Abstract
Background:
Meningiomas are the most common primary intracranial tumours, with ∼3% meeting current histopathologic criteria for malignancy.
Methods:
In this study, we explored the transcriptome of meningiomas using RNA-Seq.
Results:
Inversion-mediated fusions between two adjacent genes, NAB2 and STAT6, were detected in one malignant tumour, creating two novel in-frame transcripts that were validated by RT–PCR and Sanger sequencing.
Conclusion:
Gene fusions of NAB2-STAT6 were recently implicated in the pathogenesis of solitary fibrous tumours; our study suggested that similar fusions may also have a role in a malignant meningioma with unusual histopathologic features.
Similar content being viewed by others
Main
Meningiomas are primary central nervous system tumours that originate from the meningeal coverings of the brain and spinal cord. They represent up to 30% of all primary intracranial tumours (Riemenschneider et al, 2006). Grades I, II and III meningiomas as classified by the World Health Organisation (WHO) are typically referred to as benign, atypical and anaplastic or malignant, respectively (Perry et al, 2004). Malignant meningiomas are typically associated with early recurrence, possible metastasis and decreased overall survival (Perry et al, 1999; Mawrin and Perry, 2010). Although multimodality therapy including maximal surgical debulking, radiation and chemotherapy are frequently used to treat malignant meningiomas, outcomes for this disease remain poor.
Genomic instability is associated with progression of meningiomas. Copy number variation was found to correlate with defined grades of tumours (Lee et al, 2010). Mutations of NF2, SMO, AKT1, SMARCE1, MEG3, TRAF7 and KLF4 genes have been identified in meningiomas (Christiaans et al, 2011; Balik et al, 2013; Brastianos et al, 2013; Clark et al, 2013; Smith et al, 2013). Although expression array-based technique has revealed novel genes associated with malignant transformation (Wrobel et al, 2005), this method is incapable of detecting novel gene fusions, which have been gradually recognised as potential players in cancer progression (Mitelman et al, 2007; Maher et al, 2009a).
In our study, we explored gene fusions in meningiomas using high-throughput RNA-Seq, which represented the first investigation in malignant and benign meningiomas. In one malignant sample, we identified a gene fusion event induced by local chromosomal inversion that was recently reported as a driver of neoplasia in solitary fibrous tumours (SFTs; Chmielecki et al, 2013; Robinson et al, 2013).
Materials and methods
The study was approved by the Institutional Review Board at the University of Southern California with informed written consent obtained for all participants. Patients with benign or malignant neoplasms of the brain or spine were eligible participants.
Total RNA was extracted from fresh-frozen meningioma tissue using RNeasy Kit (Qiagen, Mainz, Germany). The quality of total RNA was assessed using Experion RNA StdSens Chip (Bio-Rad, Hercules, CA, USA). TruSeq RNA Sample Prep Kits (Illumina, San Diego, CA, USA) were used for library preparation with 2 μg of total RNA as input and eight cycles of PCR amplification. The quality of libraries was assessed using High-Sensitivity DNA Chip (Agilent, Santa Clara, CA, USA). The bar-coded libraries were sequenced using the Illumina Hi-Seq 2000 platform to produce either 90-bp or 100-bp paired-end reads.
The FASTQ files were aligned to the UCSC hg19 using TopHat 2.0.4 (Trapnell et al, 2009). The aligned reads were processed using Cufflinks 2.0.0. The GTF annotation files for UCSC hg19 were used for gene annotation. Differential expression analysis was performed with Cuffdiff. The output results were further clustered and visualised using CummeRbund (www.compbio.mit.edu/cummeRbund/). Candidate gene fusions detected by both TopHat-Fusion (Kim and Salzberg, 2011) and Chimerascan (Maher et al, 2009b) were selected for analysis. All the parameters were default.
To validate fusion transcripts, 1 μg of total RNA was reversed transcribed to cDNA using M-MuLV first-strand cDNA synthesis kit (NEB, Ipswich, MA, USA) for subsequent amplification. To identify chromosomal inversion junctions, 100 ng of genomic DNA was used for PCR reactions. Primer pairs were designed using Primer3 and synthesised from Integrated DNA Technologies (IDT; San Diego, CA, USA). The KOD hot-start DNA polymerase (EMD, Billerica, MA, USA) was used for 30 cycles of amplification. Amplified products were purified from agarose gels using Qiagen MinElute kit for Sanger sequencing.
Results
Seven meningioma tissue samples (three malignant and four benign) were subject to RNA-Seq in two batches. The sequencing data indicated that the constructed libraries were mainly mRNA, with only 0.3% to 6.6% of the total mapped reads mapped to ribosomal RNA (Supplementary Table 1). Further transcriptome-based clustering presented in the dendrogram of Jensen–Shannon distances (Figure 1) showed that the four benign meningioma samples were readily separated from the malignant ones. As noted, the malignant sample 255 is separated from the other two malignant samples, confirming stronger genomic and transcriptomic heterogeneity among malignant samples.
RNA-Seq and validation for gene fusions. (A) Dendrogram for clustering RNA-Seq data. Malignant and benign meningiomas were labelled with M and B, respectively. (B) Gene fusions of NAB2-STAT6 and STAT6-NAB2 validated by RT–PCR and Sanger sequencing. The translated amino-acid sequences at the in-frame fusion junctions are also shown. (C) Gel electrophoresis of the amplified products.
Abnormal gene fusion events have been identified in a series of tumour types (Nowell, 1962; Tomlins et al, 2005; Chmielecki et al, 2013; Robinson et al, 2013). From RNA-Seq data, two gene fusions (NAB2-STAT6 and STAT6-NAB2) were identified in the malignant sample 255. Interestingly, gene fusions of NAB2-STAT6 have been reported in recurrent SFTs, including higher-grade tumours, which in the past were considered a separate entity called hemangiopericytoma (HPC; Chmielecki et al, 2013; Robinson et al, 2013). Sample 255 had an unusual histological appearance and immunophenotype with features of both meningioma and HPC (see Discussion below). The fusion of NAB2-STAT6 in our study is localised between the end of exon 6 of NAB2 and the beginning of exon 18 of STAT6, with 98 junction reads identified from RNA-Seq data (Supplementary Table 2). The fusion of STAT6-NAB2 was supported by 126 junction reads, which connects the last nucleotide of exon 17 of STAT6 with the first nucleotide of exon 7 of NAB2. In other meningioma samples, these two gene fusions were not detected from RNA-Seq data (in the malignant sample 010, one junction read was detected and was likely due to noise). Interestingly, the normally spliced junctions between exon 6 and exon 7 of NAB2 as well as between exon 17 and exon 18 of STAT6 were also detected in sample 255, supported by 99 and 57 junction reads, respectively. Thus, the gene fusions could result from genomic alteration in one of the two alleles or a subset of the tumour cells. The gene fusion detected by RNA-Seq technique was subsequently validated using RT–PCR and Sanger sequencing. Consistent with RNA-Seq data, the gene fusions of NAB2-STAT6 and STAT6-NAB2 were only detected in sample 255 by RT–PCR using junction spanning primers (Supplementary Table 3). Sanger sequencing (Figure 2) validated the breakpoints of mRNA fusions detected in next-generation sequencing data. As noted, the exons of NAB2 and STAT6 formed in-frame fusion products. Therefore, the gene fusions occurring at the mRNA level may have resulted in novel protein products.
Both NAB2 and STAT6 are located on 12q13.3 (Figure 3) and are transcribed in opposite directions. Gene fusions of NAB2-STAT6 and STAT6-NAB2 imply potential chromosomal inversion around the q13.3 locus. Based on the gene fusion junctions identified, we hypothesise that chromosomal inversion should cover the genomic regions of exon 7 of NAB2 as well as exons 18 to 23 of STAT6. Another two sets of primer pairs (Figure 3, Supplementary Table 4) were designed to detect chromosomal inversion junctions from PCR amplification of the genomic DNA. Of all seven meningioma samples used for RNA-Seq analysis, the chromosomal inversion junctions were only detected in sample 255. Additional Sanger sequencing of PCR products pinpointed the break-points at the non-coding regions of the genome.
A similar chromosomal inversion event was also reported in SFT/HPC, resulting in a novel transcript involving fusion of NAB2 to STAT6 and expression of the NAB2-STAT6 fusion protein (Robinson et al, 2013). The transgenic Flag-tagged fusion protein binds to the promoters of EGR1 targeting genes such as NAB1, NAB2, TGFB1, CASP9 and CEBPA (Robinson et al, 2013). In the gene fusion of NAB2-STAT6 detected in our study, the fused exons of STAT6 encoded the transcription activation domain of STAT6. Interestingly, gene expression of NAB1, NAB2, TGFB1 and CASP9 was upregulated in sample 255 compared with other samples (Figure 3), suggesting a functional role of the gene fusion product in activating transcription of these genes. On the other hand, gene fusion of STAT6-NAB2 is likely to produce a protein product incapable of transcription activation. Expression of the STAT6 downstream target gene BCL2L1 is downregulated (Figure 3), implying a potential functional effect of STAT6-NAB2 fusion. Thus, the gene fusions detected in our study may have roles in malignant transformation of meningiomas.
Discussion
Genomic instability is a characteristic of malignant tumours. In our study, we discovered two fusion transcripts resulting from chromosomal inversion within the NAB2-STAT6 locus. Under normal conditions, the tumour-suppressor NAB2 regulates EGR targeting genes, whereas the transcription activator STAT6 induces expression of BCL2L1. In the malignant meningioma found to harbour gene fusions, the transcription activation domain of STAT6 fused to NAB2 is likely to activate genes targeted by the NAB2-STAT6 fusion protein. With loss of the activation domain, the fusion of STAT6-NAB2 may affect downstream genes regulated by STAT6.
Examination of clinical information revealed that sample 255 detected with NAB2-STAT6 and STAT6-NAB2 fusions was the only patient sample collected after radiation treatment and/or chemotherapy. The novel gene fusions may therefore reflect the highly aggressive nature of this tumour or potentially a genomic effect of radiation and/or chemotherapy. Indeed, the DNA damage response proteins ATM and ATR were upregulated in sample 255. Therefore, we cannot exclude the possibility that radiation therapy may have induced DNA damage resulting in inversion-mediated gene fusion of NAB2 and STAT6.
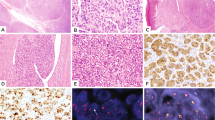
It is well known that it can be difficult in some instances to differentiate a SFT/HPC involving the dura from meningiomas, which can have many different histologic appearances, sometimes with features overlapping those of SFT/HPC. Fibroblastic meningioma and SFT may mimic one another. A highly atypical or malignant meningioma may lose its meningothelial appearance and become HPC-like displaying high cellularity, a high nuclear to cytoplasmic ratio and a patternless, sarcoma-like architecture. If a tumour does not have well-developed staghorn vessels that are characteristic of HPC then it may be difficult to decide whether it is an HPC or an unusual or higher-grade meningioma. Therefore, we performed a comprehensive survey of all the pathological records for this particular tumour sample 255 with fusion.
The tumour with fusion was initially diagnosed as a ‘mesenchymal neoplasm, favour meningiomas’ and was first excised in 1999. As a result of its HPC-like appearance, special stains and electron microscopy were performed. The special stains that have been most widely used to differentiate meningioma and SFT/HPC are epithelial membrane antigen (EMA) and CD34. Immunohistochemistry performed on the tissue from the first excision of the tumour in 1999 was positive for both EMA and CD34, and was confirmed by recent re-staining. However, neither marker is entirely specific: typical figures are that immunoreactivity for EMA is seen in 95% of meningiomas and 20–29% of meningeal SFT/HPC, yet immunoreactivity for CD34 is seen in all SFT, 60% of HPCs, 40% of fibrous meningiomas and 60% of atypical meningiomas (Hahn et al, 2006). Based on electron microscopy findings, the ultrastructural features were not well preserved and were not distinctive. Although not decisive, the consensus of three pathologists who reviewed this specimen in 1999 was that it was most likely an unusual meningioma. Despite atypical features, it did not meet the WHO (1997) criteria for atypical (grade II) meningioma. No mitotic figures were identified and immunohistochemistry for Ki-67 (MIB-1) stained only about 2% of tumour cell nuclei. The tumour did not recur until 2007 but required eight additional operations over the next 5 years. Over this 5-year period, the tumour appeared increasing aggressive, eventually showing >20 mitotic figures per 10 high-power field, a WHO (2007) criterion for anaplastic or malignant meningioma. Despite relentless local progression during the 5 years, the tumour never developed distant metastasis. Five different pathologists have looked at various samples of the tumour at different times and all favour that it is a meningioma, albeit with some uncertainty.
Over the past a few years, additional molecular diagnostic markers for meningiomas have been developed. We examined a meningioma-specific biomarker Claudin-1 (gene symbol CLDN1, Hahn et al, 2006), which is absent from all SFT but present in 53%–85% of meningiomas (Bhattacharjee et al, 2003; Hahn et al, 2006). RNA-Seq data revealed active transcription of CLDN1 in sample 255, with expression comparable to other meningioma samples (Supplementary Figure 1a). In parallel, we examined a recently generated Illumina whole-genome gene expression data set on meningiomas including sample 255, and confirmed that the expression is comparable to other meningiomas (Supplementary Figure 1b). Despite the diagnostic dilemmas occasionally encountered in differentiating meningioma from SFT/HPC, take together, these findings lead considerable support to this patient having a meningioma that underwent malignant transformation. This raised an interesting possibility that the presence of the fusion gene imparts a more mesenchymal phenotype on a meningioma, so that it has an unusual presentation and becomes difficult to be differentiated from SFT/HPC.
A recent study (Schweizer et al, 2013) explored gene fusion of NAB2-STAT6 by using strong nuclear staining of STAT6 protein as evidence for this gene fusion. Most of the 90 meningiomas showed negative, however, two tumours diagnosed as anaplastic meningioma and one as atypical meningioma were positive in immunohistochemical staining, indicating that they also carried the fusion. On review, they were found to have morphological features of both meningioma and HPC, and were recategorised as ‘mesenchymal tumour not classifiable’. Although further details of radiation treatment, EMA or CD34 status were not given, similar morphological features to sample 255 in our study suggest that they may represent a distinct type of meningeal tumour.
As a single case report, it is not possible to discern any causal relationships or even statistically significant associations between a particular gene fusion and malignancy in meningiomas. Further investigation with more samples is necessary to understand the cause of genome instability observed, and whether NAB2-STAT6 gene fusion is a cancer driver in malignant meningiomas, or simply cause a more mesenchymal phenotype.
Change history
20 August 2013
This paper was modified 12 months after initial publication to switch to Creative Commons licence terms, as noted at publication
References
Balik V, Srovnal J, Sulla I, Kalita O, Foltanova T, Vaverka M, Hrabalek L, Hajduch M (2013) MEG3: a novel long noncoding potentially tumour-suppressing RNA in meningiomas. J Neurooncol 112 (1): 1–8.
Bhattacharjee M, Adesina AM, Goodman C, Powell S (2003) Claudin-1 expression in meningiomas and schwannomas: possible role in differential diagnosis. J Neuropathol Exp Neurol 62: 581.
Brastianos PK, Horowitz PM, Santagata S, Jones RT, McKenna A, Getz G, Ligon KL, Palescandolo E, Van Hummelen P, Ducar MD, Raza A, Sunkavalli A, Macconaill LE, Stemmer-Rachamimov AO, Louis DN, Hahn WC, Dunn IF, Beroukhim R (2013) Genomic sequencing of meningiomas identifies oncogenic SMO and AKT1 mutations. Nat Genet 45 (3): 285–289.
Chmielecki J, Crago AM, Rosenberg M, O'Connor R, Walker SR, Ambrogio L, Auclair D, McKenna A, Heinrich MC, Frank DA (2013) Whole-exome sequencing identifies a recurrent NAB2-STAT6 fusion in solitary fibrous tumors. Nat Genet 45 (2): 131–132.
Christiaans I, Kenter SB, Brink HC, van Os TA, Baas F, van den Munckhof P, Kidd AM, Hulsebos TJ (2011) Germline SMARCB1 mutation and somatic NF2 mutations in familial multiple meningiomas. J Med Genet 48 (2): 93–97.
Clark VE, Erson-Omay EZ, Serin A, Yin J, Cotney J, Ozduman K, Avsar T, Li J, Murray PB, Henegariu O, Yilmaz S, Gunel JM, Carrion-Grant G, Yilmaz B, Grady C, Tanrikulu B, Bakircioglu M, Kaymakcalan H, Caglayan AO, Sencar L, Ceyhun E, Atik AF, Bayri Y, Bai H, Kolb LE, Hebert RM, Omay SB, Mishra-Gorur K, Choi M, Overton JD, Holland EC, Mane S, State NW, Bilguvar K, Baehring JM, Gutin PH, Piepmeier JM, Vortmeyer A, Brennan CW, Pamir MN, Kilic T, Lifton RP, Noonan JP, Yasuno K, Gunel M (2013) Genomic analysis of non-NF2 meningiomas reveals mutations in TRAF7, KLF4, AKT1, and SMO. Science 339 (6123): 1077–1080.
Hahn HP, Bundock EA, Hornick JL (2006) Immunohistochemical staining for claudin-1 can help distinguish meningiomas from histologic mimics. Am J Clin Pathol 125 (2): 203–208.
Kim D, Salzberg SL (2011) TopHat-Fusion: an algorithm for discovery of novel fusion transcripts. Genome Biol 12 (8): R72.
Lee Y, Liu J, Patel S, Cloughesy T, Lai A, Farooqi H, Seligson D, Dong J, Liau L, Becker D, Mischel P, Shams S, Nelson S (2010) Genomic landscape of meningiomas. Brain Pathol 20 (4): 751–762.
Maher CA, Kumar-Sinha C, Cao X, Kalyana-Sundaram S, Han B, Jing X, Sam L, Barrette T, Palanisamy N, Chinnaiyan AM (2009a) Transcriptome sequencing to detect gene fusions in cancer. Nature 458 (7234): 97–101.
Maher CA, Palanisamy N, Brenner JC, Cao X, Kalyana-Sundaram S, Luo S, Khrebtukova I, Barrette TR, Grasso C, Yu J, Lonigro RJ, Schroth G, Kumar-Sinha C, Chinnaiyan AM (2009b) Chimeric transcript discovery by paired-end transcriptome sequencing. Proc Natl Acad Sci USA 106 (30): 12353–12358.
Mawrin C, Perry A (2010) Pathological classification and molecular genetics of meningiomas. J Neurooncol 99 (3): 379–391.
Mitelman F, Johansson B, Mertens F (2007) The impact of translocations and gene fusions on cancer causation. Nat Rev Cancer 7 (4): 233–245.
Nowell PC (1962) The minute chromosome (Phl) in chronic granulocytic leukemia. Blut 8: 65–66.
Perry A, Gutmann DH, Reifenberger G (2004) Molecular pathogenesis of meningiomas. J Neurooncol 70 (2): 183–202.
Perry A, Scheithauer BW, Stafford SL, Lohse CM, Wollan PC (1999) ‘Malignancy’ in meningiomas: a clinicopathologic study of 116 patients, with grading implications. Cancer 85 (9): 2046–2056.
Riemenschneider MJ, Perry A, Reifenberger G (2006) Histological classification and molecular genetics of meningiomas. Lancet Neurol 5 (12): 1045–1054.
Robinson DR, Wu YM, Kalyana-Sundaram S, Cao X, Lonigro RJ, Sung YS, Chen CL, Zhang L, Wang R, Su F, Iyer MK, Roychowdhury S, Siddiqui J, Pienta KJ, Kunju LP, Talpaz M, Mosquera JM, Singer S, Schuetze SM, Antonescu CR, Chinnaiyan AM (2013) Identification of recurrent NAB2-STAT6 gene fusions in solitary fibrous tumor by integrative sequencing. Nat Genet 45 (2): 180–185.
Schweizer L, Koelsche C, Sahm F, Rosario PM, Capper D, Reuss DE, Pusch S, Habel A, Meyer J, Göck T, Jones DT, Mawrin C, Schittenhelm J, Becker A, Heim S, Simon M, Herold-Mende C, Mechtersheimer G, Paulus W, König R, Wiestler OD, Pfister SM, von Deimling A (2013) Meningeal hemangiopericytoma and solitary fibrous tumors carry the NAB2-STAT6 fusion and can be diagnosed by nuclear expression of STAT6 protein. Acta Neuropathologica 125 (5): 651–658.
Smith MJ, O'Sullivan J, Bhaskar SS, Hadfield KD, Poke G, Caird J, Sharif S, Eccles D, Fitzpatrick D, Rawluk D, du Plessis D, Newman WG, Evans DG (2013) Loss-of-function mutations in SMARCE1 cause an inherited disorder of multiple spinal meningiomas. Nat Genet 45 (3): 295–298.
Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, Varambally S, Cao X, Tchinda J, Kuefer R, Lee C, Montie JE, Shah RB, Pienta KJ, Rubin MA, Chinnaiyan AM (2005) Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science 310 (5748): 644–648.
Trapnell C, Pachter L, Salzberg SL (2009) TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25 (9): 1105–1111.
Wrobel G, Roerig P, Kokocinski F, Neben K, Hahn M, Reifenberger G, Lichter P (2005) Microarray-based gene expression profiling of benign, atypical and anaplastic meningiomas identifies novel genes associated with meningioma progression. Int J Cancer 114 (2): 249–256.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies this paper on British Journal of Cancer website
Supplementary information
Rights and permissions
From twelve months after its original publication, this work is licensed under the Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Gao, F., Ling, C., Shi, L. et al. Inversion-mediated gene fusions involving NAB2-STAT6 in an unusual malignant meningioma. Br J Cancer 109, 1051–1055 (2013). https://doi.org/10.1038/bjc.2013.395
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/bjc.2013.395
Keywords
This article is cited by
-
Somatic mutation landscape in a cohort of meningiomas that have undergone grade progression
BMC Cancer (2023)
-
Transcriptome signatures associated with meningioma progression
Acta Neuropathologica Communications (2019)
-
An integrated genomic analysis of anaplastic meningioma identifies prognostic molecular signatures
Scientific Reports (2018)