Abstract
Background:
Mutations in GNAQ and GNA11, encoding the oncogenic G-protein alpha subunit q and 11, respectively, occur frequently in the majority of uveal melanomas.
Methods:
Exons 4 and 5 from GNAQ and GNA11 were amplified and sequenced from 92 ciliary body and choroidal melanomas. The mutation status was correlated with disease-free survival (DFS) and other parameters.
Results:
None of the tumours harboured a GNAQ exon 4 mutation. A GNAQ mutation in exon 5 codon 209 was found in 46 out of 92 (50.0%) of the tumours. Only 1 out of 92 (1.1%) melanomas showed a mutation in GNA11 exon 4 codon 183, whereas 39 out of 92 (42.4%) harboured a mutation in exon 5 of GNA11 codon 209. Six tumours did not show any mutations in exons 4 and 5 of these genes. Univariate analyses showed no correlation between DFS and the mutation status.
Conclusion:
GNAQ and GNA11 mutations are, in equal matter, not associated with patient outcome.
Similar content being viewed by others
Main
Previous studies identified high frequencies of activating somatic mutations in the GNAQ and GNA11 genes in uveal melanoma (Onken et al, 2008; Bauer et al, 2009; Van Raamsdonk et al, 2009b, 2010). GNAQ and GNA11 encode the heterotrimeric guanine nucleotide-binding protein G subunit alpha q and 11, respectively. Mutations in GNAQ, or its paralog GNA11 (together referred to as Gα genes), occur mutually exclusively in codon 183 (exon 4) or 209 (exon 5), leading to a constitutive activation of the MAP kinase (MAPK) pathway (Van Raamsdonk et al, 2009a, 2010). Limited information is available on correlation of the mutations with survival. We examined to what extent oncogenic GNAQ and GNA11 mutations are correlated with the patient survival.
Materials and methods
Uveal melanomas were collected from enucleated patients at the Erasmus University Medical Centre and the Rotterdam Eye Hospital (Rotterdam, the Netherlands). Informed consent was obtained before the operation, and the study was performed according to the tenets of the Declaration of Helsinki. Fresh tumour material was obtained within 1 h of enucleation and processed for fluorescence in situ hybridisation as described previously (Kilic et al, 2005). Part of the tumour was snap-frozen and stored in liquid nitrogen. The remainder of the eye was embedded in paraffin. All tumours were histopathologically confirmed. Only tumours located in the ciliary body and choroid were included in this study. Fluorescence in situ hybridisation analysis was performed on directly fixated tumour cells for chromosome 1, 3, 6 and 8 using centromeric or locus-specific probes (Kilic et al, 2005). High-resolution whole-genome analysis was performed on tumour-derived DNA, using the Illumina BeadChip HumanCytoSNP-12 v2 (Illumina, San Diego, CA, USA) according to the manufacturer’s protocol. Filtering, normalisation and data analysis were done using version 6 of the Nexus software program (Biodiscovery, Inc., El Segundo, CA, USA). In total, 92 patients were selected for whom follow-up and clinical, histopathological, and cytogenetic data were available.
DNA isolation
To examine tumour content, H&E staining was conducted on a 5-μm section of snap-frozen tumour. Depending on the size of the tumour, 10–15 sections of 20 μm were used for DNA isolation using QIAmp DNA-mini kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. DNA concentration was measured with the NanoDrop ND-1000 Spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA).
GNAQ and GNA11 mutation analysis
In a previous study, our group performed mutation analysis of GNAQ exon 5 in 75 samples (Bauer et al, 2009). In the present study, we amplified GNAQ exon 5 in 17 other tumour samples with PCR using the primers 5′-ACCATTTTGCTTGGCACAGATAAGG-3′ and 5′-GTAAGTTCACTCCATTCCCCACACC-3′. GNAQ exon 4 and GNA11 exon 4 and 5 were amplified using the primers: 5′-TCTTTTTCTCCCACCCCTTGC-3′ and 5′-TTGTTTTGAAGCCTACACATGATTCC-3′ to examine GNAQ exon 4; 5′-GTGCTGTGTCCCTGTCCTG-3′ and 5′-GGCAAATGAGCCTCTCAGTG-3′ to examine GNA11 exon 4; and 5′-GATTGCAGATTGGGCCTTGG-3′ and 5′-TCTCCTCCATCCGGTTCTGG-3′ to examine GNA11 exon 5. PCR products were purified using ExoSAP-IT (USB, Staufen, Germany) and sequenced using BigDye Terminator chemistry v3.1 on an ABI Prism 3130xl Genetic Analyzer (Applied Biosystems, Foster City, CA, USA). Sequences were aligned and compared with reference sequence hg19 from the Ensemble genome database (ENST00000286548 and ENST00000078429) using SeqScape software version 2.6 (Applied Biosystems).
Statistical analysis
The primary end point for disease-free survival (DFS) was defined as the time to the development of metastatic disease, whereby death due to other causes was treated as censored. The influence of single prognostic factors on DFS was assessed using the Kaplan–Meier method (for categorical variables) or the Cox proportional hazard analysis (for continuous variables). To identify the independent value of the prognostic factors on DFS, we used a multivariate Cox proportional hazard analysis with a forward stepwise method based on likelihood ratios. An effect was considered significant if the P-value was ⩽0.05. The statistical analyses were performed with the SPSS software version 20.0 (SPSS Inc., Chicago, IL, USA).
Results
A total of 92 patients were included in the study. Forty-eight of the patients were male and 44 were female. The median age was 62 years (range 21–86); the mean largest tumour diameter was 13.3 mm (range 7.0–19.0) and the mean tumour thickness was 8.3 mm (range 1.5–22.0). On the basis of cell type, 15 tumours were classified as epithelioid, 38 as mixed, and 39 as spindle-cell tumours. Most tumours were localised in the choroid; only six were localised in the ciliary body. The mean follow-up was 74.9 months (range 5.2–200.5), and 44 patients developed metastases, from which 39 died. Sixteen patients died due to another cause, and 32 patients were still alive at the end of the study.
Molecular genetic analysis
All uveal melanomas were analysed for GNAQ and GNA11 mutations and for chromosomal aberrations in chromosome 1, 3, 6, and 8. No mutations were found in GNAQ exon 4. Forty-six tumours (50.0%) harboured a mutation in GNAQ exon 5 codon 209; details are shown in Table 1. Although only one mutated case was found in GNA11 exon 4, 39 tumours (42.4%) harboured a mutation in GNA11 exon 5. Six out of 92 tumours contained no mutations in exons 4 and 5 of both genes. One tumour (EOM-0179) showed two mutations in GNA11 exon 5 (resulting in p.Q209L and p.R214M). Tumour sample EOM-0179 was therefore subjected to deep sequencing with a custom-designed HaloPlex Target Enrichment kit for Illumina (Agilent Technologies, Santa Clara, CA, USA), and both variants were located within the same read (Koopmans et al, manuscript in preparation). No DNA from blood of this patient was available to determine whether variant R214M is a germline variant. Therefore, we isolated DNA from formalin-fixed paraffin-embedded retina tissue, and Sanger sequencing of GNA11 exon 5 revealed a wild-type status.
Statistical analysis
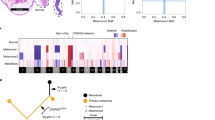
Univariate analyses showed that the DFS was significantly shorter in patients with tumours with loss of chromosome 3, loss of chromosome 8p and gain of chromosome 8q. The DFS in patients with tumours harbouring GNAQ or GNA11 mutations was not significantly less than that in the wild-type tumours (Figure 1). Correlations between the clinical and histopathological parameters, chromosomal parameters, and GNAQ and GNA11 mutations using the Fisher’s exact test and the Mann–Whitney test showed a weak association between age and both GNAQ and GNA11 mutation status (P=0.017 and 0.004, respectively; Table 2). GNA11 mutation status was also correlated with loss of chromosome 6q (P=0.045). We examined the possibility that GNAQ and GNA11 mutations may affect the prognosis of patients with monosomy 3 by constructing Kaplan–Meier curves for changes in chromosome 3, stratified for GNAQ and GNA11 mutations. Log-rank tests showed that there was no significant effect on the DFS in tumours with loss of chromosome 3 and the presence of GNAQ or GNA11 mutation (P=0.745). Multivariate models were constructed for GNAQ and GNA11 separately with positive variables from the univariate analysis. The presence of epithelioid cells, largest tumour diameter, involvement of the ciliary body, chromosome 3 loss, chromosome 8p loss, and mutations in GNAQ (P=0.587) or GNA11 (P=0.796) were rejected. Only the variable chromosome 8q gain (HR 6.562, P=0.000 for both GNAQ and GNA11 mutation status) and chromosome 6p gain (HR 0.419, P=0.014 for both GNAQ and GNA11 mutation status) were independent predictors of DFS.
Kaplan–Meier estimate of DFS in patients with tumours harbouring either a GNAQ or GNA11 mutation compared with tumours harbouring no mutation (wild type). The table shows the number of events and cases at risk over time at the respective time point. Log-rank test was used to compare survival distributions across subgroups.
Discussion
In this study, we investigated whether GNAQ and GNA11 mutations in uveal melanoma are associated with patient survival. We found that these mutations occur mutually exclusive in the majority of uveal melanomas, up to 93.4%, which is in the same range as reported previously (Onken et al, 2008; Bauer et al, 2009; Van Raamsdonk et al, 2009b, 2010). Van Raamsdonk et al (2010) suggested that GNA11 mutations might have more potent effect on melanocytes than mutations in GNAQ. Because the mutations occur in 93.4% of the tumours, it seems to be an early event in the development of a melanoma, and our study demonstrates that mutations in GNAQ and GNA11 do not contribute to the patients’ prognosis. Moreover, we conclude that GNA11 mutations are not more harmful than GNAQ mutations in uveal melanoma patients.
All mutations were localised either in codon 209 (exon 5) for both GNAQ and GNA11 or codon 183 (exon 4) for GNA11 only. Surprisingly, one tumour harboured a double mutation in GNA11 codons 209 and 214. The reported heterozygous non-synonymous variant in codon 214 results in arginine to methionine transition. A germline variant was excluded by sequencing normal retinal tissue. Using the in silico tool PolyPhen-2, both these transitions seem to be damaging on the structure and function of the protein. The tumour with the double mutation had no chromosomal alterations, and this patient has not developed any metastases at a follow-up time of 154.1 months. To our knowledge, this is the first reported double mutation in GNA11 exon 5 in uveal melanoma.
Recently, the Gα genes have been investigated in metastatic lesions, showing no difference in mutation frequency between rapidly progressive and slowly progressive lesions (Abdel-Rahman et al, 2012). This is in line with our findings that patient outcome is not influenced by the presence of mutations in GNAQ or GNA11.
GNAQ and GNA11 are involved in the MAPK pathway, and mutations in these genes lead to downstream oncogenic signalling (Van Raamsdonk et al, 2009a, 2010). Currently, new therapeutic strategies that inhibit the downstream signalling molecules are being investigated. MEK is a potential target in the MAPK pathway, and the effects of several MEK inhibitors on uveal melanoma cell lines with Gα mutations have been described (Mitsiades et al, 2011; von Euw et al, 2012). In a preclinical study, Gα-mutant uveal melanoma cells were mildly sensitive to the MEK inhibitor AZD6244, and either moderately or highly sensitive to the MEK inhibitor TAK733. Dual-pathway inhibition of the MAPK and the PI3K/AKT pathway with MEK inhibitor GSK1120212 and PI3K inhibitor GSK2126458 resulted in induction of apoptosis in Gα-mutant uveal melanoma cells (Khalili et al, 2012).
In conclusion, we confirm that mutations in GNAQ and GNA11 are, in equal matter, not associated with patient outcome. Also the newly found variant with a double mutation does not affect patient survival. Because the mutations occur in the majority of the tumours, and slowly growing as well as fast growing metastases, targeting of the downstream pathway seems promising. Even though there is no relation with development of metastatic disease, the new therapeutic options would be ideal in stabilising the disease process. At this moment, clinical studies are ongoing and the results have not yet been evaluated.
Change history
23 July 2013
This paper was modified 12 months after initial publication to switch to Creative Commons licence terms, as noted at publication
References
Abdel-Rahman MH, Cebulla CM, Verma V, Christopher BN, Carson WE 3rd, Olencki T, Davidorf FH (2012) Monosomy 3 status of uveal melanoma metastases is associated with rapidly progressive tumors and short survival. Exp Eye Res 100: 26–31.
Bauer J, Kilic E, Vaarwater J, Bastian BC, Garbe C, de Klein A (2009) Oncogenic GNAQ mutations are not correlated with disease-free survival in uveal melanoma. Br J Cancer 101: 813–815.
Khalili JS, Yu X, Wang J, Hayes BC, Davies MA, Lizee G, Esmaeli B, Woodman SE (2012) Combination small molecule MEK and PI3K inhibition enhances uveal melanoma cell death in a mutant GNAQ- and GNA11-dependent manner. Clin Cancer Res 18: 4345–4355.
Kilic E, Naus NC, van Gils W, Klaver CC, van Til ME, Verbiest MM, Stijnen T, Mooy CM, Paridaens D, Beverloo HB, Luyten GP, de Klein A (2005) Concurrent loss of chromosome arm 1p and chromosome 3 predicts a decreased disease-free survival in uveal melanoma patients. Invest Ophthalmol Vis Sci 46: 2253–2257.
Mitsiades N, Chew SA, He B, Riechardt AI, Karadedou T, Kotoula V, Poulaki V (2011) Genotype-dependent sensitivity of uveal melanoma cell lines to inhibition of B-Raf, MEK, and Akt kinases: rationale for personalized therapy. Invest Ophthalmol Vis Sci 52: 7248–7255.
Onken MD, Worley LA, Long MD, Duan S, Council ML, Bowcock AM, Harbour JW (2008) Oncogenic mutations in GNAQ occur early in uveal melanoma. Invest Ophthalmol Vis Sci 49: 5230–5234.
Van Raamsdonk CD, Barsh GS, Wakamatsu K, Ito S (2009a) Independent regulation of hair and skin color by two G protein-coupled pathways. Pigment Cell Melanoma Res 22: 819–826.
Van Raamsdonk CD, Bezrookove V, Green G, Bauer J, Gaugler L, O'Brien JM, Simpson EM, Barsh GS, Bastian BC (2009b) Frequent somatic mutations of GNAQ in uveal melanoma and blue naevi. Nature 457: 599–602.
Van Raamsdonk CD, Griewank KG, Crosby MB, Garrido MC, Vemula S, Wiesner T, Obenauf AC, Wackernagel W, Green G, Bouvier N, Sozen MM, Baimukanova G, Roy R, Heguy A, Dolgalev I, Khanin R, Busam K, Speicher MR, O’Brien J, Bastian BC (2010) Mutations in GNA11 in uveal melanoma. N Engl J Med 363: 2191–2199.
von Euw E, Atefi M, Attar N, Chu C, Zachariah S, Burgess BL, Mok S, Ng C, Wong DJ, Chmielowski B, Lichter DI, Koya RC, McCannel TA, Izmailova E, Ribas A (2012) Antitumor effects of the investigational selective MEK inhibitor TAK733 against cutaneous and uveal melanoma cell lines. Mol Cancer 11: 22.
Acknowledgements
We thank Farzia Fakhry for her contributions to the project. This study was supported by grants from the Combined Ophthalmic Research Rotterdam and the Stichting Nederlands Oogheelkundig Onderzoek.
Author information
Authors and Affiliations
Consortia
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
From twelve months after its original publication, this work is licensed under the Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Koopmans, A., Vaarwater, J., Paridaens, D. et al. Patient survival in uveal melanoma is not affected by oncogenic mutations in GNAQ and GNA11. Br J Cancer 109, 493–496 (2013). https://doi.org/10.1038/bjc.2013.299
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/bjc.2013.299
Keywords
This article is cited by
-
Natural killer cells drive 4-1BBL positive uveal melanoma towards EMT and metastatic disease
Journal of Experimental & Clinical Cancer Research (2024)
-
Clinical determinants of long-term survival in metastatic uveal melanoma
Cancer Immunology, Immunotherapy (2022)
-
Effect of oncolytic ECHO-7 virus strain Rigvir on uveal melanoma cell lines
BMC Research Notes (2020)
-
Primary sellar melanocytoma: pathological, clinical and treatment review
Journal of Endocrinological Investigation (2020)
-
Frequent and Yet Unreported GNAQ and GNA11 Mutations are Found in Uveal Melanomas
Pathology & Oncology Research (2019)