Abstract
Background:
Human papillomavirus (HPV) testing in oropharyngeal squamous cell carcinoma (OPSCC) is now advocated. Demonstration of transcriptionally active high-risk HPV (HR-HPV) in fresh tumour tissue is considered to be the analytical ‘gold standard’. Clinical testing has focused on formalin-fixed paraffin-embedded (FFPE) tissue at the expense of sensitivity and specificity. Recently, a novel RNA in situ hybridisation test (RNAscope) has been developed for the detection of HR-HPV in FFPE tissue; however, validation against the ‘gold standard’ has not been reported.
Methods:
A tissue microarray comprising FFPE cores from 79 OPSCC was tested using HR-HPV RNAscope. Analytical accuracy and prognostic capacity were established by comparison with the reference test; qRT–PCR for HR-HPV on matched fresh-frozen samples.
Results:
High-risk HPV RNAscope had a sensitivity and specificity of 97 and 93%, respectively, against the reference test. Kaplan–Meier estimates of disease-specific survival (DSS, P=0.001) and overall survival (OS, P<0.001) by RNAscope were similar to the reference test (DSS, P=0.003, OS, P<0.001) and at least, not inferior to p16 immunohistochemistry +/− HR-HPV DNA-based tests.
Conclusion:
HR-HPV RNAscope demonstrates excellent analytical and prognostic performance against the ‘gold standard’. These data suggest that the test could be developed to provide the ‘clinical standard’ for assigning a diagnosis of HPV-related OPSCC.
Similar content being viewed by others
Main
The last three decades have seen a fundamental change in the profile of oropharyngeal squamous cell carcinoma (OPSCC) within the developed world (Chaturvedi et al, 2011). The incidence of OPSCC attributable to tobacco and alcohol exposure has been gradually declining while human papillomavirus (HPV)-related OPSCC has seen a rapid increase (D’Souza et al, 2007; Sturgis and Cinciripini, 2007; Nasman et al, 2009; Schache et al, 2011). It is now clear that HPV-positive OPSCC is a biologically distinct entity (Gillison et al, 2000; Adelstein et al, 2009). Detection of high-risk HPV (HR-HPV) has profound prognostic significance as it correlates with both a disease-specific and an overall survival (OS) advantage (Fakhry et al, 2008; Lassen et al, 2009). The improved survival of these patients has prompted the formulation of clinical trials in both North America (RTOG 1016) and Europe (DeESCALaTE-HPV) to test the feasibility of de-escalation of therapy to reduce acute and late toxicity. Despite a consensus within the head and neck oncology community that acknowledges the importance of HPV testing (Adelstein et al, 2009; Nasman et al, 2009; Marur et al, 2010; Mehanna et al, 2012), a validated reference test to establish a diagnosis of HPV-related OPSCC in clinical practice has yet to be established (Braakhuis et al, 2009; Robinson et al, 2010).
As both the initiation and maintenance of an HPV-driven carcinoma requires persistent viral oncogene expression (Marur et al, 2010), the detection of E6/E7 mRNA transcripts by quantitative reverse transcriptase PCR (qRT–PCR) has been proposed as the most appropriate ‘gold standard’ or reference test (Smeets et al, 2007). As a consequence of RNA instability, this testing has relied on both the analysis of fresh-frozen tissue and specialist research laboratory techniques, thus limiting its translation to routine clinical diagnostics (Smeets et al, 2007). For clinical utility, HPV testing strategies have necessarily focused on formalin-fixed paraffin-embedded (FFPE) tissue, but this has been at the expense of reduced sensitivity and specificity for oncogenic HPV (Smeets et al, 2007; Schache et al, 2011). Diagnostic algorithms that combine different HPV tests have been proposed as a strategy to compensate for the known limitations of individual tests (Westra, 2009). However, the inclusion of multiple analytical stages to achieve an accurate and reliable HPV status is technically cumbersome, may produce discordant results across the different tests employed and inevitably increases costs.
Recent evidence suggests that a novel RNA-based chromogenic in situ hybridisation (ISH) technique (RNAscope, Advanced Cell Diagnostics Inc., Hayward, CA, USA) is capable of reliably detecting transcriptionally active genes, including HR-HPV E6/E7 oncogenes, in FFPE tissue samples (Wang et al, 2012). The technique has shown promising results by comparison with other FFPE-based HPV diagnostic tests in head and neck cancer (Ukpo et al, 2011; Bishop et al, 2012; Lewis et al, 2012) and also informs prognosis in OPSCC (Ukpo et al, 2011); however, for this test to be considered for broader clinical usage its efficacy should be measured against an analytical ‘gold standard’. Using a cohort of well-characterised OPSCC, comprising matched fresh-frozen and FFPE tumour samples, we sought to validate HR-HPV RNAscope against the reference test for oncogenic HPV; qRT–PCR for HPV-16, -18 and -33 E6/E7 transcripts.
Materials and methods
Case selection
Biopsy or excision material was available for 79 cases originating from a cohort of stringently classified OPSCC cases (Schache et al, 2011). All samples had been sourced in compliance with previously granted ethical approval (South Sefton Research Ethics Committee; EC.47.01-6 & North West 5 Research Ethics Committee; EC.09.H1010.5) from individuals treated in the Liverpool Head and Neck Oncology Service, a multidisciplinary unit serving a geographically stable population of approximately two million individuals within Merseyside and Cheshire, UK. All cases had been treated using primary surgery and where required, adjuvant radiotherapy or chemoradiotherapy.
Samples corresponding to each case had previously been tested using a portfolio of HPV tests (HPV-16, -18, -33 qRT–PCR and DNA qPCR performed on fresh tissue: p16 immunohistochemistry (IHC) and HR-HPV DNA ISH performed on FFPE tissue, capable of detecting HR-HPV types -16, -18, -31, -33, -35, -39, -45, -51, -52, -56, -58 and -66), and were associated with clinically validated demographic and outcome data (Schache et al, 2011).
Briefly, tissue microarrays (TMAs) consisting of triplicate tumour cores from corresponding FFPE tumour donor blocks were constructed. Both p16 IHC and HR-HPV DNA ISH analysis was conducted using proprietary kits (CINtec Histology, mtm laboratories AG, Heidelberg, Germany; Inform HPV III Family 16 Probe B, Ventana Medical Systems Inc., Tucson, AZ, USA) on a Ventana Benchmark Autostainer (Ventana Medical Systems Inc.).
Duplicated real-time DNA and RNA (cDNA) qPCR reactions were conducted using fresh tumour tissue-derived nucleic acid samples on an Applied Biosystems 7500 FAST system (Foster City, CA, USA). Further assay details including primer/probe sequences, PCR conditions and endogenous references have been published previously (Schache et al, 2011).
RNA ISH for HR-HPV
Detection of HR-HPV E6/E7 mRNA in TMA cores was performed using the HR-HPV RNAscope kit (Advanced Cell Diagnostics Inc.) as previously described (Ukpo et al, 2011) and in accordance with the manufacturers instructions. Briefly, 4 μm TMA sections were deparaffinised and pretreated with heat and protease before hybridisation with target-specific probes for the E6 and E7 genes of seven HR-HPV genotypes (HPV-16, -18, -31, -33, -35, -52 and -58). Ubiquitin C (UBC, a constitutively expressed endogenous gene) and the bacterial gene, dapB, were used as positive and negative controls, respectively.
Whole-tissue sections for selected cases (see below) were stained for HR-HPV RNA, UBC and dapB by a fully automated RNAscope assay (RNAscopeVS) using the Ventana Discovery XT slide autostaining system (Ventana Medical Systems Inc.).
Interpretation of tests
Test assessment was conducted independently by two pathologists (M.R. and P.S.) for all tissue-based analyses.
Scoring of p16 IHC status was assessed using the widely used threshold of strong and diffuse nuclear and cytoplasmic staining in ⩾70% of the tumour (Singhi and Westra, 2010) and the recently proposed and validated H score for p16 IHC, with an H score of >60 defined as p16 positive (Jordan et al, 2012).
High-risk HPV DNA in situ hybridisation was scored using a binary classification (positive vs negative) to reflect any detectable chromogen in any of the malignant cells, as described previously (Schache et al, 2011).
For RNAscope, the UBC test was used to assess the presence of hybridisable RNA and was defined as adequate if there was strong staining in the majority of cells in the section. The dapB test was used to assess nonspecific staining; only those cases that were negative or weakly stained were considered for HPV scoring. A positive HPV test result was defined as punctate staining that co-localised to the cytoplasm and/or nucleus of any of the malignant cells and, where staining was present in the control, was at least twice as strong as the dapB test.
The results were collated by the study coordinator (A.G.S) and discordant scores were re-examined at a meeting between the pathologists to establish a consensus interpretation. In order to quality assure the results, cases that had discordant scores between the pathologists and/or variable scores between cores from the same tumour were additionally subjected to analysis of whole-tumour sections as described above.
Statistical analysis
The χ2 and Kruskal–Wallis tests were used for comparison of demographic and tumour-specific features between HPV-positive and -negative groups as defined by the reference test; HPV-16, -18, -33 qRT–PCR. Only tumours displaying viral oncogene expression in duplicate runs of qRT–PCR were deemed as harbouring oncogenic HPV. Analytical sensitivity and specificity, measured against the reference test, was calculated for HR-HPV RNAscope and for other single and combined diagnostic tests (HPV-16 DNA qPCR, p16 IHC and HR-HPV DNA ISH). Positive and negative predictive values were similarly generated for each test. Median follow-up was estimated using the Kaplan–Meir potential follow-up method (Schemper and Smith, 1996). Kaplan–Meier estimates of survival were generated for single tests (high-risk qRT–PCR, HR-HPV RNAscope, HPV-16 DNA qPCR, HR-HPV DNA ISH, p16 IHC;) and combined analysis tests (p16 IHC/HR-HPV qRT–PCR, p16 IHC/HPV-16 DNA qPCR, p16 IHC/HR-HPV DNA ISH). The log-rank (Mantel–Cox) test was used for comparison between survival curves according to each of the diagnostic methods. Disease-specific survival (DSS) was defined as death from or owing to OPSCC, and OS was defined as death from any cause. Both DSS and OS were calculated at 36 months follow-up beyond the date of initial diagnosis.
To ensure that the ability to amplify target sequences for the defined reference test (HR-HPV qRT–PCR) was not adversely affected by duration of tissue storage, RNA quality was assessed by a Kruskal–Wallis test of the ΔCT of endogenous reference gene (β-actin).
Results
Tissue sample quality and consistency
Interpretable results were available for all 79 cases identified; however, one case had insufficient staining for UBC and was excluded from further analysis, leaving 78 of 79 (99%) cases for HPV analysis. Seventeen cases (22%) had discordant scores following TMA analysis, due either to inter-observer variation or inter-core variation, and were subject to further testing and independent scoring using whole FFPE sections. A resultant Kappa scope of 0.948 (95% CI 0.88–1.0) for inter-observer analysis of scoring was evident following complete analysis. The tumour cell proportion within fresh-frozen tissue samples was estimated to be >50% for all cases and >80% for two-thirds of the cohort. There was no evidence of statistically significant variation in reference gene detection (cycle to threshold for β-actin) over time suggesting sample storage had no effect on detection of viral oncogene expression.
Cohort characteristics
The entire cohort had a median follow-up of 27 months (95% CI 27–37). The characteristics of the OPSCC cohort as a whole and subdivided by HPV status, defined by HR-HPV qRT–PCR, are shown in Table 1. The age of patient at diagnosis conformed to a normal distribution as signified by a one-sample Kolmogorov–Smirnov test (P=0.999). Individuals within the HPV-positive group were statistically significantly younger than those in the HPV-negative group (mean 54.2 vs 61.3 years of age at diagnosis, P=0.003). Of the 69 cases for which reliable risk factor data were available, those individuals who were either non-smokers or who had smoked <20 pack-years were statistically more likely to have HPV-positive OPSCC (P=0.004). Similarly, there was a trend towards lower alcohol exposure in the HPV-positive group. There were no statistical differences between the groups by sex, tumour subsite or nodal category.
Test analysis
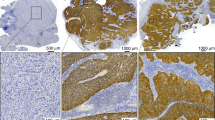
Photomicrographs of cases classified as HPV positive by HR-HPV RNAscope are shown in Figure 1. The HR-HPV RNAscope test had a sensitivity of 97% and a specificity of 93% against the reference test, with positive and negative predictive values of 91 and 98%, respectively (Table 2). Sensitivity values for other HPV tests when used as single tests were comparable; p16 IHC 97%, HR-HPV ISH 94% and to a lesser extent HPV-16 DNA qPCR 91%; however, lower levels of specificity for oncogenic HPV were apparent for two of these tests; p16 IHC 82%, HPV-16 DNA qPCR 87%. Interpretation of more than one test per sample, in a diagnostic algorithm, appeared to improve specificity, but at the expense of sensitivity, exemplified by combined p16 IHC/HPV-16 DNA qPCR; sensitivity 91% and specificity 93%.
Photomicrographs of OPSCC stained using RNAscope with probes for high-risk HPV, dapB (negative control) and UBC (positive control). The cases demonstrate a range of positive staining patterns for high-risk HPV. Cases 109 and 97 showed strong and moderate staining, respectively, and contained HPV-16 E6/E7 mRNA by qRT–PCR. Case 87 showed strong staining and contained HPV-18 E6 mRNA by qRT–PCR. Case 95 showed weak staining and was negative for HPV-16 E6/E7 mRNA, HPV-18 E6 mRNA and HPV-33 E6 mRNA by qRT–PCR (false-positive result). Scale bars are equivalent to 200 μm for cases 109 and 87, and 50 μm for cases 97 and 95.
A comparison of the two p16 IHC scoring techniques revealed no difference in p16 status (positive or negative), either at the level of individual TMA cores or specific tumour cases.
The Kaplan–Meier survival estimates illustrated in Figures 2 and 3 and detailed in Tables 3 and 4 show the prognostic capacity of all HPV tests. High-risk HPV RNAscope displayed an encouraging capacity to discriminate survival, both in terms of OS (P<0.001) and DSS (P=0.001), and this was comparable to the reference test (OS P<0.001, DSS P=0.003).
False-positive and false-negative reporting
High-risk HPV RNAscope conferred positive results for three cases (4%) where there was an absence of detectable HPV mRNA by qRT–PCR. Corresponding test results for these cases indicated that they were also positive by p16 IHC, HR-HPV DNA ISH, HPV-16 DNA qPCR and consequently combinations of these tests.
High-risk HPV RNAscope classified as negative, one case (1%) that displayed evidence of HPV-16 transcripts by qRT–PCR. The case was also classified as negative by p16 IHC, HR-HPV DNA ISH, HPV-16 DNA qPCR and consequently combinations of these tests.
Discussion
HPV analysis of OPSCC in clinical practice is becoming a fundamental requirement to provide both adequate prognostic information for patients and to facilitate entry into appropriately stratified clinical trials, including those investigating the potential to de-escalate the intensity of curative therapies. Paradoxically, there is no ‘international standard’ for defining HPV-related OPSCC in clinical practice and an adequately validated diagnostic standard for FFPE tissue has yet to be defined (Braakhuis et al, 2009). To demonstrate the efficacy of any test, it must be appraised against a ‘reference’ or ‘gold standard’ test. In the context of HPV-driven malignancy, it is the expression of the viral oncogenes (E6 and E7) that is the prerequisite for carcinogenesis and this has been hypothesised as the most appropriate analytical standard (Wiest et al, 2002; Smeets et al, 2007). While acknowledging that HPV oncogene expression is only part of a complex process of altered molecular pathways in viral-driven cancer, it is against quantitative detection of transcriptionally active virus that we sought to measure a novel HPV test, HR-HPV RNAscope.
High-risk HPV RNAscope has previously shown promising capability when compared with other HPV diagnostic tests (Ukpo et al, 2011; Bishop et al, 2012) yet validation against an analytical standard had not been possible to date. The Mersey Head and Neck Oncology Research Group tissue collection benefits from a large series of matched fresh-frozen and FFPE tumour samples. The cohort was treated with primary surgical excision producing an abundant legacy of reserve tissue for biomarker studies. By contrast, other cancer centres treating OPSCC by primary chemoradiotherapy are usually restricted to analysis of a limited resource of small FFPE diagnostic biopsies.
Previous evaluations of clinical outcomes in OPSCC based on HPV status have shown that individuals with HPV-positive malignancy have appreciably improved outcomes by comparison with their HPV-negative counterparts (Licitra et al, 2006; Lindquist et al, 2007; Fakhry et al, 2008; Lassen et al, 2009; Ang et al, 2010). High-risk HPV RNAscope replicated these findings and demonstrated a similar capacity to predict outcomes by comparison with the ‘gold standard’, as have other clinically applicable tests (Shaw and Robinson, 2011).
It is, however, the high sensitivity (97%) and specificity (93%) of HR-HPV RNAscope that offers considerable potential as a diagnostic test for HPV-related OPSCC, particularly as these results are achieved with a single test format. The incorporation of control tests (UBC and dapB) on parallel sections enhances the quality control of test interpretation. The only other single test to have demonstrated comparable sensitivity in previous comparison with viral oncogene expression is p16 IHC (94–100%) (Smeets et al, 2007; Schache et al, 2011). The level of specificity (79–82%) demonstrated by p16 under the same conditions, however, is somewhat lower, due mostly to alternative, and as yet unexplained, elevations of p16 expression in HPV-negative malignancy (Bishop et al, 2012; Harris et al, 2011). It is for this reason that algorithms combining both p16 IHC- and HPV-specific tests have been advocated (Westra, 2009). Investigation of the potential differences between p16 IHC scoring techniques was undertaken by application of both the currently applied standard for p16 IHC analysis (Singhi and Westra, 2010), strong and diffuse nuclear and cytoplasmic staining in >70% of the tumour, and the recently described H score analysis (Jordan et al, 2012), derived from the cross-product of staining intensity and proportion of tumour. Interestingly, both of the p16 IHC scoring techniques gave identical results and therefore had no apparent bearing on either the sensitivity/specificity of p16 IHC or its prognostic capacity.
High-risk HPV RNAscope classified three cases as HPV positive in the absence of detectable HPV mRNA. It is conceivable that the samples might harbour other HR-HPV genotypes not included in the reference test, which was restricted to the analysis of the three most common HPV genotypes isolated from OPSCC (HPV-16, -18 and -33). It is interesting to note that one of the three cases had high levels of HPV-16 E2 expression detected by qRT–PCR (unpublished data). E2 is a known transcriptional repressor of E6 and E7 genes, and its influence may have been sufficient to reduce E6 and E7 transcript levels below the detection threshold of qRT–PCR while remaining within the detection range of HR-HPV RNAscope. Alternatively, the mismatch between the qRT–PCR result and the tests on FFPE raises the possibility of methodological flaws, despite the use of stringent experimental design and detection protocols to quality assure test results.
The solitary case reported as negative by HR-HPV RNAscope demonstrated an expression level for both E6 and E7 that was low by comparison with other samples; however, it was not the lowest and remained within the threshold for detection set before analysis of the samples. Interestingly, this case was similarly ‘misclassified’ by both p16 IHC and HR-HPV DNA ISH. It is possible that fixation and processing parameters may have resulted in suboptimal preservation of the target molecules; however, given that the FFPE samples were all derived from the same diagnostic service this seems unlikely.
The clinical significance of HPV testing may reach considerably further than merely guiding expected prognosis. Should successful primary outcomes be apparent from prospective randomised clinical trials seeking to modify treatment on the basis of HPV status, then further de-escalation of therapy may be advocated. Any such move will necessitate ultimate stringency for the diagnostic testing to avoid incorrect classification of HPV-positive and -negative cases. The risks of inaccurate molecular diagnosis include missed opportunities to allocate cases to the novel intervention arm of a trial or, of greater importance, the delivery of potentially subtherapeutic treatment to individuals with HPV-negative OPSCC. It is questionable whether the levels of sensitivity and specificity demonstrated to date by single or combined-test algorithms for HR-HPV diagnostic testing are adequate to guard against such eventualities; however, RNAscope seems to offer a substantial improvement. Several criteria for an acceptable test of HPV status in OPSCC must be met before recommending a specific HPV testing strategy. The test or combination of tests must have high sensitivity and specificity against an accepted analytical gold standard and have excellent prognostic capacity. Ideally, the test would be feasible as part of the routine diagnostic process, reproducible between laboratories and cost effective. In this study, HR-HPV RNAscope showed a high degree of accuracy against the most appropriate analytical gold standard and was the best discriminator of disease-specific and OS. Before adoption of HR-HPV RNAscope into clinical practice could be formally advocated, this test requires mandatory approval as an in vitro diagnostic device; however, the impending application and availability of HR-HPV RNAscope to a widely available automated staining platform (Ventana Medical Systems Inc.) will facilitate standardisation of test conditions and reproducibility between laboratories. These features raise the possibility that HR-HPV RNAscope could be developed to provide the ‘clinical standard’ for assigning a diagnosis of HPV-related OPSCC.
Change history
02 April 2013
This paper was modified 12 months after initial publication to switch to Creative Commons licence terms, as noted at publication
References
Adelstein DJ, Ridge JA, Gillison ML, Chaturvedi AK, D’Souza G, Gravitt PE, Westra W, Psyrri A, Kast WM, Koutsky LA, Giuliano A, Krosnick S, Trotti A, Schuller DE, Forastiere A, Ullmann CD (2009) Head and neck squamous cell cancer and the human papillomavirus: summary of a National Cancer Institute State of the Science Meeting, November 9–10, 2008, Washington, D.C. Head Neck 31: 1393–1422
Ang KK, Harris J, Wheeler R, Weber R, Rosenthal DI, Nguyen-Tan PF, Westra WH, Chung CH, Jordan RC, Lu C, Kim H, Axelrod R, Silverman CC, Redmond KP, Gillison ML (2010) Human papillomavirus and survival of patients with oropharyngeal cancer. N Engl J Med 363: 24–35
Bishop JA, Ma XJ, Wang H, Luo Y, Illei PB, Begum S, Taube JM, Koch WM, Westra WH (2012) Detection of transcriptionally active high-risk HPV in patients with head and neck squamous cell carcinoma as visualized by a novel E6/E7 mRNA in situ hybridization method. Am J Surg Pathol 36 (12): 1874–1882
Braakhuis BJ, Brakenhoff RH, Meijer CJ, Snijders PJ, Leemans CR (2009) Human papilloma virus in head and neck cancer: the need for a standardised assay to assess the full clinical importance. Eur J Cancer 45: 2935–2939
Chaturvedi AK, Engels EA, Pfeiffer RM, Hernandez BY, Xiao W, Kim E, Jiang B, Goodman MT, Sibug-Saber M, Cozen W, Liu L, Lynch CF, Wentzensen N, Jordan RC, Altekruse S, Anderson WF, Rosenberg PS, Gillison ML (2011) Human papillomavirus and rising oropharyngeal cancer incidence in the United States. J Clin Oncol 29 (32): 4294–4301
D’Souza G, Kreimer AR, Viscidi R, Pawlita M, Fakhry C, Koch WM, Westra WH, Gillison ML (2007) Case-control study of human papillomavirus and oropharyngeal cancer. N Engl J Med 356: 1944–1956
Fakhry C, Westra WH, Li S, Cmelak A, Ridge JA, Pinto H, Forastiere A, Gillison ML (2008) Improved survival of patients with human papillomavirus-positive head and neck squamous cell carcinoma in a prospective clinical trial. J Natl Cancer Inst 100: 261–269
Gillison ML, Koch WM, Capone RB, Spafford M, Westra WH, Wu L, Zahurak ML, Daniel RW, Viglione M, Symer DE, Shah KV, Sidransky D (2000) Evidence for a causal association between human papillomavirus and a subset of head and neck cancers. J Natl Cancer Inst 92: 709–720
Harris SL, Thorne LB, Seaman WT, Neil Hayes D, Couch ME, Kimple RJ (2011) Association of p16(INK4a) overexpression with improved outcomes in young patients with squamous cell cancers of the oral tongue. Head Neck 33 (11): 1622–1627
Jordan RC, Lingen MW, Perez-Ordonez B, He X, Pickard R, Koluder M, Jiang B, Wakely P, Xiao W, Gillison ML (2012) Validation of methods for oropharyngeal cancer HPV status determination in US cooperative group trials. Am J Surg Pathol 36: 945–954
Lassen P, Eriksen JG, Hamilton-Dutoit S, Tramm T, Alsner J, Overgaard J (2009) Effect of HPV-associated p16INK4A expression on response to radiotherapy and survival in squamous cell carcinoma of the head and neck. J Clin Oncol 27: 1992–1998
Lewis JS, Ukpo OC, Ma XJ, Flanagan JJ, Luo Y, Thorstad WL, Chernock RD (2012) Transcriptionally-active high-risk human papillomavirus is rare in oral cavity and laryngeal/hypopharyngeal squamous cell carcinomas – a tissue microarray study utilizing E6/E7 mRNA in situ hybridization. Histopathology 60: 982–991
Licitra L, Perrone F, Bossi P, Suardi S, Mariani L, Artusi R, Oggionni M, Rossini C, Cantu G, Squadrelli M, Quattrone P, Locati LD, Bergamini C, Olmi P, Pierotti MA, Pilotti S (2006) High-risk human papillomavirus affects prognosis in patients with surgically treated oropharyngeal squamous cell carcinoma. J Clin Oncol 24: 5630–5636
Lindquist D, Romanitan M, Hammarstedt L, Nasman A, Dahlstrand H, Lindholm J, Onelov L, Ramqvist T, Ye W, Munck-Wikland E, Dalianis T (2007) Human papillomavirus is a favourable prognostic factor in tonsillar cancer and its oncogenic role is supported by the expression of E6 and E7. Mol Oncol 1: 350–355
Marur S, D’Souza G, Westra WH, Forastiere AA (2010) HPV-associated head and neck cancer: a virus-related cancer epidemic. Lancet Oncol 11: 781–789
Mehanna H, Beech T, Nicholson T, El-Hariry I, McConkey C, Paleri V, Roberts S (2012) Prevalence of human papillomavirus in oropharyngeal and nonoropharyngeal head and neck cancer-systematic review and meta-analysis of trends by time and region. Head Neck; e-pub ahead of print 20 January 2012 doi:10.1002/hed.22015
Nasman A, Attner P, Hammarstedt L, Du J, Eriksson M, Giraud G, Ahrlund-Richter S, Marklund L, Romanitan M, Lindquist D, Ramqvist T, Lindholm J, Sparen P, Ye W, Dahlstrand H, Munck-Wikland E, Dalianis T (2009) Incidence of human papillomavirus (HPV) positive tonsillar carcinoma in Stockholm, Sweden: an epidemic of viral-induced carcinoma? Int J Cancer 125: 362–366
Robinson M, Sloan P, Shaw R (2010) Refining the diagnosis of oropharyngeal squamous cell carcinoma using human papillomavirus testing. Oral Oncol 46: 492–496
Schache AG, Liloglou T, Risk JM, Filia A, Jones TM, Sheard J, Woolgar JA, Helliwell TR, Triantafyllou A, Robinson M, Sloan P, Harvey-Woodworth C, Sisson D, Shaw RJ (2011) Evaluation of human papilloma virus diagnostic testing in oropharyngeal squamous cell carcinoma: sensitivity, specificity, and prognostic discrimination. Clin Cancer Res 17: 6262–6271
Schemper M, Smith TL (1996) A note on quantifying follow-up in studies of failure time. Control Clin Trials 17: 343–346
Shaw R, Robinson M (2011) The increasing clinical relevance of human papillomavirus type 16 (HPV-16) infection in oropharyngeal cancer. Br J Oral Maxillofac Surg 49 (6): 423–429
Singhi AD, Westra WH (2010) Comparison of human papillomavirus in situ hybridization and p16 immunohistochemistry in the detection of human papillomavirus-associated head and neck cancer based on a prospective clinical experience. Cancer 116: 2166–2173
Smeets SJ, Hesselink AT, Speel EJ, Haesevoets A, Snijders PJ, Pawlita M, Meijer CJ, Braakhuis BJ, Leemans CR, Brakenhoff RH (2007) A novel algorithm for reliable detection of human papillomavirus in paraffin embedded head and neck cancer specimen. Int J Cancer 121: 2465–2472
Sturgis EM, Cinciripini PM (2007) Trends in head and neck cancer incidence in relation to smoking prevalence: an emerging epidemic of human papillomavirus-associated cancers? Cancer 110: 1429–1435
Ukpo OC, Flanagan JJ, Ma XJ, Luo Y, Thorstad WL, Lewis JS (2011) High-risk human papillomavirus E6/E7 mRNA detection by a novel in situ hybridization assay strongly correlates with p16 expression and patient outcomes in oropharyngeal squamous cell carcinoma. Am J Surg Pathol 35: 1343–1350
Wang F, Flanagan J, Su N, Wang LC, Bui S, Nielson A, Wu X, Vo HT, Ma XJ, Luo Y (2012) RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J Mol Diagn 14: 22–29
Westra WH (2009) The changing face of head and neck cancer in the 21st century: the impact of HPV on the epidemiology and pathology of oral cancer. Head Neck Pathol 3: 78–81
Wiest T, Schwarz E, Enders C, Flechtenmacher C, Bosch FX (2002) Involvement of intact HPV16 E6/E7 gene expression in head and neck cancers with unaltered p53 status and perturbed pRb cell cycle control. Oncogene 21: 1510–1517
Acknowledgements
This work received funding from the Wellcome Trust through the Wellcome-FDS Research Training Fellowship (held by AGS), Faculty of Dental Surgery of the Royal College of Surgeons of England, Liverpool Tissue Bank (LTB) and McGreavy Family (in memory of Paul McGreavy). Access to, and costs associated with, RNAscope testing was provided by Advanced Cell Diagnostics Inc. X-JM, HW and YL have stock in this company and stand to profit by any use of this testing through their company. For the remaining authors no conflict of interest exists.
Author information
Authors and Affiliations
Corresponding author
Additional information
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
Rights and permissions
From twelve months after its original publication, this work is licensed under the Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Schache, A., Liloglou, T., Risk, J. et al. Validation of a novel diagnostic standard in HPV-positive oropharyngeal squamous cell carcinoma. Br J Cancer 108, 1332–1339 (2013). https://doi.org/10.1038/bjc.2013.63
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/bjc.2013.63
Keywords
This article is cited by
-
HPV-Associated Squamous Cell Carcinoma of the Eyelid: Diagnostic Utility of p16 Immunohistochemistry and mRNA In Situ Hybridization
Head and Neck Pathology (2023)
-
Findings from a Malaysian multicentre study on oropharyngeal squamous cell carcinoma
Infectious Agents and Cancer (2023)
-
Human papillomavirus E6E7 mRNA and TERC lncRNA in situ detection in cervical scraped cells and cervical disease progression assessment
Virology Journal (2022)
-
Evaluation of the Suitability of RNAscope as a Technique to Measure Gene Expression in Clinical Diagnostics: A Systematic Review
Molecular Diagnosis & Therapy (2022)
-
HPV E6/E7 mRNA in situ hybridization in endocervical adenocarcinoma: implications for prognosis and diagnosis
Cancer Cell International (2021)