Abstract
Background:
MiR-221/-222 are frequently overexpressed in breast cancer and are associated with increased malignancy. The specific modification of microRNAs (miRNAs) expression could be a promising strategy in breast cancer therapy, leading to the suppression of tumourigenic processes in tumour cells.
Methods:
MiR-221/-222 expressions were analysed in 86 breast cancer tissues by quantitative RT–PCR and tested for correlation with immunohistochemistry data and clinical follow-up. In vitro assays were conducted using human breast cancer cell lines with lentiviral overexpression of miR-221/-222.
Results:
In tumour tissues, miR-221/-222 were associated with the occurrence of distant metastases. In particular, high levels of miR-221 were revealed to have a high prognostic impact for the identification of significantly different groups with advanced tumours. MiR-221/-222 overexpression strongly increased cell proliferation and invasion in vitro. Following miR-221/-222 overexpression an increased uPAR expression and cell invasion were observed.
Conclusion:
This study demonstrates a significant role for highly expressed miR-221/-222 in advanced breast cancers allowing for the identification of significantly different prognostic groups, particularly for HER2-positive and lymph-node-positive breast cancers. Considering that miR-221/-222 are strongly involved in cell invasion, these miRNAs may be promising markers for breast cancer prognosis and therapy.
Similar content being viewed by others
Main
In breast cancer, advices for adjuvant therapy are based on traditional factors like age, tumour size, grade of differentiation and steroid hormone receptor status. The expression levels of oestrogen receptor (ER) and HER2 (also called ErbB2) are currently the best-known predictive and prognostic biomarkers for personalised breast cancer therapy (Early Breast Cancer Trialists' Collaborative G, 2005). The HER2 oncogene is amplified or overexpressed in ∼30% of human breast cancers and its overexpression is related to the number of lymph node (LN) metastasis and poor prognosis in patients (Golubnitschaja et al, 2013). Despite much advancement in knowledge from researching metastasis, the molecular mechanisms are still not completely understood. Several reports have described the involvement of a recently discovered class of noncoding regulatory RNAs, termed microRNAs (miRNAs) in the regulation of cancer (Wang and Wang, 2012). MiRNAs are small (18–24 nucleotides) noncoding RNAs that regulate the stability and translational efficiency of targeted messenger RNA (mRNA) in a sequence-specific manner (Wang and Wang, 2012). Depending on the complementarity of the miRNA to its target, one miRNA may bind several regions on the same mRNA or on different mRNAs (Filipowicz et al, 2008). The expression of several miRNAs has been shown to promote tumourigenesis, whereas other miRNAs exert tumour suppressor properties (Iorio and Croce, 2009). Upregulated miR-21 and miR-103/107 are associated with advanced malignancy and poor prognosis in breast cancer (Yan et al, 2008; Martello et al, 2010; Anastasov et al, 2012). In contrast, downregulated miR-126, miR-335 and miR-210 are inversely correlated with metastasis (Foekens et al, 2008; Tavazoie et al, 2008). In breast cancer cell lines, overexpression of miR-221 and miR-222 has been shown to be involved in the EGFR–RAS–RAF–MEK pathway and to downregulate p27Kip1 and PTEN leading to elevated cell proliferation (Miller et al, 2008; Garofalo et al, 2009). Moreover, there is evidence that miR-221 and miR-222 directly target ERα and that such targeting negatively correlates with its expression in breast cancer (Cochrane et al, 2010). MiR-221 and miR-222 are involved in the promotion of an aggressive basal-like breast cancer phenotype, and both miRNAs are highly expressed in triple-negative breast cancer (TNBC) (Howe et al, 2012). The overexpression of miR-221 and miR-222 results in a poorly differentiated mesenchymal-like phenotype (Howe et al, 2012), and increasing the levels of these miRNAs elevate the levels of the mesenchymal marker vimentin in the MCF10A cell line (Tang et al, 2012). Antagonising miR-221 and miR-222 in the metastatic MDA-MB-231 breast cancer cell line induced a reverse phenotype (Tang et al, 2012). A possible mechanism through which miR-221- and miR-222-positive cells overcome apoptosis may be by directly repressing pro-apoptotic regulators such as PUMA, BMF or p27Kip1 (le Sage et al, 2007; Gramantieri et al, 2009; Zhang et al, 2010). However, some controversial studies have demonstrated different roles for overexpressed miR-221 and miR-222 in breast cancer tissue indicating that overexpression of miR-221 is associated with a more aggressive phenotype than upregulated miR-222 (Radojicic et al, 2011).
In primary breast cancer, independent prognostic factors such as the serine protease urokinase-type plasminogen activator (uPA) indicate a level of the tumour invasion and metastatic disease that is of particular value for therapy decision (Golubnitschaja et al, 2013). Potential clinical utilisation of novel biomarkers, such as Ki67 and uPA/PAI-1 is in the perspective (Harbeck et al, 2002; Li et al, 2010). Urokinase-type plasminogen activator initiates extracellular matrix (ECM) degradation by activating plasminogen to plasmin. Plasminogen activation by uPA is tightly regulated by its cell surface receptor uPAR, and two specific fast-acting plasminogen activator inhibitors, PAI-1 and PAI-2 (Foekens et al, 2000). The uPA–uPAR interaction has an essential role in the proteolytical degradation of ECM and the membrane surrounding the primary tumour, which may favour cancer invasion and metastasis (Li et al, 2010). More detailed investigations concerning the impact of miR-221 and miR-222 and the uPA system (uPAS) in breast cancer development are necessary and the determination of their expression levels may represent an attractive therapeutic or predictive tool.
In this study, we investigated the expression of miR-221 and miR-222 in 86 formalin-fixed paraffin-embedded (FFPE) tissues from invasive ductal breast carcinomas (IDC) to determine the clinical impact of altered miRNA expression. We show, for the first time, that both miRNAs are of significant prognostic value, and that miR-221, in particular, is highly significant in distinguishing prognostic subgroups in advanced (HER2-positive, LN-positive) tumours. To support our findings in tumour tissues and to further explore the impact of these miRNAs for malignancy in breast cancer, we overexpressed them in human breast cancer cell lines (T47D, SKBR3 and MDA-MB-361) and analysed the effects on signalling proteins, proliferation, migration, invasion and uPAR expression changes in particular.
Materials and methods
Tumour tissues and patient data
Formalin-fixed paraffin-embedded tissues from 86 randomly selected IDC were analysed for the expression of the miRNAs miR-221 and miR-222. Forty-eight tumours were LN-negative and 59 tumours were small in size (⩽2 cm). Nine of the tumours were histological grade 1, 55 were grade 2 and 22 were grade 3 (Elston and Ellis, 1991). The hormone receptor status was evaluated immunohistochemically and revealed that 22 of the tumours were ER-positive. The median patient’s age at the time of diagnosis was 66 years (please see also Supplementary Table 1). All of the patients were surgically treated, and no patient received preoperative treatment. Postoperatively, radiotherapy was administered to 29 patients, anti-hormonal treatment to 3, chemotherapy to 2 and radio- and chemotherapy to 4 patients. Detailed long-term clinical follow-up was available for all of the patients with a median follow-up period of 113 months (min. 5 months, max. 468 months). Forty patients relapsed with distant metastases within the total follow-up period. The ethical approval for the study was obtained from the Ethics Committee of the Medical Faculty of the Technical University of Munich (TUM) and the inform consent was given by all patients from TUM and from Karolinska Institute and Hospital, Stockholm, Sweden.
Immunohistochemistry
In addition to the histopathological and follow-up data, immunohistochemistry (IHC) data were available from the tumours detailing the expression of human epidermal growth factor receptors (HER1, HER2, HER3 and HER4), ER and progesterone receptor (PR) and PTEN (Aubele et al, 2007). For IHC of the cell cycle inhibitor p27Kip1 and the cell proliferation marker Ki67, two primary antibodies were used on the tissue microarray (TMA) sections: anti-p27Kip1 (610242, 1 : 500, BD Bioscience, Heidelberg, Germany) and anti-Ki67 (15580, 1 : 1.000, Abcam, Cambridge, UK). The deparaffinization of the TMA sections, antigen retrieval and incubation of the primary antibodies was performed in general as previously described (Aubele et al, 2007). The staining and counterstaining were done by an automated immunostainer (Ventana Medical System, Inc., Tucson, AZ, USA) using 3, 3′-diaminobenzidine for detection and hematoxylin for counterstaining. The tissue staining intensities were scored blind by two investigators describing the number of positively stained tumour cells (in per cent). In case of discrepancy consensus between the two observers was found.
Growth and maintenance of cell lines
The T47D cell line (HTB-133) was acquired from American Type Culture Collection (ATCC) and maintained in RPMI 1640 with GlutaMAX (Roswell Park Memorial Institute medium) supplemented with bovine insulin (10 μg ml−1, Sigma, St. Lois, MO, USA). The SKBR3 (HTB-30 was acquired from ATCC) and MDA-MB-361 breast cancer cell lines (a kind gift from Professor M Schmitt, Clinical Research Unit, Department of Obstetrics and Gynecology, Technische Universität München) were cultured in DMEM with GlutaMAX (Dulbecco Modified Eagles medium). Both media were supplemented with 10% fetal calf serum (FCS, Invitrogen, Carlsbad, CA, USA) and 0.25% each of penicillin and streptomycin (Life Technologies, Darmstadt, Germany). The cell cultures were maintained in a water humidified 37 °C incubator with 5% CO2. The cells used were authenticated by genetic profiling service (Eurofins MWG Operon, Ebersberg, Germany), and the obtained profiles were subsequently verified by the online STR matching analysis database (DSMZ, Heidelberg, Germany).
Lentivirus production and infection of breast cancer cell lines
Replication-defective lentiviral particles were produced by transient co-transfection of HEK293T with the packaging plasmids pMDLg/pRRE, pRSV. Rev and pMD2.G (a kind gift from Prof. D Trono, École polytechnique fédérale de Lausanne) and the lentiviral precursor expression vectors for miR-221 and miR-222 overexpression (Cat. Nr. pMIRH221PA-1-GVO-SB and pMIRH222PA-1-GVO-SB; Biocat, Heidelberg, Germany) as previously described (Anastasov et al, 2012). The lentiviral transduction vector pGreenPuro (pGP; System Biosciences, Mountain View, CA, USA) was used as control vector for lentiviral infections. The virus particles were harvested 48 h after transfection, cleared and concentrated as reported previously (Anastasov et al, 2009). According to the virus titre determination, the virus production ranged between 108 and 109 TU ml−1 (Transduction Units per ml). The viral infection of breast cancer cells was performed as previously described (Anastasov et al, 2010, 2012).
RNA isolation for miRNA expression analysis
Paraffin-embedded tissue was microdissected with a sterile needle from 5 μm thick sections using a stereo microscope (Stemi 2000, Zeiss, Germany). A consecutive H&E-stained section was used for guidance. The tumour cell material (containing at least >80% tumour cells) was collected from all cases. In addition, histologically normal tissue was collected from five cases and pooled as control (normal tissue). Total RNA was isolated from the microdissected tissues, digested, purified and dissolved in 20 μl RNase-free water. Five microlitres (100 ng) of RNA were reverse-transcribed using MultiScribeTM reverse transcriptase (Applied Biosystems, Foster City, CA, USA) (Chen et al, 2005; Anastasov et al, 2012). Further processing and evaluation of the results was performed according to the manufacturer’s instructions. Total RNA was isolated from each of the breast cancer cell lines (T47D, SKBR3 and MDA-MB-361) after viral transduction. The cells were pelleted by centrifugation at 1500 r.p.m. for 5 min, and washed with 1 ml Dulbecco’s phosphate-buffered saline without MgCl2 and CaCl2 (Invitrogen). Small RNAs (<200 nucleotides) were isolated from the cells using the mirVana miRNA isolation kit (Applied Biosystems) following the protocol for total RNA isolation. The quantity and quality of the total RNA and miRNA was measured with the Nanodrop spectrophotometer (PeqLab Biotechnology, Erlangen, Germany).
TaqMan-miRNA assays and data analyses
Specific single TaqMan-miRNA assays (Applied Biosystems) were used for miR-221 and miR-222 expression analysis (Cat. Nr. 4427975; Assay ID 000524 and ID 002276) in total RNA isolations from FFPE samples and from cells. Quantitative PCR was performed using the StepOnePlus Detection System (Applied Biosystems) according to the manufacturer’s instructions. The relative expression values of specific miRNA were calculated by using the 2−ΔΔCT method (Anastasov et al, 2012) and normalised to the control miRNA (RNU43 and RNU44, no. 4427975; Assay ID 001094 and 001095) and to the FFPE control (normal) tissue sample for tumour FFPE samples or to the SKBR3 control cells (used as calibrator) after subsequent lentiviral transductions in three different cell lines. All of the reactions were performed in duplicate and three independent experiments were conducted.
In silico analysis of miR-221 and miR-222 target sequences
The 3′ untranslated region (UTR) sequences of the relevant components of the uPAS (uPA, its receptor uPAR and its inhibitor PAI-1) were analysed for potential miR-221 and miR-222 binding sites based on the miRanda algorithm target prediction using the microRNA.org and Target Scan (release 6.2 miRNA target) prediction website.
Western blot analysis
The isolation of proteins, immunoblotting and quantifications were performed as previously described (Ludyga et al, 2013). The respective target proteins regulated by the miRNAs were detected with the following primary antibodies: cyclin D1 (sc-753), p-cdc2 (sc-6248), ER (sc-8002, Santa Cruz Biotechnology, Heidelberg, Germany), HER1 (M7298), HER2 (A0485, DAKO, Glostrup, Denmark), HER3 (MS-201-P1, Thermo Scientific, Fremont, CA, USA), HER4 (4795), (phospho, 9554) PTEN (9559), (phospho 4051) Akt (9272), (phospho, 4376) MAPK (4695), (phospho, 9211) p38MAPK (9212), phospho-STAT3 (9134), MMP9 (3852, Cell Signaling Technology, Beverly, MA, USA), STAT3 (610190), Cyclin A (611268), p27Kip1 (610242, BD Transduction Laboratories, Lexington, KY, USA), uPAR (IID7) a kind gift from Professor M Schmitt and Professor V Magdolen (Clinical Research Unit, Department of Obstetrics and Gynecology, Technische Universität München) and tubulin as a loading control (T5168, Sigma). The following peroxidase-conjugated secondary antibodies were used: anti-rabbit (NA934V) and anti-mouse (NA931V) (GE Healthcare, Chalfont St. Giles, UK).
WST-1 cell proliferation assay
Cell proliferation was determined using water-soluble tetrazolium WST-1 (4-[3-(4-Iodophenyl)-2-(4-nitrophenyl)-2H-5-tetrazolio]-1,3-benzene disulfonate) for the spectrophotometric assay according to the manufacturer’s protocol (05015944001, Roche Diagnostics, Mannheim, Germany). The T47D, SKBR3 and MDA-MB-361 cells were seeded at a concentration of 1 × 104 cells per well in a 96-well tissue culture plate and incubated for 48 h at 37 °C. The measurements were performed on three independent experiments as described previously (Ludyga et al, 2013).
Wound scratch migration assay
A wound was scratched into a confluent monolayer of T47D and SKBR3 cells, the MDA-MB-361 cells are not suitable for this assay owing to a non-confluent growth. The migration assay and quantification were performed as previously described (Ludyga et al, 2013). These experiments were performed three times.
Matrigel invasion assay
Control or infected T47D, SKBR3 and MDA-MB-361 cells were seeded at a concentration of 5 × 104 in serum-reduced medium onto BD BioCoat Matrigel Invasion Chambers (354480, BD Biosciences, Bedford, MA, USA), inserted into 24-well cell culture plates and incubated at cell line-specific conditions for 24 h (T47D and MDA-MB-361 cells) or 48 h (SKBR3 cells) at 37 °C. The invasion assay was performed according to the manufacturer’s protocol as previously described (Ludyga et al, 2013). The assays were performed three times.
Statistics
In the tumour tissues, the correlations among the markers and the miRNA expressions were examined by Spearman’s rank correlation test. For univariate survival analysis, Kaplan–Meier curves were calculated and the differences between strata were determined with the log-rank Chi-Square test. Optimal cut-offs for miR-221 or miR-222 expression in univariate survival analysis were determined by the method of the maximally selected log-rank statistic according to Lausen and Schumacher. For this, bivariate Kaplan–Meier analysis for HER2-positive or -negative cases yielded a maximum log-rank chi-square of 22.9 (df=3) for the 66.7% quantile cut-off of the miR-221 expression (log2-fold −0.15, fold-value 0.90, minimum P-value <0.0001, adjusted P-value 0.005); accordingly, for the nodal (LN+, LN−) status, the maximum log-rank chi-square was 25.3 (df=3) for the 33.3% quantile cut-off of the miR-221 expression values (log2-fold −1.2, fold-value 0.435, minimum P-value<0.0001, adjusted P-value 0.0031). Multivariate analysis was performed by Cox regression analysis. Here, only covariates reaching significance with P⩽0.10 were allowed in the regression model. In all other analyses, the statistical significance was considered at the P⩽0.05 level. For illustration of the multivariate result, the selected parameters, weighted by their coefficients, were used to calculate a new prognostic variable (prognostic factor, PF) (Aubele et al, 2007) and patients were grouped according to their PF for Kaplan–Meier analysis. For statistical analyses of the in vitro proliferation, migration and invasion assays, as well as for western blots, Student’s t-test was used.
Results
The expression of miR-221 and miR-222 is inversely correlated with p27Kip1 in tumour tissues
We identified a significant association between miR-221 expression and the expression of HER4 (inverse, P=0.006), and between miR-222 and the expression of HER1 (P=0.003) and HER3 (P=0.003). The distribution of the parameters from individual samples is available as Supplementary Figure 1. An inverse correlation was found between miR-222 and the expression of the ER (P=0.016) and with the expression of p27Kip1 protein (P=0.048), but there was no significant correlation between the expression of miR-221 or miR-222 and PTEN in the tumour tissues. The expression of the cell cycle inhibitor p27Kip1 inversely correlated with the proliferation marker Ki67 (P=0.012, Figure 1), which was evaluated in this study to confirm the proliferation at low p27Kip1 expression. The histological grade was significantly associated with the Ki67 IHC (P⩽0.001) and was inversely associated with p27Kip1 IHC (P=0.03).
The expression of miR-221 and miR-222 is highly associated with metastases’ risk in advanced breast cancers
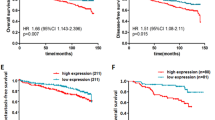
In univariate survival analyses, considering the occurrence of distant metastases in patients, high miR-221 expression showed a significant correlation (P=0.02) with a worse prognosis for the patients (Table 1). Considering the subgroups of the tumours, in terms of nodal (LN+, LN−) or HER2 status (HER2-negative (0, 1+) or HER2-positive (2+, 3+)), miR-221 allows for highly significant differentiation of the prognostic groups within the HER2-positive tumours (minimal P=0.0013, Figure 2A) and in the LN-positive tumours (minimal P=0.012, Figure 2C), but not in the HER2-negative (Figure 2B) or the LN-negative tumours (Figure 2D). Therefore, we consider miR-221 to be a highly significant prognostic marker for distinguishing prognostic subgroups particularly in advanced (LN+, HER2+) breast cancers (Figure 2, Table 1). MiR-222 was of prognostic significance in the LN-negative tumours (minimal P=0.02), but not in LN-positive tumours, and therefore, may be of high impact in differentiating between different LN-negative prognostic groups (Table 1).
MiR-221 expression highly correlates with an increased risk for metastases in advanced breast cancers. Kaplan–Meier curves were calculated for subgroups of the cohort, as described in the statistic part. (A) HER2-positive tumours only, (B) HER2-negative tumours, (C) lymph-node-positive and (D) lymph-node-negative tumours. The number of patients in a group, the number of events, the corresponding minimal P-values as well as the adjusted P-values are given.
In multivariate analyses, all parameters reaching a significance level of P⩽0.10 in the univariate approaches were offered (tumour size P=0.004, LN status P=0.003, HER2 P=0.083, miR-221 P=0.026). These all remained significant as independent covariates in the Cox regression model (Table 1). According to this result, we calculated the PF (Aubele et al, 2007) by the linear combination of the variables weighted by their coefficients: PF=0.993*LN status+0.943*miR-221 expression+0.552*tumour size+0.308*HER2-IHC. This calculation gives us a continuous variable for each patient with a range of 0.9–4.6, representing the patients’ risk for development of distant metastases. To visualise the patients’ classification with this multivariate risk factor, we calculated Kaplan–Meier curves (Figure 3) showing the mean distant metastases-free survival curve of all patients (plotted in grey), patients with a low (PF⩽2.79, n=46, 12 events) and those with a high PF (> 2.79, 31, 22 events; P⩽0.0001).
MiR-221 expression is of high prognostic value in a multivariate analysis for a distant metastases-free survival of patients. Kaplan–Meier curves for a distant metastases-free survival of patients of >400 months. Patients were grouped into two different risk groups according to their multivariate PF: low PF (⩽2.79) and high PF (>2.79). The number of patients in a group, the number of events and the P-value are given (grey line: mean survival curve of all patients).
To summarise our results in the tumour tissues, significantly different prognostic groups could be identified by miR-221 expression within advanced (LN-positive or HER2-positive) tumours, or when calculating the multivariate PF, which is a combination of LN status, tumour size, HER2 and miR-221 expression.
Overexpression of miR-221 and miR-222 leads to increased proliferation of T47D, SKBR3 and MDA-MB-361 breast cancer cells
In comparison with the pooled normal tissue samples, the mean expression levels of miR-221 and miR-222 in tumour tissues are shown in Figure 4A. In the breast cancer cell lines, the expression of miR-221 and miR-222 was low with small differences in miRNA expression levels between three cell lines (Figure 4B). The SKBR3 cell line showed the highest detection level of miR-221 and miR-222 expression and was used as control (calibrator) in subsequent qRT–PCR reactions. To strengthen our results from the tumour tissue samples, we overexpressed both miRNAs using the lentiviral approach in three breast cancer cell lines (T47D, SKBR3 and MDA-MB-361) and analysed the effects on tumourigenicity in vitro. The elevated expression of the miRNAs in the cell lines was quantified by qRT–PCR. In the T47D cell line ∼100-fold (Figure 4C) overexpression was detected in comparison with the control SKBR3 cells used as calibrator (control cell line). In the SKBR3 cells ∼30-fold (miR-221) and 80-fold (miR-222) miRNA expression was detected after lentiviral infection with specified miRNAs (Figure 4C). In the MDA-MB-361 cells ∼95-fold (miR-221) and 115-fold (miR-222) overexpression was detected in comparison with the control (Figure 4C). As a consequence of the miR-221 or miR-222 upregulation, we observed a reduction in the protein expression of the cell cycle inhibitor p27Kip1 (Figure 5A). Concerning the expression levels of the HER receptors (HER1, HER3 and HER4) and the cell cycle proteins cyclin A, D1, E and p-cdc2, no change was observed, HER2 is shown as example (Figure 5A).
MiR-221 and miR-222 are differentially expressed in tumour tissues and breast cancer cell lines. (A) The boxplots show the distribution of miRNA levels in individual tumour samples normalised by the miRNA level of normal tissues (where RNA was pooled from five cases). The cohort means of miR-221 and of miR-222 are significantly less than the miRNA levels of the pooled controls (P=0.0003 and P<0.0001, respectively), which were set as one. (B) The expression levels (mean±of miR-221 and miR-222) in the human breast cancer cell lines T47D, SKBR3 and MDA-MB-361 compared with normal tissue. (C) The expression levels of miR-221 and miR-222 following overexpression in the cell lines T47D, SKBR3 and MDA-MB-361 (where control SKBR3 sample was used as calibrator (control) for all different lentiviral infections).
The overexpression of miR-221 and miR-222 affect the protein levels of direct targets and elevate cell proliferation in vitro. (A) Western blot analysis of HER2, p27Kip1, ER and PTEN following the overexpression of miR-221 and miR-222 in the T47D, SKBR3 (ER-negative) and MDA-MB-361 cells compared with the controls. Tubulin was used as the loading control. The mean values of two independent experiments are shown. (B) The graph represents the amount of viable T47D, SKBR3 and MDA-MB-361 cells following miR-221 and miR-222 overexpression related to the controls. The mean values of three independent experiments, the standard deviations and the P-values (Student’s t-test) are shown.
However, we identified cell line-specific effects on the expression levels of ER and the tumour suppressor PTEN (Figure 5A). In the T47D and MDA-MB-361 cells, following miR-221 and miR-222 overexpression, we detected strongly decreased ER expression, whereas in the SKBR3 cell line, the expression of PTEN was decreased, in particular following the overexpression of miR-222. The PTEN levels were not changed in the T47D cells and only slightly in the MDA-MB-361 cells; the SKBR3 cells are ER-negative (Figure 5A).
The phosphorylation and expression of the signal transduction proteins STAT3, Akt, MAPK 1/3 (ERK 1/2) and p38MAPK were unchanged following the overexpression of both miRNAs (data not shown).
Consistent with the reduced cell cycle inhibitor p27Kip1 expression, we further observed a statistically significant increase in cell proliferation following the overexpression of both miRNAs, with P=0.029 (miR-221) and P=0.036 (miR-222) in the T47D cells, with P=0.003 (miR-221) and P<0.001 (miR-222) in the SKBR3 cells, and with P=0.004 (miR-221) in the MDA-MB-361 cells compared with the respective controls (Figure 5B).
Overexpression of miR-221 and miR-222 leads to increased invasion of breast cancer cells
To further investigate the effects on migration in breast cancer cells following miR-221 and miR-222 overexpression, we performed a wound scratch assay. In both T47D and SKBR3 cell lines, we did not observe any changes following the overexpression of both miRNAs compared with controls and the MDA-MB-361 were not suitable for this scratch assay (data not shown).
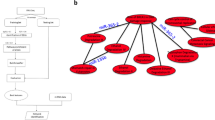
However, in vitro, the invasion of SKBR3 cells expressing high levels of miRNA-221 led to a significantly increased invasion (P=0.02) in comparison with the respective control, and the elevated miR-221 level seems to affect the invasive potential of the SKBR3 cells more than miR-222 (Figure 6A). In the MDA-MB-361 cells, the invasion was significantly elevated (P=0.029) following the overexpression of miR-222; however, the effect was lower than in the SKBR3 cells after miR-221 overexpression. Based on this observation, we examined in more detail how the overexpression of miR-221 and miR-222 is associated with proteins of the uPAS, which strongly promotes cell invasion (Li et al, 2010). Using in silico sequence alignments, the 3′ UTRs of uPA, uPAR and PAI-1 were analysed for putative binding sites for miR-221 and miR-222. The 3′ UTR of the uPAR isoform 2 [PLAUR NM_001005376] was predicted to have binding sites for both miRNAs (Figure 6B). Within the 3′ UTRs of uPA or PAI-1, no binding sites for these miRNAs were detected.
The overexpression of miR-221 and miR-222 elevates cell invasion in vitro and enhances the uPAR expression. (A) The relative amount of invasive cells based on the Matrigel invasion assay after 24 h (T47D and MDA-MB-361 cells) and after 48 h (SKBR3 cells). The graphs show the mean values±s.d. of three independent experiments. (B) An example of miR-221 and miR-222 target prediction using the microRNA.org website. The target predictions are based on the miRanda algorithm with high score matches of seven complementary bases within the 3' UTR of the PLAUR [NM_001005376] sequence. (C) Western blot analysis of uPAR following the overexpression of miR-221 and miR-222 in SKBR3 cells compared with the control. Tubulin was used as the loading control. (D) The graphs represent the quantification of protein levels relative to the control. The mean values of three independent infections, the SDs and the significant P-values are shown.
uPAR may exhibit different isoforms, which in turn may be glycosylated post-translationally showing multiple molecular weight forms between 35 and 60 kDa (Behrendt et al, 1990). The uPAR isoform 2 differs from the full-length uPAR in that it lacks the glycosylphosphatidylinositol (GPI) anchor leading to a soluble uPAR (Stewart and Sayers, 2009). Based on their results, we performed western blot analyses for the detection of uPAR in the SKBR3 cells. In the T47D and MDA-MB-361 cells, the uPAR was not expressed (data not shown). In the SKBR3 cells, following miR-221 and miR-222 overexpression, the uPAR level in particular at ∼40 kDa was significantly increased (P=0.044 and P=0.008, respectively, example is shown in Figure 6C and D). Following miR-221 overexpression, the uPAR protein levels increased more than after miR-222 overexpression, supporting the invasion assay results.
Discussion
Regarding breast cancer, several miRNAs were shown to promote tumourigenesis or act as tumour suppressors and thus are potential therapy targets (Iorio and Croce, 2009). In this study, we have revealed a prognostic value for miR-221 and miR-222 in invasive breast cancers. Further, for the first time, we show the impact of miR-221 expression on the occurrence of distant metastases, particularly in advanced breast cancers. MiR-221 and miR-222 regulate cell proliferation through silencing of the cell cycle inhibitor p27Kip1 (Miller et al, 2008), which has already been described as a direct target of these miRNAs (Howe et al, 2012). This was confirmed in our study where, in tumour tissues, there is an inverse association between miR-222 and p27Kip1 expression, as well as in our in vitro analyses showing reduced p27Kip1 protein levels following the overexpression of the miRNAs. It was also reported that miR-221 and miR-222 directly target ERα and negatively correlate with its status and that both miRNAs were highly expressed in TNBC (Zhao et al, 2008). Consistently, we have also found an inverse correlation between miR-221 and ER expression in the tumour tissues and for both miRNAs by western blot analysis, while miR-222 expression did not show an association with ER status in the tumours. Interestingly, a positive correlation of miR-221 levels with ER status in the breast cancer samples was reported, whereas miR-222 expression did not change between the ER-positive and the ER-negative cases (Yoshimoto et al, 2011). These partially controversial data present high importance of improved scientific analysis and significance of using distinct miRNAs as prognostic biomarkers in the future. It was previously shown that elevated miR-221 or miR-222 expression levels considerably increase cell invasion of a gastric carcinoma cell line, possibly via direct modulation of PTEN expression, which was also shown in prostate carcinomas (Chun-Zhi et al, 2010; Zheng et al, 2012). In our tumour cohort, PTEN expression was not significantly associated with the expression of miR-221 or miR-222. Using immunoblots, we detected slightly altered PTEN protein levels (in the MDA-MB-361 cells) or reduced expressions in the SKBR3 cells, indicating a cell-specific regulation.
Further, in our tumour cohort, we observed a significant inverse correlation between p27Kip1 expression and the histological grade of the tumours, as well as a direct correlation of Ki67 and the histological grade. However, no significant association could be identified between the analysed miRNAs and the LN status, the tumour size or the histological grade. High miR-221 or miR-222 levels have been found to be associated with increased tumour grade in urothelial carcinomas and in human gliomas (Lu et al, 2009; Veerla et al, 2009), but in breast cancer specimens no correlation between miR-221 or miR-222 and the tumour grade has been found.
In our present study, we show that the high expression of miR-221 and miR-222 is significantly associated with the occurrence of distant metastases in breast cancer. We further demonstrate that high miR-221 expression in the tumour tissues allows for significant distinguishing of different prognostic groups, in particular, in advanced (LN+, HER2+) breast cancers. The number of cases available for this study in each subgroup (in particular in the HER2-positive subgroup) is extremely low. Therefore, the discrimination between prognostic groups in Kaplan–Meier analysis should be interpreted with some caution. Nevertheless, this result gives a first indication of the potential clinical impact of miR-221 expression analysis. Based on these results, we consider the expression of miR-221 as an important prognostic and therapeutic marker of the advanced stages of this disease. To the best of our knowledge, this is the first evidence that miR-221 expression may be a promising marker for the identification of patients with advanced breast cancer but a still better prognosis when it is low expressed in tumour samples.
Previous studies demonstrated that MDA-MB-231 and SKBR3 cells (over) express uPAR with (high) invasive and metastasis potential (Li et al, 2010). We have shown that the SKBR3 cell invasion may be associated with the upregulation of miR-221 and miR-222. Owing to the fact that T47D and MDA-MB-361 cells do not express uPAR (data not shown), whereas none or slightly increased invasion following miR-221/-222 overexpression was detected, we suggest a cell line-specific association of uPAR with miR-221 and miR-222, which may also be influenced by additional factors. Based on in silico analyses, a soluble uPAR isoform was predicted as putative miR-221 and miR-222 target. To date, post-transcriptional mechanisms for uPAR regulation through HuR, hnRNPC and p53 have been described (Tran et al, 2003; Shetty, 2005; Shetty et al, 2007). According to our results, miR-221 and miR-222 seem to positively influence the uPAR expression. In general, miRNAs negatively regulate their direct targets via mRNA degradation or translational repression (Tang et al, 2012). A positive role of miRNAs, such as miR-369-3, that upregulates the translation of tumour necrosis factor-α or xlmiR16 regulating Myt1 kinase, together with co-factors such as microribonucleoproteins have also been described (Vasudevan et al, 2007; Mortensen et al, 2011). Multiple uPAR isoforms were identified (Stewart and Sayers, 2009) including a soluble variant with an alternative exon 7b resulting in the loss of the domain III and the GPI anchor with a molecular weight of ∼37 kDa (Pyke et al, 1993; Stewart and Sayers, 2013) that correlates with expression changes observed in our study. The role of this soluble uPAR is not fully understood. However, a chemotactic function in lung cells was described that may bind uPA or integrins and therefore may enhance the tumourigenic potential of cancer cells (Stewart and Sayers, 2009, 2013).
Consistent with our results in tumour tissues demonstrating a correlation between miR-221 overexpression in advanced tumours and the clinical course of the disease, together with increased SKBR3 cell invasion following overexpression of these miRNAs in vitro, we hypothesise an influence of miR-221/-222 on uPAR expression. Detection of the soluble uPAR was dependent on miR-221 and miR-222 overexpression and we observed effects on previously described direct targets, such as p27Kip1, ER or PTEN (Garofalo et al, 2009; Cochrane et al, 2010) but not on signalling proteins, such as Akt, MAPK or STAT3. However, the precise function and uPAR expression regulation directly or indirectly through additional mechanisms has to be clarified in further experiments. These results, along with elevated cell invasion in vitro may suggest that the increased level of the soluble uPAR rather acts extracellularly as a chemoattractant leading to an increased tumourigenesis. Further approval of miR-221/-222 for their prognostic and therapeutic value may be promising candidates to more precisely assess the individual risk of patients and future development of therapy regimens.
Change history
12 November 2013
This paper was modified 12 months after initial publication to switch to Creative Commons licence terms, as noted at publication
References
Anastasov N, Bonzheim I, Rudelius M, Klier M, Dau T, Angermeier D, Duyster J, Pittaluga S, Fend F, Raffeld M, Quintanilla-Martinez L (2010) C/EBPbeta expression in ALK-positive anaplastic large cell lymphomas is required for cell proliferation and is induced by the STAT3 signaling pathway. Haematologica 95 (5): 760–767.
Anastasov N, Hofig I, Vasconcellos IG, Rappl K, Braselmann H, Ludyga N, Auer G, Aubele M, Atkinson MJ (2012) Radiation resistance due to high expression of miR-21 and G2/M checkpoint arrest in breast cancer cells. Radiat Oncol 7: 206.
Anastasov N, Klier M, Koch I, Angermeier D, Hofler H, Fend F, Quintanilla-Martinez L (2009) Efficient shRNA delivery into B and T lymphoma cells using lentiviral vector-mediated transfer. J Hematopathol 2 (1): 9–19.
Aubele M, Auer G, Walch AK, Munro A, Atkinson MJ, Braselmann H, Fornander T, Bartlett JM (2007) PTK (protein tyrosine kinase)-6 and HER2 and 4, but not HER1 and 3 predict long-term survival in breast carcinomas. Br J Cancer 96 (5): 801–807.
Behrendt N, Ronne E, Ploug M, Petri T, Lober D, Nielsen LS, Schleuning WD, Blasi F, Appella E, Dano K (1990) The human receptor for urokinase plasminogen activator. NH2-terminal amino acid sequence and glycosylation variants. J Biol Chem 265 (11): 6453–6460.
Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, Lao KQ, Livak KJ, Guegler KJ (2005) Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res 33 (20): e179.
Chun-Zhi Z, Lei H, An-Ling Z, Yan-Chao F, Xiao Y, Guang-Xiu W, Zhi-Fan J, Pei-Yu P, Qing-Yu Z, Chun-Sheng K (2010) MicroRNA-221 and microRNA-222 regulate gastric carcinoma cell proliferation and radioresistance by targeting PTEN. BMC Cancer 10: 367.
Cochrane DR, Cittelly DM, Howe EN, Spoelstra NS, McKinsey EL, LaPara K, Elias A, Yee D, Richer JK (2010) MicroRNAs link estrogen receptor alpha status and Dicer levels in breast cancer. Horm Cancer 1 (6): 306–319.
Early Breast Cancer Trialists' Collaborative G (2005) Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: an overview of the randomised trials. Lancet 365 (9472): 1687–1717.
Elston CW, Ellis IO (1991) Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long-term follow-up. Histopathology 19 (5): 403–410.
Filipowicz W, Bhattacharyya SN, Sonenberg N (2008) Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nature Rev Genet 9 (2): 102–114.
Foekens JA, Peters HA, Look MP, Portengen H, Schmitt M, Kramer MD, Brunner N, Janicke F, Meijer-van Gelder ME, Henzen-Logmans SC, van Putten WL, Klijn JG (2000) The urokinase system of plasminogen activation and prognosis in 2780 breast cancer patients. Cancer Res 60 (3): 636–643.
Foekens JA, Sieuwerts AM, Smid M, Look MP, de Weerd V, Boersma AW, Klijn JG, Wiemer EA, Martens JW (2008) Four miRNAs associated with aggressiveness of lymph node-negative, estrogen receptor-positive human breast cancer. Proc Natl Acad Sci USA 105 (35): 13021–13026.
Garofalo M, Di Leva G, Romano G, Nuovo G, Suh SS, Ngankeu A, Taccioli C, Pichiorri F, Alder H, Secchiero P, Gasparini P, Gonelli A, Costinean S, Acunzo M, Condorelli G, Croce CM (2009) miR-221&222 regulate TRAIL resistance and enhance tumorigenicity through PTEN and TIMP3 downregulation. Cancer Cell 16 (6): 498–509.
Golubnitschaja O, Yeghiazaryan K, Costigliola V, Trog D, Braun M, Debald M, Kuhn W, Schild HH (2013) Risk assessment, disease prevention and personalised treatments in breast cancer: is clinically qualified integrative approach in the horizon? EPMA J 4 (1): 6.
Gramantieri L, Fornari F, Ferracin M, Veronese A, Sabbioni S, Calin GA, Grazi GL, Croce CM, Bolondi L, Negrini M (2009) MicroRNA-221 targets Bmf in hepatocellular carcinoma and correlates with tumor multifocality. Clin Cancer Res 15 (16): 5073–5081.
Harbeck N, Kates RE, Schmitt M (2002) Clinical relevance of invasion factors urokinase-type plasminogen activator and plasminogen activator inhibitor type 1 for individualized therapy decisions in primary breast cancer is greatest when used in combination. J Clin Oncol 20 (4): 1000–1007.
Howe EN, Cochrane DR, Richer JK (2012) The miR-200 and miR-221/222 microRNA families: opposing effects on epithelial identity. J Mammary Gland Biol Neoplasia 17 (1): 65–77.
Iorio MV, Croce CM (2009) MicroRNAs in cancer: small molecules with a huge impact. J Clin Oncol 27 (34): 5848–5856.
le Sage C, Nagel R, Egan DA, Schrier M, Mesman E, Mangiola A, Anile C, Maira G, Mercatelli N, Ciafre SA, Farace MG, Agami R (2007) Regulation of the p27(Kip1) tumor suppressor by miR-221 and miR-222 promotes cancer cell proliferation. EMBO J 26 (15): 3699–3708.
Li C, Cao S, Liu Z, Ye X, Chen L, Meng S (2010) RNAi-mediated downregulation of uPAR synergizes with targeting of HER2 through the ERK pathway in breast cancer cells. Int J Cancer 127 (7): 1507–1516.
Lu X, Zhao P, Zhang C, Fu Z, Chen Y, Lu A, Liu N, You Y, Pu P, Kang C (2009) Analysis of miR-221 and p27 expression in human gliomas. Mol Med Rep 2 (4): 651–656.
Ludyga N, Anastasov N, Rosemann M, Seiler J, Lohmann N, Braselmann H, Mengele K, Schmitt M, Hofler H, Aubele M (2013) Effects of simultaneous knockdown of HER2 and PTK6 on malignancy and tumor progression in human breast cancer cells. Mol Cancer Res 11 (4): 381–392.
Martello G, Rosato A, Ferrari F, Manfrin A, Cordenonsi M, Dupont S, Enzo E, Guzzardo V, Rondina M, Spruce T, Parenti AR, Daidone MG, Bicciato S, Piccolo S (2010) A MicroRNA targeting dicer for metastasis control. Cell 141 (7): 1195–1207.
Miller TE, Ghoshal K, Ramaswamy B, Roy S, Datta J, Shapiro CL, Jacob S, Majumder S (2008) MicroRNA-221/222 confers tamoxifen resistance in breast cancer by targeting p27Kip1. J Biol Chem 283 (44): 29897–29903.
Mortensen RD, Serra M, Steitz JA, Vasudevan S (2011) Posttranscriptional activation of gene expression in Xenopus laevis oocytes by microRNA-protein complexes (microRNPs). Proc Natl Acad Sci USA 108 (20): 8281–8286.
Pyke C, Eriksen J, Solberg H, Nielsen BS, Kristensen P, Lund LR, Dano K (1993) An alternatively spliced variant of mRNA for the human receptor for urokinase plasminogen activator. FEBS Lett 326 (1-3): 69–74.
Radojicic J, Zaravinos A, Vrekoussis T, Kafousi M, Spandidos DA, Stathopoulos EN (2011) MicroRNA expression analysis in triple-negative (ER, PR and Her2/neu) breast cancer. Cell Cycle 10 (3): 507–517.
Shetty S (2005) Regulation of urokinase receptor mRNA stability by hnRNP C in lung epithelial cells. Mol Cell Biochem 272 (1-2): 107–118.
Shetty S, Velusamy T, Idell S, Shetty P, Mazar AP, Bhandary YP, Shetty RS (2007) Regulation of urokinase receptor expression by p53: novel role in stabilization of uPAR mRNA. Mol Cell Biol 27 (16): 5607–5618.
Stewart CE, Sayers I (2009) Characterisation of urokinase plasminogen activator receptor variants in human airway and peripheral cells. BMC Mol Biol 10: 75.
Stewart CE, Sayers I (2013) Urokinase receptor orchestrates the plasminogen system in airway epithelial cell function. Lung 191 (2): 215–225.
Tang J, Ahmad A, Sarkar FH (2012) The role of MicroRNAs in breast cancer migration, invasion and metastasis. Int J Mol Sci 13 (10): 13414–13437.
Tavazoie SF, Alarcon C, Oskarsson T, Padua D, Wang Q, Bos PD, Gerald WL, Massague J (2008) Endogenous human microRNAs that suppress breast cancer metastasis. Nature 451 (7175): 147–152.
Tran H, Maurer F, Nagamine Y (2003) Stabilization of urokinase and urokinase receptor mRNAs by HuR is linked to its cytoplasmic accumulation induced by activated mitogen-activated protein kinase-activated protein kinase 2. Mol Cell Biol 23 (20): 7177–7188.
Vasudevan S, Tong Y, Steitz JA (2007) Switching from repression to activation: microRNAs can up-regulate translation. Science 318 (5858): 1931–1934.
Veerla S, Lindgren D, Kvist A, Frigyesi A, Staaf J, Persson H, Liedberg F, Chebil G, Gudjonsson S, Borg A, Mansson W, Rovira C, Hoglund M (2009) MiRNA expression in urothelial carcinomas: important roles of miR-10a, miR-222, miR-125b, miR-7 and miR-452 for tumor stage and metastasis, and frequent homozygous losses of miR-31. Int J Cancer 124 (9): 2236–2242.
Wang L, Wang J (2012) MicroRNA-mediated breast cancer metastasis: from primary site to distant organs. Oncogene 31 (20): 2499–2511.
Yan LX, Huang XF, Shao Q, Huang MY, Deng L, Wu QL, Zeng YX, Shao JY (2008) MicroRNA miR-21 overexpression in human breast cancer is associated with advanced clinical stage, lymph node metastasis and patient poor prognosis. RNA 14 (11): 2348–2360.
Yoshimoto N, Toyama T, Takahashi S, Sugiura H, Endo Y, Iwasa M, Fujii Y, Yamashita H (2011) Distinct expressions of microRNAs that directly target estrogen receptor alpha in human breast cancer. Breast Cancer Res Treat 130 (1): 331–339.
Zhang CZ, Zhang JX, Zhang AL, Shi ZD, Han L, Jia ZF, Yang WD, Wang GX, Jiang T, You YP, Pu PY, Cheng JQ, Kang CS (2010) MiR-221 and miR-222 target PUMA to induce cell survival in glioblastoma. Mol Cancer 9: 229.
Zhao JJ, Lin J, Yang H, Kong W, He L, Ma X, Coppola D, Cheng JQ (2008) MicroRNA-221/222 negatively regulates estrogen receptor alpha and is associated with tamoxifen resistance in breast cancer. J Biol Chem 283 (45): 31079–31086.
Zheng C, Yinghao S, Li J (2012) MiR-221 expression affects invasion potential of human prostate carcinoma cell lines by targeting DVL2. Med Oncol 29 (2): 815–822.
Acknowledgements
The authors thank Katrin Lindner and Stefanie Winkler for their excellent technical assistance, and Professor V Magdolen for providing antibodies targeting the uPAS. This study was partly financed by the Wilhelm-Sander Stiftung (M.A. and M.S. 2012.028.1) and AW and MS gratefully acknowledge the financial support of the Deutsche Forschungsgemeinschaft (WA 1656/3-1).
Author's contributions
NF: participated in the conception and design, analysed and interpreted the data and drafted the manuscript; NA: designed, performed, analysed, interpreted the data and drafted the manuscript; KR: carried out the qRT–PCR experiments and participated in the analysis of the data; HB: performed statistical analyses of the data; GA: contributed the essential tumour material for the study and participated in interpretation of the data; AW: participated in the acquisition and interpretation of the tumour tissue data; MH and IH: participated in the in vitro assays and interpreted the data; MS and MJA: participated in the study supervision; HH: participated in the supervision and the design of the study; MA: conceived and supervised the study, analysed the tumour tissue data, interpreted the data and drafted the manuscript. All of the authors substantially contributed to the study, critically read and approved the final manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
Supplementary Information accompanies this paper on British Journal of Cancer website
Rights and permissions
From twelve months after its original publication, this work is licensed under the Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Falkenberg, N., Anastasov, N., Rappl, K. et al. MiR-221/-222 differentiate prognostic groups in advanced breast cancers and influence cell invasion. Br J Cancer 109, 2714–2723 (2013). https://doi.org/10.1038/bjc.2013.625
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/bjc.2013.625
Keywords
This article is cited by
-
Evaluating the effects of curcumin nano-chitosan on miR-221 and miR-222 expression and Wnt/β-catenin pathways in MCF-7, MDA-MB-231 and SKBR3 cell lines
Diagnostic Pathology (2024)
-
Prognostic value of miR-221 in human malignancy: evidence from 3041 subjects
BMC Cancer (2019)
-
HNRNPA2/B1 is upregulated in endocrine-resistant LCC9 breast cancer cells and alters the miRNA transcriptome when overexpressed in MCF-7 cells
Scientific Reports (2019)
-
Serum microRNA-21 predicted treatment outcome and survival in HER2-positive breast cancer patients receiving neoadjuvant chemotherapy combined with trastuzumab
Cancer Chemotherapy and Pharmacology (2019)
-
Changes in miR-221/222 Levels in Invasive and In Situ Carcinomas of the Breast: Differences in Association with Estrogen Receptor and TIMP3 Expression Levels
Molecular Diagnosis & Therapy (2016)