aggressiveness of HCC in a poorly oxygenated microenvironment include high rates of cell proliferation and aberrant blood vessel formation (Pouyssegur et al, 2006). The principal mechanism by which hypoxia promotes the lethal cancer phenotype is the induction of hypoxia-inducible factor-1 (HIF-1), and the expression of HIF-1 was increased in a variety of human tumors, including bladder, breast, liver, ovarian, pancreatic, prostate and renal cancers (Harris, 2002; Semenza, 2003). HIF-1 is a heterodimeric protein that is composed of O2-regulated HIF-1α and constitutively expressed HIF-1β subunits (Semenza, 2003). Under normoxic conditions, HIF-1α is hydroxylated on key proline residues by proline hydroxylase domain protein 2 (PHD2), which allows for recognition by von-Hippel-Lindau protein (pVHL), the substrate recognition component of an E3 ubiquitin ligase complex that targets HIF-α for proteasomal degradation (Epstein et al, 2001; Ivan et al, 2001). Under the hypoxic condition, PHD2 activity is inhibited, and HIF-1α accumulates, translocates to the nucleus and dimerises with HIF-1β to form a complex capable of DNA binding at the hypoxia response elements (HREs), which controls the expression of the target proteins that have a key role in many aspects of cancer biology, such as angiogenesis, proliferation, metastasis and differentiation (Semenza, 2003).
The potential relationship between HBx and HIF-α has prompted widespread concern. Previous studies have indicated that wild-type HBx increases the level of HIF-1α via two mechanisms. HBx may bind directly to the bHLH/PAS domain of HIF-1α to inhibit the interaction between pVHL and HIF-1α, thus preventing degradation of HIF-1α protein (Yoo et al, 2003; Yoo and Lee, 2004). In addition, HBx can upregulate HIF-1α expression by stimulating metastasis-associated protein 1, histone deacetylase and the mitogen-activated protein kinase pathway (Yoo et al, 2008). It is worth noting that the HBx constructs used in all of the above studies were wild type. However, most of HBx in HCC tissues contained mutations, which may change the function of HBx. Indeed, artificial point mutations at codons 61, 69 and 137 of HBx have been shown to result in a marked loss of transactivation capability (Kumar et al, 1996), whereas C-terminally truncated HBx mutants were capable of enhancing the activation of cell proliferation and transformation (Ma et al, 2008). As a result, although the relationship between wild-type HBx and HIF-1α has been investigated, the role played by naturally occurring HBx mutants in HIF-1α expression and functions has not been clearly elucidated. The aim of the study reported herein was thus to explore the possible impact of natural HBx mutants on HIF-1α.
Materials and methods
Patient characteristics and tissue sample collection
One hundred and twenty liver tumor tissues were collected from patients with HCC who underwent the surgical resection of their tumors in our hospital. All patients were tested positive for HBsAg and negative for antibodies to the hepatitis C virus (anti-HCV) and human immunodeficiency virus (anti-HIV). The study was carried out with the approval of the Chinese University of Hong Kong Ethical Committee, and the informed consent was obtained from all patients recruited.
Amplification and sequence analysis
DNA was extracted from the tissues, and HBx DNA was amplified by nested PCR. Gene sequencing was performed on an ABI PRISM 377TM DNA Sequencer (Applied Biosystems, Foster City, CA, USA) according to the manufacturer’s instructions. More detailed information on the amplification and sequence analysis of HBx was presented in the Supplementary Materials and Methods.
Plasmids and site-directed mutagenesis
The wild-type HBx plasmid, FLAG-154X, was a kind gift from Dr M. J. Bouchard. HA 154 HBx and the mutants were constructed through amplifying the HBx sequence with primers described in Supplementary Table S1 and inserting it into the vector pSG5L-HA, which encodes HA-tagged fusion protein. Site-directed mutagenesis was performed on HA 154 HBx using a PCR-based strategy through applying QuikChange Lightning Multi Site Directed Mutagenesis kit (Agilent Technologies, Santa Clara, CA, USA). The pcDNA-HIF-1α plasmid was constructed through cutting the sequence of HIF-1α from HA-tagged HIF-1α, a kind gift from Drs L. Neckers and J. S. Isaacs, and inserting it into vector pcDNA 3.1. More detailed information on the plasmids and site-directed mutagenesis was described in the Supplementary Materials and Methods.
Cell culture and reagents
The human hepatocarcinoma cell line, HepG2 (HB 8065), was obtained from the American Type Culture Collection and cultured in Minimum Essential Medium (Sigma, St Louis, MO, USA) supplemented with 10% fetal bovine serum (FBS). The cells were maintained at 37 °C in a humid atmosphere of 5% CO2. Cycloheximide (CHX) was obtained from Sigma-Aldrich (St Louis, MO, USA). The primary antibodies for HA, HBx, HIF-1α and β-actin were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). The primary antibody for Hydroxy-HIF-1α was purchased from Cell Signaling Technology (Danvers, MA, USA).
Western blotting and determination of HBx protein half-life
Western blotting was performed as described previously (Liu et al, 2012). To measure the half-life of HBx protein, HepG2 cells were cultured as above, and after 24 h cycloheximide (CHX, 30 μg ml−1) was added to the media. At indicated time points, cells were removed and protein abundance was measured by immunoblot analysis. The levels of HBx and these mutants at each time points are expressed as a percentage of their abundance at time zero.
Transient transfection and luciferase assays
Transient transfection and luciferase assays were performed as described previously (Liu et al, 2012).
Immunohistochemical analysis
Immunohistochemical analysis was performed as described previously (Liu et al, 2012).
Statistical analysis
Statistical analysis was performed using SPSS version 13.0. Continuous variables were expressed as a mean with s.e. and analysed using the Student’s t-test (two-tailed). Finally, a Spearman correlation analysis was carried out to determine the correlation between two rank-order variables. A P–value <0.05 was considered to be statistically significant.
Results
HBx mutations occur frequently in HCC
HBV DNA strands were successfully isolated and amplified by nested PCR in 101 (84.16%) cases, from a total of 120 HCC patients. The second round products of the nested PCR for HBx were purified and subjected to gene sequencing analysis. Three types of HBx mutations, including point mutations, distal carboxyl-terminal truncations and deletion mutations, were identified as shown in Supplementary Figure S2. A total of 39 point mutation hotspots were discovered (Supplementary Table S2). The top four were at nucleotides 1630, 1721, 1762 and 1764 and occurred in 51.49%, 48.51%, 53.47% and 50.50% of the 101 cases, respectively. It was particularly interested to find that mutants A1630G and G1721A always appeared concurrently and that mutant A1762T constantly accompanied mutant G1764A. As shown in Supplementary Table S3, a total of 20 mutation hotspots of HBx proteins were identified when the codons were translated into sequences of amino acids. The dual mutations at nucleotides A1762T/G1764A affected the codons of HBx, resulting in a lysine to methionine change at codon 130 and a valine to isoleucine change at codon 131 (K130M/V131I). Of the double mutations A1630G/G1721A, the mutation G1721A does not lead to the alteration of HBx codon, and thus the double mutations affect only the codon 86 of the X protein (H86Y). In addition to these point mutations, distal C-terminal truncations have been discovered in 31.68% (32/101) of HCC cases. Deletion mutations at the C-terminus were found in three (2.97%) of them, with two cases from codon 128 to 135 and one from codon 144 to 150. It is worth noting that C-terminal truncated HBx was always coexistent with the full-length HBx with point mutations, although the truncated HBx accounted for only a smaller portion of them (Supplementary Figure S2). This finding may be attributable to the mixture of both free and integrated HBV genomes in hepatocytes (Chen and Yang, 2011).
Dual point mutations K130M/V131I upregulate HIF-1α
Among various HBx mutations, we firstly analysed several representative point mutation hotspots and created the engineered mutants through site-directed mutagenesis. To this end, six artificial HBx mutants have been established, including HA-A12T-HBx, HA-V44I-HBx, HA-A66T-HBx, HA-H86Y-HBx, HA-E109D-HBx and HA-K130M/V131I-HBx (Figure 1A). Equal amounts of wild-type HBx plasmid and the engineered mutants were transfected into HepG2 cells and confirmed by real-time PCR (Supplementary Figure S3A). To define the role of HBx mutations on the expression of HIF-1α, both total and hydroxyprolin formation of HIF-1α were determined by western blotting with specific antibodies to each of them. It was found that their expression was increased remarkably in cells transfected with both wild-type HBx and mutants. Especially, the mutant HA-K130M/V131I-HBx induced the highest expression of them, compared with wild-type HBx and the other five HBx mutants (Figure 1B). No significant difference was found between the expression of total HIF-1α and hydroxylated HIF-1α. Therefore, these findings indicated that the dual point mutations A1762T/G1764A, which result in mutated K130M/V131I HBx proteins, enhanced the activity of HBx to regulate HIF-1α expression. In accordance with HIF-1α protein expression, there was also a remarked increase in the transcriptional activity of HIF-1α in HepG2 cells transfected with both wild-type HBx and the mutants. Furthermore, the mutant HA-K130M/V131I-HBx has been demonstrated to possess the ability to enhance the transcriptional activity of HIF-1α, compared with wild-type HBx and the other mutants (P<0.05). Compared with the effects of HBx on HIF-1α, a similar tendency of changes in NF-κB, a well defined target of HBx, was also observed (Figure 1C). The nuclear HIF-1α was demonstrated to be activated and the wild-type HBx was shown to induce the nuclear localization of HIF-1α (Yoo et al, 2003; Yoo and Lee, 2004). In line with these reports, we found that the expression of HIF-1α in the nucleus was significant higher in cells transfected with wild-type HBx (Supplementary Figure S4). We further showed that the similar situation occurred in cells with HBx mutants. We next examined the effect of these mutations on themselves localization, which might have an impact on their functions. As shown in Supplementary Figure S5, both wild-type HBx and the mutated HBx displayed a similar, predominately cytosolic localization. This finding suggested that HBx mutations remain in the cytosol in HepG2 cells. Together, these results revealed that HBx point mutants retained the functionality in upregulating the expression and activity of HIF-1α, and that among these HBx point mutants, HA-K130M/V131I-HBx possesses the greatest effect on HIF-1α.
The effect of HBx point mutations on HIF-1 α. (A) Schematic of the full-length HBx and HBx point mutations used in this study. The vertical bars in orange indicated the position of mutations (B) HepG2 cells were transfected with the indicated plasmids. After 24 h transfection, the expression of protein was detected by western blot analysis (WB) with indicated antibodies. (C) HepG2 cells were cotransfected with the indicated plasmids and the luciferase reporter gene constructs pBI-GL (vector), pBI-GL-V6L and pBI-GL-NF-κB. After 24 h transfection, the activity was detected by luciferase assays. The error bars represent s.e. * indicates P<0.05, compared with HA or HA 154 HBx plasmids transfected groups. The experiments were repeated three times with similar results.
Carboxyl-terminal truncations in HBx weaken their ability to stimulate HIF-1α
HBV DNAs from 31.68% (32/101) of HCC cases were found with C-terminally truncated HBx. Meanwhile, HBx mutants with distal 14 amino acids or 35 amino acids deletion were discovered in the present study as well as in previous investigation (Yip et al, 2011), suggesting that these two mutants may exist universally. We thus established two HBx constructs containing these two representative mutations for further experiments (Figure 2A). Equal amounts of wild-type HBx plasmid and these mutants were transfected into HepG2 cells and confirmed by real-time PCR (Supplementary Figure S3B). Further studies showed that C-terminal truncations suppressed the ability of HBx to increase the expression and transcriptional activity of HIF-1α (Figure 2B and C). This finding is supported by an early observation in which mutants with C-terminal truncations abrogated both the antiproliferative and transactivation effects of HBx (Ma et al, 2008). We further found that the length as well as the position of deletion affected the properties of HBx. A deletion of 14 amino acids slightly impaired the function of HBx, whereas a deletion of 35 amino acids remarkably impaired its function. In line with these results, C-terminal truncations also weakened the effect of HBx to promote the nuclear localization of HIF-1α, although these mutations did not affect the cellular localization of themselves (Supplementary Figures S4 and S5). Therefore, the region of amino acid 119 to 140 appeared to be essential for functionality of HBx. The dual point mutations K130M/V131I in this region enhanced the properties of HBx to regulate HIF-1α, whereas the truncations of this region abrogated this effect.
C-terminal truncations inhibit the expression and transcriptional activity of HIF-1 α. (A) Schematic of the full-length HBx and mutants with C-terminal truncations created for this study. (B) HepG2 cells were transfected with the indicated plasmids. After 24 h transfection, the expression of protein was detected by WB with indicated antibodies. (C) HepG2 cells were cotransfected with the indicated plasmids and the luciferase reporter gene constructs pBI-GL (vector), pBI-GL-V6L and pBI-GL-NF-κB. After 24 h transfection, the activity was detected by luciferase assays. The error bars represent s.e. * indicates P<0.05. The experiments were repeated three times with similar results.
Distal C-terminal deletion mutations of HBx are less powerful in the upregulation of HIF-1α
Three (2.97%) of 101 HCC cases were also found with deletion mutations at the C-terminal. Of these three cases of deletion mutations, two of them (No. 6 and No. 10) generated deletions of seven amino acids from codon 128 to codon 135, and one of them (No. 93) created a reading-frame shift, leading to a deletion of the last 11 amino acids from 144. The effect of this deletion is thought to be similar to that of the C-terminal truncations, as they retain the same backbone construction. Therefore, we would like to focus on the mutants with deletion of seven amino acids from codon 128 to codon 135 of HBx. We isolated the natural occurring deletion mutants and cloned them into the vector of pcDNA 3.1. It is difficult to analyse the function of these deletion mutants because they coexist with some point mutations, which may interfere with the outcome. To address this issue, a full-length HBx fragment that contains the same point mutations as the deletion mutants was constructed and used as the control (Figure 3A). Equal amounts of wild-type HBx plasmid and these mutants were transfected into HepG2 cells and confirmed by real-time PCR (Supplementary Figure S3C). Because the deleted HBx constructs were just seven amino acids less than the full-length HBx, it was impossible to detect a visible change in the electrophoretic mobility with SDS-PAGE in western blotting assays (Figure 3B). We found that the mutation with deletions from codon 128 to codon 135 changed the function of HBx by decreasing the expression and the transcriptional activity of HIF-1α (Figure 3B and C). This is a novel finding as the effect of HBx deletion mutations on the regulation of HIF-1α has not been reported. Our results confirm the hypothesis that the regions of amino acid 119 to 140 are crucial for maintaining the function of HBx. Furthermore, we have narrowed down the critical regions of HBx from codon 128 to 135 in terms of their ability to regulate HIF-1α. Interestingly, these critical regions also contained point mutations with high frequency.
C-terminal deletion mutations decreased the expression and the transcriptional activity of HIF-1 α. (A) Schematic of wild-type full-length HBx (wt 154 HBx) or full-length HBx containing point mutations (mt 154 HBx), and the mutants with C-terminal deletions (deleted HBx) created for this study. (B) HepG2 cells were transfected with the indicated plasmids. After 24 h transfection, the expression of protein was detected by western blot analysis (WB) with indicated antibodies. (C) HepG2 cells were cotransfected with the indicated HBx plasmids and the luciferase reporter gene constructs, pBI-GL (vector), pBI-GL-V6L and pBI-GL-NF-κB. After 24 h transfection, the activity was detected by luciferase assays. The error bars represent s.e. * indicates P<0.05. The experiments were repeated three times with similar results.
The C-terminal is essential for the stable and function of HBx
We noticed that the protein levels of HBx with C-terminal truncations or deletion mutations were obviously less than the levels of full-length HBx, although the same amount of HBx plasmids was transfected, as determined in the real-time PCR assays (Supplementary Figure S3B). These findings may indicate that the C-terminus is essential for the stability of HBx. To further confirm this hypothesis, the half-life of HBx and its mutants were measured as described in the Materials and Methods. In brief, 24 h post transfection, cycloheximide was added to inhibit the protein synthesis and then the HBx protein was monitored by western blotting at an array of time points (Figure 4A). As our expectation, the half-life of HBx mutants with C-terminal truncation was significantly shorter than that of wild-type HBx. As shown in Figure 4B, the half-life of wild-type HBx was about 105 min, whereas that of HBx mutants HA 1–140 HBx and HA 1–119 HBx were only approximately 50 min and 35 min, respectively. Therefore, distal C-terminal truncations can impair the stability of HBx, and this finding may, at least partly, account for the reduced ability of the distal C-terminal truncation HBx in the upregulation of the expression and transcriptional activity of HIF-1α. Whether the C-terminal truncations could also impair the transactivational activity of HBx was another important issue to be addressed. Due to the short half-life of C-terminally truncated HBx, we thus transfected an array of amounts of these plasmids to make sure the protein levels of HBx mutants reached that of wild-type HBx. As shown in Figure 4C and D, despite the same, even more, protein levels of HBx mutants, the transcriptional activity of HIF-1α in these cells was less than that of wild-type HBx transfected. Therefore, in addition to the impairment of the stability, C-terminal truncations can also abrogate the HBx transactivational activity in the regulation of HIF-1α.
C-terminal truncations lead to the decreased half-life and transactivation activity of HBx. (A) HepG2 cells in six-wells plates at 80% confluenc were transfected with 0.5 μg HA 154 HBx, 1 μg HA 1–140 HBx and 3 μg HA 1–119 HBx constructs. After 24 h transfection, cycloheximide (CHX, 30 μg ml−1) was added to the medium to inhibit protein synthesis, and then the HBx protein was monitored by western blotting with an anti-HA antibody at the indicated time points. (B) The schematic illustration shows the densitometric quantification of the results in which the expression of HA 154 HBx, HA 1–140 HBx, or HA 1–119 HBx at time zero was set as 100%. The expression levels of these mutants at subsequent time points are expressed as a percentage of the levels of each relative to their expression at time zero. The error bars represent s.e. (C) HepG2 cells were cotransfected with the luciferase reporter gene constructs, pBI-GL-V6L, and an array of amounts of the indicated HBx plasmids. After 24 h transfection, the activity was detected by luciferase assays. The error bars represent s.e. (D) The protein levels of HBx of these samples were detected via western blotting analysis with an anti-HA antibody. The experiments were repeated three times with similar results.
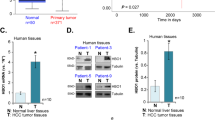
Expression of HBx protein is positively related to HIF-1α in HCC tissues
As described above, most of HBx point mutants retained the ability to upregulate HIF-1α, and especially the dual mutations K130M/V131I enhanced this function. C-terminal truncations and deletion mutations, however, impaired the stability and transactivation activation of HBx. As a result, the effect of HBx mutations on HIF-1α in a given case could be complicated because several different point mutations and/or C-terminal truncations often coexist in the same case. The overall effects of HBx mutations should be dependent on the ratio of these mutations, as schematised in Figure 5A. To confirm this inference, we determined the correlation between HBx protein and HIF-1α expression in HCC tissues, in which HBx mutants occurred with the mixture of these mutations. Immunohistochemical analysis was employed to determine the expression of these two proteins. The monoclonal anti-HBx antibody that was able to recognise the full-length HBx was selected so that the full-length, truncated and deleted HBx could be detected (Yip et al, 2011). As shown in Figure 5B, the levels of HBx proteins were positively correlated with HIF-1α protein (P<0.05). This finding suggests that despite the fact that different types of HBx mutations may possess differentiated effects toward HIF-1α, the overall impact of HBx mutants appear to enhance HIF-1α. As the HIF-1α protein level is an indicator of hypoxia, which promotes the progression of HCC (Harris, 2002; Yoo et al, 2008; Liu et al, 2010), we wondered whether HIF-1α expression was associated with patients’ prognosis. As shown in Figure 6, the expression of HIF-1α protein was negatively correlated with the disease-free survival rate and cumulative survival rate in this study. Taken together, our results suggest that there is a positive correlation between HBx mutants and HIF-1α and that an increased HIF-1α level signals a poor outcome for patients.
Expression of HBx protein is positively related to HIF-1 α in HCC tissues. (A) Schematic illustration shows the different effects of HBx mutations on the regulation of HIF-1α. Although most of point mutations in HBx did not affect their ability to regulate HIF-1α, dual mutations K130M/V131I enhanced HBx ability to upregulate HIF-1α, whereas C-terminal truncations and deletion mutations reduced their ability to stimulate HIF-1α. The overall effects of HBx mutants on HIF-1α depend on the balance of these mutations. (B) The protein levels of HBx and HIF-1α were detected by immunohistochemical analysis (upper panel), and the relationship between them was determined by Spearman’s rank correlation analysis (lower panel). The experiments were repeated three times with similar results.
The levels of HIF-1 α protein in HCC tissues are correlated with poor outcomes of HCC patients. The protein levels of HIF-1α were detected by ELISA and the Kaplan–Meier survival analysis was carried out to determine the overall survival rate and disease-free survival rate. The difference between the HIF-1α <250 ng ml−1 group and the HIF-1α >250 ng ml−1 group was significant in both the disease-free survival rate and cumulative survival rate.
Discussion
Wild-type HBx has been demonstrated to upregulate the expression and transcriptional activity of HIF-1α (Yoo et al, 2003), whereas the effects of HBx mutants on HIF-1α have not been fully elucidated. In the present studies, we thus aimed to uncover the potential relationship between HBx mutations and the activation of HIF-1α. Here, we revealed that the dual mutations K130M/V131I can enhance the function of HBx in upregulating HIF-1α. It is worth noting that these dual mutations account for approximate 50% of HCC cases in our studies and even higher incidence rate reported by others (Asim et al, 2010). Dual mutations K130M/V131I have also been demonstrated to associate with an increased risk of HCC independent for viral loading and other risk factor (Yang et al, 2008). Our results suggest that the enhanced function of HBx with these dual mutations in upregulating HIF-1α may, at least partly, contribute to the mechanism that accounts for the risk of HCC development.
The other point mutations, however, were not been shown to enhance the effect of HBx on HIF-1α. The underlying mechanisms accounted for the difference of the function between dual mutations K130M/V131I and other mutations may be dependent on their secondary structures and the target molecules. HBx appears to be an unstructured protein that can gain a secondary structure under specific conditions, and can be folded and acquire a specific function through its interaction with target proteins (Rui et al, 2005). This flexibility may account for the large array of HBx activity. With the dual mutations K130M/V131I, HBx would gain more potent secondary structures to interact with HIF-1α, and thus increase the expression and the transcriptional activity of HIF-1α. Indeed, HBx mutants with dual mutations K130M/V131I have been shown to possess the increased capability of binding to p53 compared with the wild-type HBx (Iyer and Groopman, 2011). Whether the dual mutations K130M/V131I exert more potent activity in the upregulation of HIF-1α than other mutations is due to its greater ability to bind to HIF-1α would be an important issue for further investigations. We have also noticed that no significant difference was found between the expression of total and hydroxylated HIF-1α induced by HBx. PHD2 promotes the formation of hydroxylated HIF-1α, which allows for recognition by pVHL and thus targets HIF-1α for proteasomal degradation (Epstein et al, 2001; Ivan et al, 2001). These results suggested that HBx might not inhibit the function of PHD2 in forming hydroxylated HIF-1α but suppressed the proteasomal degradation of hydroxylated HIF-1α through binding to them.
Carboxyl-terminal has been demonstrated to be essential for HBx to enhance the replication of HBV (Lucifora et al, 2011), and C-terminal truncations of HBx decrease the transcription activation of NF-κB as well as the HBV replication (Lizzano et al, 2011). In accordance with these findings, we demonstrated here that C-terminal truncation of 14 and 35 amino acids impaired the function of HBx in upregulating the expression and the transcriptional activity of HIF-1α. This finding was also supported by the report that HBx mutants with C-terminal truncations abrogated both the antiproliferative and transactivation effects of HBx (Ma et al, 2008). The size of the truncation at the C-terminus of HBx appeared to affect the HBx function, as the truncation of 14 amino acids (HA 1–140 HBx) only slightly inhibited the function of HBx, whereas the truncation of 35 amino acids (HA 1–119 HBx) suppressed the function of HBx tremendously. Therefore, the region of amino acids 119–140 is supposed to be essential for the function of HBx. This inference was further confirmed by the findings that HBx mutants with deletions of amino acids 128–135 also significantly abrogated the function of HBx in upregulating HIF-1α. Surprisingly, we have also noticed that the expression of HBx mutants with C-terminal truncations or deletion mutations was significantly less than that of wild-type HBx, although the same amount of plasmids were used. Further studies demonstrated that C-terminal truncation of 14 and 35 amino acids decreased the half-life of HBx from 105 min to 50 min and 35 min, respectively. This tendency was consistent with previous report showing that the half-life of 1–131 HBx and 1–140 HBx was less than the wild-type HBx (Lizzano et al, 2011). The decreased stability of the truncated HBx mutants suggested that we had to overcome decreased protein levels of HBx mutants to test the functionality of the C-terminal truncated HBx mutants. This was accomplished by increasing the amounts of transfected C-terminal truncation plasmids relative to the wild-type HBx plasmid. Although the truncated HBx mutants were expressed at levels that were similar to or greater than the level of wild-type HBx, neither HA 1–140 HBx nor HA 1–119 HBx could stimulate HIF-1α transcriptional activity to the level achieved by wild-type HBx. Therefore, the C-terminus, especially the region of amino acids 119–140, appears to be essential for the stability and the transactivation of HBx. Truncations or deletions of C-terminus may disrupt the structure of HBx, leading to degradation and impairment of their capability in interacting with target proteins.
Altogether, most of HBx point mutants retained the ability to upregulate HIF-1α, and especially the dual mutations K130M/V131I enhanced this function. C-terminal truncations and deletion mutations, however, impaired the stability and transactivation activation of HBx. It is noted that the HBx mutants with point mutations and the mutants with C-terminal truncation or deletion frequently coexist in the same HCC case. Therefore, the overall effect of HBx mutants in HCC should depend on the ratio of these mutants. In line with this inference, the level of total HBx protein is positively correlated with the expression of HIF-1α in the HCC tissue samples that contain various types of HBx mutants, and thus the effect of naturally occurring HBx mutants may dominantly upregulate HIF-1α. Indeed, it has been noted that in HBV-positive cells only a small amount of HBx is found, whereas transfected cells would gain strong overexpression of HBx protein, although a small amount of plasmids was used in our experiments. Further investigations should focus on the establishment of HBx expression systems that can be used to mimic the natural HBV infection in HCC cells.
In conclusion, the carboxyl-terminus, especially amino acids 119–140, is essential for the stability and transactivation of HBx. The dual point mutations K130M/V131I in this region result in enhanced functionality of HBx, whereas mutations of C-terminal truncation or deletion may weaken the function of HBx in upregulating HIF-1α. However, the overall function of HBx in a given case appears to enhance HIF-1α, as the protein levels of HBx are positively correlated with the expression of HIF-1α in HCC tissues containing various types of HBx mutants.
Change history
18 February 2014
This paper was modified 12 months after initial publication to switch to Creative Commons licence terms, as noted at publication
References
Asim M, Malik A, Sarma MP, Polipalli SK, Begum N, Ahmad I, Khan LA, Husain SA, Akhtar N, Husain S, Thayumanavan L, Singla R, Kar P (2010) Hepatitis B virus BCP, Precore/core, X gene mutations/genotypes and the risk of hepatocellular carcinoma in India. J Med Virol 82: 1115–1125.
Chen CJ, Yang HI (2011) Natural history of chronic hepatitis B REVEALed. J Gastroenterol Hepatol 26: 628–638.
Chen CJ, Yang HI, Su J, Jen CL, You SL, Lu SN, Huang GT, Iloeje UH (2006) Risk of hepatocellular carcinoma across a biological gradient of serum hepatitis B virus DNA level. JAMA 295: 65–73.
Epstein AC, Gleadle JM, McNeill LA, Hewitson KS, O’Rourke J, Mole DR, Mukherji M, Metzen E, Wilson MI, Dhanda A, Tian YM, Masson N, Hamilton DL, Jaakkola P, Barstead R, Hodgkin J, Maxwell PH, Pugh CW, Schofield CJ, Ratcliffe PJ (2001) C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell 107: 43–54.
Harris AL (2002) Hypoxia-a key regulatory factor in tumour growth. Nat Rev 2: 38–47.
Ivan M, Kondo K, Yang H, Kim W, Valiando J, Ohh M, Salic A, Asara JM, Lane WS, Kaelin WG Jr (2001) HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science 292: 464–468.
Iyer S, Groopman JD (2011) Interaction of mutant hepatitis B X protein with p53 tumor suppressor protein affects both transcription and cell survival. Mol Carcinog 50: 972–980.
Kumar V, Jayasuryan N, Kumar R (1996) A truncated mutant (residues 58–140) of the hepatitis B virus X protein retains transactivation function. Proc Natl Acad Sci USA 93: 5647–5652.
Liu LP, Ho RL, Chen GG, Lai P (2012) Sorafenib inhibits hypoxia-inducible factor-1alpha synthesis: implications for anti-angiogenic activity in hepatocellular carcinoma. Clin Cancer Res 18: 5662–5671.
Liu LP, Liang HF, Chen XP, Zhang WG, Yang SL, Xu T, Ren L (2010) The role of NF-kappaB in Hepatitis b virus X protein-mediated upregulation of VEGF and MMPs. Cancer Invest 28: 443–451.
Lizzano RA, Yang B, Clippinger AJ, Bouchard MJ (2011) The C-terminal region of the hepatitis B virus X protein is essential for its stability and function. Virus Res 155: 231–239.
Lucifora J, Arzberger S, Durantel D, Belloni L, Strubin M, Levrero M, Zoulim F, Hantz O, Protzer U (2011) Hepatitis B virus X protein is essential to initiate and maintain virus replication after infection. J Hepatol 55: 996–1003.
Ma NF, Lau SH, Hu L, Xie D, Wu J, Yang J, Wang Y, Wu MC, Fung J, Bai X, Tzang CH, Fu L, Yang M, Su YA, Guan XY (2008) COOH-terminal truncated HBV X protein plays key role in hepatocarcinogenesis. Clin Cancer Res 14: 5061–5068.
Neuveut C, Wei Y, Buendia MA (2010) Mechanisms of HBV-related hepatocarcinogenesis. J Hepatol 52: 594–604.
Ng SA, Lee C (2011) Hepatitis B virus X gene and hepatocarcinogenesis. J Gastroenterol 46: 974–990.
Pouyssegur J, Dayan F, Mazure NM (2006) Hypoxia signalling in cancer and approaches to enforce tumour regression. Nature 441: 437–443.
Rui E, Moura PR, Goncalves Kde A, Kobarg J (2005) Expression and spectroscopic analysis of a mutant hepatitis B virus onco-protein HBx without cysteine residues. J Virol Methods 126: 65–74.
Semenza GL (2003) Targeting HIF-1 for cancer therapy. Nat Rev 3: 721–732.
Siegel R, Naishadham D, Jemal A (2012) Cancer statistics, 2012. CA Cancer J Clin 62: 10–29.
Yang HI, Yeh SH, Chen PJ, Iloeje UH, Jen CL, Su J, Wang LY, Lu SN, You SL, Chen DS, Liaw YF, Chen CJ (2008) Associations between hepatitis B virus genotype and mutants and the risk of hepatocellular carcinoma. J Natl Cancer Inst 100: 1134–1143.
Yip WK, Cheng AS, Zhu R, Lung RW, Tsang DP, Lau SS, Chen Y, Sung JG, Lai PB, Ng EK, Yu J, Wong N, To KF, Wong VW, Sung JJ, Chan HL (2011) Carboxyl-terminal truncated HBx regulates a distinct microRNA transcription program in hepatocellular carcinoma development. PLoS One 6: e22888.
Yoo YG, Lee MO (2004) Hepatitis B virus X protein induces expression of Fas ligand gene through enhancing transcriptional activity of early growth response factor. J Biol Chem 279: 36242–36249.
Yoo YG, Na TY, Seo HW, Seong JK, Park CK, Shin YK, Lee MO (2008) Hepatitis B virus X protein induces the expression of MTA1 and HDAC1, which enhances hypoxia signaling in hepatocellular carcinoma cells. Oncogene 27: 3405–3413.
Yoo YG, Oh SH, Park ES, Cho H, Lee N, Park H, Kim DK, Yu DY, Seong JK, Lee MO (2003) Hepatitis B virus X protein enhances transcriptional activity of hypoxia-inducible factor-1alpha through activation of mitogen-activated protein kinase pathway. J Biol Chem 278: 39076–39084.
Acknowledgements
We thank Drs L Neckers and JS Isaacs (Center for Cancer Research, National Cancer Institute, Rockville, MD) for providing the HA-HIF-1α construct and Dr MJ Bouchard (Department of Biochemistry and Molecular Biology, Drexel University College of Medicine, Philadelphia, PA, USA) for providing the FLAG-HBx construct. We also gratefully acknowledge the excellent technical assistance provided by SY Chun, and ECW Chak. This study was supported by a direct grant (2008.1.099) from the Chinese University of Hong Kong. The contents are solely the responsibility of the authors and do not necessarily represent the official view of CUHK.
Disclaimer
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
Supplementary Information accompanies this paper on British Journal of Cancer website
Supplementary information
Rights and permissions
From twelve months after its original publication, this work is licensed under the Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Liu, Lp., Hu, Bg., Ye, C. et al. HBx mutants differentially affect the activation of hypoxia-inducible factor-1α in hepatocellular carcinoma. Br J Cancer 110, 1066–1073 (2014). https://doi.org/10.1038/bjc.2013.787
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/bjc.2013.787
Keywords
This article is cited by
-
Hypoxic Hepatocellular Carcinoma Cells Acquire Arsenic Trioxide Resistance by Upregulating HIF-1α Expression
Digestive Diseases and Sciences (2022)
-
Oxygen: viral friend or foe?
Virology Journal (2020)
-
Hypoxic gene expression in chronic hepatitis B virus infected patients is not observed in state-of-the-art in vitro and mouse infection models
Scientific Reports (2020)
-
Pro-oncogenic, intra host viral quasispecies in Diffuse large B cell lymphoma patients with occult Hepatitis B Virus infection
Scientific Reports (2019)
-
USP16 Downregulation by Carboxyl-terminal Truncated HBx Promotes the Growth of Hepatocellular Carcinoma Cells
Scientific Reports (2016)