Abstract
Background:
Polymorphic variation at the 5p15.33 (TERT–CLPTM1L) locus is associated with the risk of many cancers but a relationship with colorectal cancer (CRC) risk has yet to be defined.
Methods:
We used data from six genome-wide association studies (GWAS) of CRC, linkage disequilibrium mapping and imputation, to examine the relationship between 73 single-nucleotide polymorphisms at 5p15.33 and CRC risk in detail.
Results:
rs2736100, which localises to intron 2 of TERT, provided the strongest evidence of an association with CRC (P=2.28 × 10−4). The association was also shown in an independent series of 10 047 CRC cases and 6918 controls (P=0.02). A meta-analysis of all seven studies (totalling 16 039 cases, 16 430 controls) provided increased evidence of association (P=2.49 × 10−5; per allele odds ratio=1.07). The association of rs2736100 on CRC risk was shown to be independent of 15 low-penetrance variants previously identified.
Conclusion:
The rs2736100 association demonstrates an influence of variation at 5p15.33 on CRC risk and further evidence that the 5p15.33 (TERT–CLPTM1L) locus has pleiotropic effects (reflecting generic or lineage-specific effects) on cancer risk.
Similar content being viewed by others
Main
Many colorectal cancers (CRCs) develop in genetically susceptible individuals most of whom are not carriers of germ-line mismatch repair or APC mutations (Lichtenstein et al, 2000; Aaltonen et al, 2007). It is likely that much of the unexplained heritable risk is attributable to a combination of multiple low-/moderate-penetrance genetic variants, which are associated with relatively small effects on risk in the individual but contribute substantially to the overall risk in the population (Fletcher and Houlston, 2010).
Genome-wide association studies (GWAS), using large sets of cases and controls, have proven to be an effective strategy to identify common single-nucleotide polymorphisms (SNPs) associated with cancer risk without prior knowledge of position or function (Fletcher and Houlston, 2010). This approach has successfully identified novel loci for most of the common cancers including CRC (Fletcher and Houlston, 2010). The majority of SNP associations identified to date have been tumour specific, which is consistent with the epidemiological studies of familial cancer risks (Fletcher and Houlston, 2010). Evidence for pleiotropic effects, reflecting generic or lineage-specific effects, is provided by variation at 5p15.33 (TERT–CLPTM1L) that is associated with the risk of many tumours including breast, testicular, bladder and lung cancers (McKay et al, 2008; Wang et al, 2008; Rafnar et al, 2009; Shete et al, 2009; Van Dyke et al, 2009; Hsiung et al, 2010; Turnbull et al, 2010; Beesley et al, 2011; Gago-Dominguez et al, 2011; Haiman et al, 2011; Kratz et al, 2011; Law et al, 2011; Peters et al, 2012).
Although many cancer associations at 5p15.33 have been identified with rs2736100, which localises to intron 2 of TERT (McKay et al, 2008; Wang et al, 2008; Shete et al, 2009; Hsiung et al, 2010; Turnbull et al, 2010; Gago-Dominguez et al, 2011), the existence of other SNP associations within the region supports the existence of multiple risk loci with different tumour specificities. Recently, an association between the SNP rs2853668, which maps centromeric to TERT but is only weakly correlated with rs2736100, has been reported for CRC risk (Peters et al, 2012). Given the ubiquitous necessity for tumours to avoid replicative senescence through shortened telomere repeat length, a process that is often mediated through expression of telomerase (Hanahan and Weinberg, 2000), a variant at TERT associated with CRC would be biologically plausible.
Using data from six GWAS of CRC, linkage disequilibrium (LD) mapping and imputation, we have studied the relationship between variation at 5p15.33 and CRC risk in detail. To further characterise the impact of 5p15.33 variation on CRC risk, we genotyped an additional 10 047 CRC cases and 6918 controls.
Materials and methods
Ethics
Collection of blood samples and clinico-pathological information from subjects was undertaken with informed consent and ethical review board approval at all sites in accordance with the tenets of the Declaration of Helsinki.
GWAS datasets
London 1 (LP1) comprised 940 cases with colorectal neoplasia (47% male) ascertained through the Colorectal Tumour Gene Identification (CoRGI) consortium. All had at least one first-degree relative affected by CRC and one or more of the following phenotypes: CRC at age 75 years or less; any colorectal adenoma (CRAd) at age 45 or less; ⩾3 colorectal adenomas at age 75 or less; or a large (>1 cm diameter) or aggressive (villous and/or severely dysplastic) adenoma at age 75 years or less. The 965 controls (45% males) were spouses or partners unaffected by cancer and without a personal family history (to second-degree relative level) of colorectal neoplasia. Known dominant polyposis syndromes, HNPCC/Lynch syndrome or bi-allelic MUTYH mutation carriers were excluded. All cases and controls had self-reported European ancestry. Both cases and controls were genotyped using Illumina HumanHap550 BeadChip Arrays (Teo et al, 2007).
Scotland1 (SP1) included 1012 CRC cases (51% male; mean age at diagnosis 49.6 years, s.d.±6.1) and 1012 cancer-free population controls (51% male; mean age 51.0 years; s.d.±5.9). Cases were selected for early age at onset (age ⩽55 years). Known dominant polyposis syndromes, HNPCC/Lynch syndrome or bi-allelic MUTYH mutation carriers were excluded. Control subjects were sampled from the Scottish population NHS registers, matched by age (±5 years), gender and area of residence within Scotland. Both cases and controls were genotyped using Illumina HumanHap300 and HumanHap240S arrays (Gunderson et al, 2006; Abraham et al, 2008).
VQ58 (VQ) comprised 1800 CRC cases (1099 males, mean age of diagnosis 62.5 years; s.d.±10.9) from the VICTOR and QUASAR2 (http://www.octo-oxford.org.uk/alltrials/infollowup/q2.html) trials. Genotyping of cases was conducted using Illumina Hap300 and Hap370 arrays (Gunderson et al, 2006; Howarth et al, 2009). The 2690 controls, typed on the Illumina Human 1.2M-Duo Custom_v1 Array BeadChips, were from the UK population-based 1958 birth cohort, for which genotype data are publicly available from the Wellcome Trust Case–Control Consortium 2 (Power and Elliott, 2006; The Wellcome Trust Case-Control Consortium, 2007).
The Colon Cancer Family Registry (CFR1) data set comprised 1290 familial CRC cases and 1055 controls CFR (Colon-CFR) (http://epi.grants.cancer.gov/CFR/about_colon.html). The cases were recently diagnosed CRC cases reported to population complete cancer registries in the United States (Puget Sound, WA, USA) recruited by the Seattle Familial Colorectal Cancer Registry; in Canada (ON) recruited by the Ontario Familial Cancer Registry; and in Australia (Melbourne, VIC) recruited by the Australasian Colorectal Cancer Family Study. Controls were population-based and for this analysis were restricted to those without a family history of CRC (Newcomb et al, 2007). Cases and controls were genotyped using Illumina HumanHap550 and 1M and 1Mduo BeadChip Arrays.
CFR2 comprised an additional 796 cases ascertained through the CFR (http://epi.grants.cancer.gov/CFR/about_colon.html). Cases were genotyped using 1M Omni-Express BeadChip Arrays. Illumina HumanHap550 BeadChip data on 2304 individuals from the Cancer Genetic Markers of Susceptibility (CGEMS) studies served as control genotypes (Hunter et al, 2007; Yeager et al, 2007).
The Australian (AUS) study (Tie et al, 2010) comprised 591 patients treated for CRC at the Royal Melbourne, Western and St Francis Xavier Cabrini Hospitals in Melbourne from 1999 to 2009. The 2353 controls were derived from Queensland or Melbourne: for the former, the controls came from the Brisbane Twin Nevus Study (Duffy et al, 2010); for the latter, individuals were participants in the Genes in Myopia study (Baird et al, 2010). There was no overlap between the CFR and Australian datasets. Both cases and controls were genotyped using Illumina HumanHap550 BeadChip Arrays.
Each of these six GWAS datasets was subjected to extensive quality control procedures. Specifically, the exclusion of samples and SNPs with call rates <95%, non-European (CEU) ancestry, relatedness (duplicates or related within or between each case–control series) and sex discrepancy. Furthermore, there was no evidence of systematic inflation of the test statistic in any study, as assessed using the genomic overdispersion factor, λGC, which ranged from 1.00 to 1.04.
Replication series
In total, 10 488 CRC cases, aged <80 years at diagnosis, were ascertained between March 2003 and October 2011 through the National Study of Colorectal Cancer Genetics (NSCCG) (Penegar et al, 2007) (n=9268); the Study of the Genetic Epidemiology of Colorectal Cancer (n=581) and the Royal Marsden Hospital National Health Services Trust (RMHNHST) family history DNA database (n=639). Controls (n=7137) were the spouses of cancer cases and were ascertained through the NSCCG (n=3047); the Genetic Lung Cancer Predisposition Study (n=1637); the Colorectal Adenoma Gene-Environment Interactions Study (n=711); the Study of the Genetic Epidemiology of Colorectal Cancer (n=344); and the RMHNHST family history DNA database (n=1398). None of the controls had a personal history of malignancy at ascertainment. All subjects were British residents with self-reported European ethnicity and there were no obvious demographic differences between cases and controls.
Statistical and bioinformatic analysis
A P-value (two-sided)⩽0.05 was considered significant. We applied a Bonferoni correction to adjust for multiple testing. Statistical analyses were undertaken using SNPtest/META (The Wellcome Trust Case-Control Consortium, 2007), and STATA v.10 (StataCorp LP, College Station, TX, USA) software. The association between each SNP and risk of CRC was assessed by the Cochran–Armitage trend test. Odds ratios and associated 95% confidence intervals (CIs) were calculated by unconditional logistic regression. Patterns of risk for associated SNPs were investigated by logistic regression, coding the SNP genotypes according to additive, dominant and recessive models. We then compared models by calculating the Akaike information criterion and Akaike weights for each mode of inheritance.
Interaction between SNP and genotypes was evaluated by likelihood ratio tests comparing an additive model to a model with an interaction term. Prediction of the non-genotyped SNPs within the 119.3-kb region of 5p15.33 (TERT–CLPTM1L) (1 243 475–1 362 793 bps, NCBI build b37) was carried out using IMPUTEv2 based on the June 2011 release of 1000 Genomes Project data (Howie et al, 2009, 2011; The 1000 Genomes Project Consortium, 2010). Association testing of genotyped and imputed imputed data were analysed using SNPTEST v2 to account for uncertainties in SNP prediction. Imputed genotypes were only called if they had a probability >0.90. Association meta-analyses only included markers with proper_info scores >0.9, imputed call rates/SNP>0.9 and Hardy–Weinberg>0.01. To condition by SNP, the SNP was added as a covariate. Meta-analysis was performed using a fixed-effects model, estimating Cochran’s Q statistic to test for heterogeneity and the I2 statistic to quantify the proportion of the total variation between studies.
Analysis of LD was performed using the Broad Institute SNP Annotation and Proxy (SNAP) Search utilising 1000 Genomes Project data. Transcription factor-binding prediction was performed with TFSearch. Cross-species evolutionary conservation was assessed with the deCode ECR browser. The UCSC genome browser was used to examine H3K4Me1, H3K4Me3 and DNase-I hypersensitivity in publicly available cell line data.
Association between rs2736100 and tumour site (colon-ICD International Classification of Diseases 9th revision (ICD9)-153; rectal cancer: ICD9-154), Dukes stage (A+B; C+D), grade (poorly; moderate/well differentiated), sex, age at diagnosis (⩽55, >55), family history of CRC in a first-degree relative and MSI status was evaluated by case-only analysis.
Molecular analysis
DNA was extracted from EDTA venous blood samples using conventional methodologies and PicoGreen quantified (Invitrogen Corporation, Carlsbad, CA, USA). We selected 15 SNPs that have been reported to be associated with CRC from 14 chromosomal regions: rs6691170 (1q41), rs10936599 (3q26.2), rs16892766 (8q23.3), rs6983267 (8q24.21), rs10795668 (10p14), rs3802842 (11q23.1), rs11169552 (12q13.13), rs4444235 (14q22.2), rs4779584 (15q13.33), rs11632715 (15q13.3) rs9929218 (16q22.1), rs4939827 (18q21.1), rs10411210 (19q13.1), rs961253 (20p12.3) and rs4925386 (20q13.33) (Tomlinson et al, 2007; Houlston et al, 2008; Jaeger et al, 2008; Tenesa et al, 2008; Tomlinson et al, 2008; Houlston et al, 2010; Tomlinson et al, 2011). Genotyping of these SNPs and rs2736100 was conducted using KASPar competitive allele-specific PCR chemistry (KBiosciences Ltd, Hoddesdon, UK; primer sequences and conditions available on request). To monitor quality control, duplicate samples were included in assays and a subset of samples sequenced. Concordance between duplicate samples was (>99%).
Tumour MSI status in CRCs was determined as described previously (Penegar et al, 2007) using the mononucleotide microsatellite loci BAT25 and BAT26, which are highly sensitive MSI markers. Briefly, 10-mm sections were cut from formalin-fixed paraffin-embedded CRC tumours, lightly stained with toluidine blue and regions containing at least 60% tumour microdissected. Tumour DNA was extracted using the QIAamp DNA Mini kit (Qiagen, Crawley, UK) according to the manufacturer’s instructions and genotyped for the BAT25 and BAT26 loci. Samples showing novel alleles, when compared with normal DNA, at either or both markers were assigned as MSI-H (corresponding to MSI-high) (Boland et al, 1998).
Results
Descriptive data
Table 1 provides summary information on the clinico-pathological characteristics and demographic information on each of the six GWAS datasets and the replication case–control series.
LD structure of the 5p15.33 region
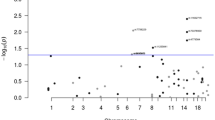
The six GWA studies of CRC provided genotype data for 12–45 SNPs (depending on study) mapping to the 119.3-kb region of 5p15.33 in a total of 6007 cases and 9520 controls (Figure 1). To further investigate the relationship between genetic variation at this region and CRC risk, using 1000 genomes data, we imputed the genotypes of 22 SNPs not directly genotyped in one or more studies and 45 SNPs not genotyped in any study. Including 6 SNPs directly genotyped in all studies, a total of 73 SNPs were imputed (Figure 1). In a combined analysis of these data, the strongest association was shown by the directly typed SNP, rs2736100 (P=2.28 × 10−4; Figure 1). The per allele OR of CRC associated with rs2736100-T genotype was 1.10 (95% CI:1.05–1.15). The association between rs2853668, a SNP previously reported to be associated with CRC(Peters et al, 2012), and CRC risk was non-significant (P=0.06). To explore the possibility of secondary associations with CRC, we conducted pairwise conditional analyses of the eight SNPs showing the best evidence for an association with risk. rs2736100 genotype was shown to be sufficient to capture the 5p15.33 association with CRC risk (Table 2).
Regional plot of SNP association with CRC across the 5p15.33 locus. Association results of both genotyped (triangles) and imputed (circles) SNPs in the GWAS samples and recombination rates. −log10 P-values (y-axis) of the SNPs are shown according to their chromosomal positions (x-axis). rs2736100 is represented by a large triangle. The grayscale intensity of each symbol reflects the extent of LD with rs2736100: white (r2=0) through to dark grey (r2=1.0). Genetic recombination rates (cM/Mb), estimated using HapMap CEU samples, are shown with a light grey line. Physical positions are based on NCBI build 37 of the human genome. Also shown are the relative positions of genes and transcripts mapping to each region of association. Genes have been redrawn to show the relative positions; therefore, maps are not to physical scale. For SNPs where r2 data were unavailable, these values were set to 0. The colour reproduction of this figure is available at British Journal of Cancer online.
Replication of the rs2736100 association
To provide further independent replication of the rs2736100 association with CRC risk, we genotyped an additional 10 488 cases and 7137 controls. Genotypes were obtained for 96% of cases (n=10 047) and 97% of controls (n=6918) (Table 1). There was no evidence of population stratification in controls as the genotype distribution satisfied Hardy–Weinberg equilibrium (P=0.76). As with the GWAS data there was a significant over-representation of the rs2736100-T genotype in CRC cases (P=0.019; Supplementary Table 1). Respective ORs of CRC associated with heterozygosity and homozygosity for rs2736100-T were 1.09 and 1.11, respectively, (Supplementary Table 1). The impact of rs2736100 genotype on CRC was thus comparable to that shown in the combined analysis of the six GWAS datasets. Although the pattern of risk for CRC was most parsimonious with a dominant model, a multiplicative model was equally favoured (P=0.27). To enhance our ability to demonstrate a relationship between 5p15.33 variation and CRC risk, we conducted a combined analysis of all datasets (Figure 2). In this meta-analysis, the per allele OR was 1.07 (95% CI: 1.04–1.11; P=2.49 × 10−5; Bonferroni-adjusted P-value was 1.82 × 10−3) and there was no evidence for between-study heterogeneity (Phet=0.7, I2=0%).
Forest plot of allelic odds ratio associated with rs2736100 genotype and CRC in the six genome-wide association studies and in the replication series. Horizontal lines represent 95% CIs. Each box represents the allelic OR point estimate, with the area being proportional to the weight of the study. The diamond (and broken line) denotes the overall summary estimate, with CIs given by its width. The unbroken vertical line is at the null value (OR=1.0).
Relationship between rs2736100 genotype and phenotype
To explore the relationship between rs2736100 genotype and CRC phenotype, we performed a case-only analysis using the replication series. This analysis provided no statistically significant evidence that the CRC association was modified by age, sex, or family history of CRC (Supplementary Table 1). For 3200 of the cases with known MSI status, rs2736100 genotypes were successfully generated on 2981 (93%), allowing us to calculate CRC risks stratified by MSI status. This analysis did not provide evidence for a relationship between SNP genotype and MSI status (Supplementary Table 1).
Interaction between rs2736100 and other common CRC risk variants
Using logistic regression analysis, we tested for an interaction between rs2736100 and each of 15 SNPs shown previously to be associated with CRC, namely, rs6691170, rs10936599, rs16892766, rs6983267, rs10795668, rs3802842, rs11169552, rs4444235, rs4779584, rs9929218, rs4939827, rs10411210, rs961253, rs4925386 and rs11632715. No evidence of statistical interaction between any of the 15 SNPs and rs2736100 was shown (Supplementary Table 2).
Discussion
Here we have demonstrated a statistically significant association between rs2736100 genotype and risk of CRC. We have also been able to show that variation at 5p15.33 influences CRC risk independently of other previously identified common risk variants. This is consistent with a model in which rs2736100 is acting additively with other common risk variants in mediating CRC susceptibility. As the risk allele of rs2736100 is common, the variant is likely to underscore ∼7% of all CRC in European populations.
Intriguingly rs2736100-T is associated with elevated risk of testicular cancer (Turnbull et al, 2010), but reduced risk of glioma (Shete et al, 2009), lung adenocarcinoma (McKay et al, 2008; Wang et al, 2008) and bladder cancer (Gago-Dominguez et al, 2011). These differential effects of genotype are likely to be reflective of tumour- and lineage-specific effect.
Although rs2736100 localises to intron 2 of TERT, this does not exclude the possibility of long-range effects as the functional basis for the 5p15.33 cancer association. However, although the 5p15.33 locus includes the TERT and CLPTM1L genes, these essentially map to two distinct regions of LD, making it likely rs2736100 impacts either directly or indirectly on TERT.
A recent study reported an association between the TERT SNP rs2853668 and CRC risk (Peters et al, 2012). In our study, rs2736100 provided superior evidence for an association with CRC than rs2853668. As rs2736100 and rs2853668 are correlated, albeit weakly (r2=0.15, D′=0.69), it is likely that the association reported by Peters et al, (2012) reflects the impact of rs2736100 genotype, or a hitherto unidentified correlated variant on CRC risk.
Functional variants in TERT have been shown to affect telomerase expression through modulating promoter activity (Beesley et al, 2011), and such a mechanism offers a possible explanation for how a putative functional variant at TERT affects CRC risk (Aisner, 2002). Although sequence conservation in non-coding regions has been shown to be a good predictor of cis-regulatory sequences (Gomez-Skarmeta et al, 2006), there is little evidence for high conservation directly at rs2736100 (Figure 3). ENCODE project data does not show evidence for DNAse hypersensitivity sites (indicating open chromatin), or histone H3KMe1/H3KMe3 methylation (often near regulatory elements) at rs2736100. Although these data do not support rs2736100 being directly functional, in an analysis of putative binding sites at rs2736100 using TFSearch, SRY and Hfh-2 sites are only predicted for rs2736100-T and not rs2736100-G. SRY (sex determining region Y) is a male-expressed gene involved in sex determination (Wallis et al, 2008). Although speculative, rs2736100-T-mediated SRY recruitment to TERT might lead to increased expression of telomerase in germ cells, thereby providing an explanation for the increased risk of testicular cancer associated with rs2736100-T (Turnbull et al, 2010). It is unknown whether Hfh-2 (also known as FOXD3) regulates telomerase expression, however, this forkhead transcription factor has a role in early cell development, thus suggesting another biological basis for the 5p15.33 association (Guo et al, 2002). Such speculations are predicated on the assumption that rs2736100 underscores the 5p15.33 association. Although our imputation provided no evidence for a stronger signal at 5p15.33 than that afforded by rs2736100, it is possible the association is mediated through one or more rare disease-causing variants, which are not adequately catalogued by the 1000 Genomes project data. High-depth coverage sequencing of a large series of CRC cases for 5p15.33 variation would allow this possibility to be explored.
Evolutionary conservation and chromatin status of a 14.8-kb region of 5p15.33 encompassing rs2736100. Regions of DNase-I hypersensitivity are shown by greyscale bars. Also shown are regions prone to H3K4Me1 or H3K4Me3 modifications, often found near regulatory elements and promoters respectively. Evolutionarily conserved regions (ECRs) between humans and chimpanzee (Pan troglodytes), mouse (Mus musculus) and fugu fish (Takifugu rubripes) are denoted by light grey horizontal lines relative to TERT (darkest grey, coding exons; lightest grey, untranslated regions; darker grey, intergenic regions; light grey, intronic regions; medium grey, transposons and simple repeats of the gene). The colour reproduction of this figure is available at British Journal of Cancer online.
In conclusion, our data demonstrate that polymorphic variation at 5p15.33 is a determinant of CRC risk. It has recently been shown that polymorphisms in TERC (telomerase RNA component) are associated with CRC risk and increased telomere length (Codd et al, 2010; Houlston et al, 2010; Jones et al, 2012); collectively these data extend the role of genetic variation in telomere elongation mechanisms in defining cancer risk per se.
Change history
18 August 2012
This paper was modified 12 months after initial publication to switch to Creative Commons licence terms, as noted at publication
References
Aaltonen L, Johns L, Jarvinen H, Mecklin JP, Houlston R (2007) Explaining the familial colorectal cancer risk associated with mismatch repair (MMR)-deficient and MMR-stable tumors. Clin Cancer Res 13: 356–361
Abraham R, Moskvina V, Sims R, Hollingworth P, Morgan A, Georgieva L, Dowzell K, Cichon S, Hillmer AM, O'Donovan MC, Williams J, Owen MJ, Kirov G (2008) A genome-wide association study for late-onset Alzheimer's disease using DNA pooling. BMC Medical Genomics 1: 44
Aisner D (2002) Telomerase regulation: not just flipping the switch. Curr Opin Genet Dev 12: 80–85
Baird PN, Schache M, Dirani M (2010) The GEnes in Myopia (GEM) study in understanding the aetiology of refractive errors. Prog Retin Eye Res 29: 520–542
Beesley J, Pickett HA, Johnatty SE, Dunning AM, Chen X, Li J, Michailidou K, Lu Y, Rider DN, Palmieri RT, Stutz MD, Lambrechts D, Despierre E, Lambrechts S, Vergote I, Chang-Claude J, Nickels S, Vrieling A, Flesch-Janys D, Wang-Gohrke S, Eilber U, Bogdanova N, Antonenkova N, Runnebaum IB, Dork T, Goodman MT, Lurie G, Wilkens LR, Matsuno RK, Kiemeney LA, Aben KK, Marees T, Massuger LF, Fridley BL, Vierkant RA, Bandera EV, Olson SH, Orlow I, Rodriguez-Rodriguez L, Cook LS, Le ND, Brooks-Wilson A, Kelemen LE, Campbell I, Gayther SA, Ramus SJ, Gentry-Maharaj A, Menon U, Ahmed S, Baynes C, Pharoah PD, kConFab Investigator, Australian Ovarian Cancer Study Group, Muir K, Lophatananon A, Chaiwerawattana A, Wiangnon S, Macgregor S, Easton DF, Reddel RR, Goode EL, Chenevix-Trench G, Ovarian Cancer Association Consortium (2011) Functional polymorphisms in the TERT promoter are associated with risk of serous epithelial ovarian and breast cancers. PLoS One 6: e24987
Boland CR, Thibodeau SN, Hamilton SR, Sidransky D, Eshleman JR, Burt RW, Meltzer SJ, Rodriguez-Bigas MA, Fodde R, Ranzani GN, Srivastava S (1998) A National Cancer Institute workshop on microsatellite instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res 58: 5248–5257
Codd V, Mangino M, van der Harst P, Braund PS, Kaiser M, Beveridge AJ, Rafelt S, Moore J, Nelson C, Soranzo N, Zhai G, Valdes AM, Blackburn H, Mateo Leach I, de Boer RA, Kimura M, Aviv A, Wellcome Trust Case Control Consortium, Goodall AH, Ouwehand W, van Veldhuisen DJ, van Gilst WH, Navis G, Burton PR, Tobin MD, Hall AS, Thompson JR, Spector T, Samani NJ (2010) Common variants near TERC are associated with mean telomere length. Nat Genet 42: 197–199
The 1000 Genomes Project Consortium (2010) A map of human genome variation from population-scale sequencing. Nature 467: 1061–1073
The Wellcome Trust Case-Control Consortium (2007) Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 447: 661–678
Duffy DL, Iles MM, Glass D, Zhu G, Barrett JH, Hoiom V, Zhao ZZ, Sturm RA, Soranzo N, Hammond C, Kvaskoff M, Whiteman DC, Mangino M, Hansson J, Newton-Bishop JA, Bataille V, Hayward NK, Martin NG, Bishop DT, Spector TD, Montgomery GW (2010) IRF4 variants have age-specific effects on nevus count and predispose to melanoma. Am J Hum Genet 87: 6–16
Fletcher O, Houlston RS (2010) Architecture of inherited susceptibility to common cancer. Nat Rev Cancer 10: 353–361
Gago-Dominguez M, Jiang X, Conti DV, Castelao JE, Stern MC, Cortessis VK, Pike MC, Xiang YB, Gao YT, Yuan JM, Van Den Berg DJ (2011) Genetic variations on chromosomes 5p15 and 15q25 and bladder cancer risk: findings from the Los Angeles-Shanghai bladder case-control study. Carcinogenesis 32: 197–202
Gomez-Skarmeta JL, Lenhard B, Becker TS (2006) New technologies, new findings, and new concepts in the study of vertebrate cis-regulatory sequences. Dev Dyn 235: 870–885
Gunderson KL, Kuhn KM, Steemers FJ, Ng P, Murray SS, Shen R (2006) Whole-genome genotyping of haplotype tag single nucleotide polymorphisms. Pharmacogenomics 7: 641–648
Guo Y, Costa R, Ramsey H, Starnes T, Vance G, Robertson K, Kelley M, Reinbold R, Scholer H, Hromas R (2002) The embryonic stem cell transcription factors Oct-4 and FoxD3 interact to regulate endodermal-specific promoter expression. Proc Natl Acad Sci USA 99: 3663–3667
Haiman CA, Chen GK, Vachon CM, Canzian F, Dunning A, Millikan RC, Wang X, Ademuyiwa F, Ahmed S, Ambrosone CB, Baglietto L, Balleine R, Bandera EV, Beckmann MW, Berg CD, Bernstein L, Blomqvist C, Blot WJ, Brauch H, Buring JE, Carey LA, Carpenter JE, Chang-Claude J, Chanock SJ, Chasman DI, Clarke CL, Cox A, Cross SS, Deming SL, Diasio RB, Dimopoulos AM, Driver WR, Dünnebier T, Durcan L, Eccles D, Edlund CK, Ekici AB, Fasching PA, Feigelson HS, Flesch-Janys D, Fostira F, Försti A, Fountzilas G, Gerty SM, Giles GG, Godwin AK, Goodfellow P, Graham N, Greco D, Hamann U, Hankinson SE, Hartmann A, Hein R, Heinz J, Holbrook A, Hoover RN, Hu JJ, Hunter DJ, Ingles SA, Irwanto A, Ivanovich J, John EM, Johnson N, Jukkola-Vuorinen A, Kaaks R, Ko YD, Kolonel LN, Konstantopoulou I, Kosma VM, Kulkarni S, Lambrechts D, Lee AM, Marchand LL, Lesnick T, Liu J, Lindstrom S, Mannermaa A, Margolin S, Martin NG, Miron P, Montgomery GW, Nevanlinna H, Nickels S, Nyante S, Olswold C, Palmer J, Pathak H, Pectasides D, Perou CM, Peto J, Pharoah PD, Pooler LC, Press MF, Pylkäs K, Rebbeck TR, Rodriguez-Gil JL, Rosenberg L, Ross E, Rüdiger T, Silva Idos S, Sawyer E, Schmidt MK, Schulz-Wendtland R, Schumacher F, Severi G, Sheng X, Signorello LB, Sinn HP, Stevens KN, Southey MC, Tapper WJ, Tomlinson I, Hogervorst FB, Wauters E, Weaver J, Wildiers H, Winqvist R, Van Den Berg D, Wan P, Xia LY, Yannoukakos D, Zheng W, Ziegler RG, Siddiq A, Slager SL, Stram DO, Easton D, Kraft P, Henderson BE, Couch FJ, Gene Environment Interaction and Breast Cancer in Germany (GENICA) Consortium (2011) A common variant at the TERT-CLPTM1L locus is associated with estrogen receptor-negative breast cancer. Nat Genet 43: 1210–1214
Hanahan D, Weinberg RA (2000) The hallmarks of cancer. Cell 100: 57–70
Houlston RS, Cheadle J, Dobbins SE, Tenesa A, Jones AM, Howarth K, Spain SL, Broderick P, Domingo E, Farrington S, Prendergast JGD, Pittman AM, Theodoratou E, Smith CG, Olver B, Walther A, Barnetson RA, Churchman M, Jaeger EEM, Penegar S, Barclay E, Martin L, Gorman M, Mager R, Johnstone E, Midgley R, Niittymaki I, Tuupanen S, Colley J, Idziaszczyk S, Thomas HJW, Lucassen AM, Evans DGR, Maher ER, Maughan T, Dimas A, Dermitzakis E, Cazier JB, Aaltonen LA, Pharoah P, Kerr DJ, Carvajal-Carmona LG, Campbell H, Dunlop MG, Tomlinson IPM, Consortium, C Grp, CC (2010) Meta-analysis of three genome-wide association studies identifies susceptibility loci for colorectal cancer at 1q41, 3q26.2, 12q13.13 and 20q13.33. Nat Genet 42: 973–U89
Houlston RS, Webb E, Broderick P, Pittman AM, Di Bernardo MC, Lubbe S, Chandler I, Vijayakrishnan J, Sullivan K, Penegar S, Carvajal-Carmona L, Howarth K, Jaeger E, Spain SL, Walther A, Barclay E, Martin L, Gorman M, Domingo E, Teixeira AS, Kerr D, Cazier JB, Niittymaki I, Tuupanen S, Karhu A, Aaltonen LA, Tomlinson IP, Farrington SM, Tenesa A, Prendergast JG, Barnetson RA, Cetnarskyj R, Porteous ME, Pharoah PD, Koessler T, Hampe J, Buch S, Schafmayer C, Tepel J, Schreiber S, Volzke H, Chang-Claude J, Hoffmeister M, Brenner H, Zanke BW, Montpetit A, Hudson TJ, Gallinger S, Campbell H, Dunlop MG (2008) Meta-analysis of genome-wide association data identifies four new susceptibility loci for colorectal cancer. Nat Genet 40: 1426–1435
Howarth K, Ranta S, Winter E, Teixeira A, Schaschl H, Harvey JJ, Rowan A, Jones A, Spain S, Clark S, Guenther T, Stewart A, Silver A, Tomlinson I (2009) A mitotic recombination map proximal to the APC locus on chromosome 5q and assessment of influences on colorectal cancer risk. BMC Med Genet 10: 54
Howie B, Marchini J, Stephens M (2011) Genotype imputation with thousands of genomes. G3: Genes Genomes Genet 1: 457–470
Howie BN, Donnelly P, Marchini J (2009) A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet 5: e1000529
Hsiung CA, Lan Q, Hong YC, Chen CJ, Hosgood HD, Chang IS, Chatterjee N, Brennan P, Wu C, Zheng W, Chang GC, Wu T, Park JY, Hsiao CF, Kim YH, Shen H, Seow A, Yeager M, Tsai YH, Kim YT, Chow WH, Guo H, Wang WC, Sung SW, Hu Z, Chen KY, Kim JH, Chen Y, Huang L, Lee KM, Lo YL, Gao YT, Kim JH, Liu L, Huang MS, Jung TH, Jin G, Caporaso N, Yu D, Kim CH, Su WC, Shu XO, Xu P, Kim IS, Chen YM, Ma H, Shen M, Cha SI, Tan W, Chang CH, Sung JS, Zhang M, Yang TY, Park KH, Yuenger J, Wang CL, Ryu JS, Xiang Y, Deng Q, Hutchinson A, Kim JS, Cai Q, Landi MT, Yu CJ, Park JY, Tucker M, Hung JY, Lin CC, Perng RP, Boffetta P, Chen CY, Chen KC, Yang SY, Hu CY, Chang CK, Fraumeni JF, Chanock S, Yang PC, Rothman N, Lin D (2010) The 5p15.33 locus is associated with risk of lung adenocarcinoma in never-smoking females in Asia. PLoS Genet 6: e1001051
Hunter DJ, Kraft P, Jacobs KB, Cox DG, Yeager M, Hankinson SE, Wacholder S, Wang Z, Welch R, Hutchinson A, Wang J, Yu K, Chatterjee N, Orr N, Willett WC, Colditz GA, Ziegler RG, Berg CD, Buys SS, McCarty CA, Feigelson HS, Calle EE, Thun MJ, Hayes RB, Tucker M, Gerhard DS, Fraumeni JF, Hoover RN, Thomas G, Chanock SJ (2007) A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat Genet 39: 870–874
Jaeger E, Webb E, Howarth K, Carvajal-Carmona L, Rowan A, Broderick P, Walther A, Spain S, Pittman A, Kemp Z, Sullivan K, Heinimann K, Lubbe S, Domingo E, Barclay E, Martin L, Gorman M, Chandler I, Vijayakrishnan J, Wood W, Papaemmanuil E, Penegar S, Qureshi M, Farrington S, Tenesa A, Cazier JB, Kerr D, Gray R, Peto J, Dunlop M, Campbell H, Thomas H, Houlston R, Tomlinson I, Consortium C (2008) Common genetic variants at the CRAC1 (HMPS) locus on chromosome 15q13.3 influence colorectal cancer risk. Nat Genet 40: 26–28
Jones AM, Beggs AD, Carvajal-Carmona L, Farrington S, Tenesa A, Walker M, Howarth K, Ballereau S, Hodgson SV, Zauber A, Bertagnolli M, Midgley R, Campbell H, Kerr D, Dunlop MG, Tomlinson IP (2012) TERC polymorphisms are associated both with susceptibility to colorectal cancer and with longer telomeres. Gut 61: 248–254
Kratz CP, Han SS, Rosenberg PS, Berndt SI, Burdett L, Yeager M, Korde LA, Mai PL, Pfeiffer R, Greene MH (2011) Variants in or near KITLG, BAK1, DMRT1, and TERT-CLPTM1L predispose to familial testicular germ cell tumour. J Med Genet 48: 473–476
Law MH, Montgomery GW, Brown KM, Martin NG, Mann GJ, Hayward NK, MacGregor S (2011) Meta-analysis combining new and existing data sets confirms that the TERT-CLPTM1L locus influences melanoma risk. J Invest Dermatol 132: 485–487
Lichtenstein P, Holm NV, Verkasalo PK, Iliadou A, Kaprio J, Koskenvuo M, Pukkala E, Skytthe A, Hemminki K (2000) Environmental and heritable factors in the causation of cancer-analyses of cohorts of twins from Sweden, Denmark, and Finland. N Engl J Med 343: 78–85
McKay JD, Hung RJ, Gaborieau V, Boffetta P, Chabrier A, Byrnes G, Zaridze D, Mukeria A, Szeszenia-Dabrowska N, Lissowska J, Rudnai P, Fabianova E, Mates D, Bencko V, Foretova L, Janout V, McLaughlin J, Shepherd F, Montpetit A, Narod S, Krokan HE, Skorpen F, Elvestad MB, Vatten L, Njolstad I, Axelsson T, Chen C, Goodman G, Barnett M, Loomis MM, Lubinski J, Matyjasik J, Lener M, Oszutowska D, Field J, Liloglou T, Xinarianos G, Cassidy A, Study E, Vineis P, Clavel-Chapelon F, Palli D, Tumino R, Krogh V, Panico S, Gonzalez CA, Ramon Quiros J, Martinez C, Navarro C, Ardanaz E, Larranaga N, Kham KT, Key T, Bueno-de-Mesquita HB, Peeters PH, Trichopoulou A, Linseisen J, Boeing H, Hallmans G, Overvad K, Tjonneland A, Kumle M, Riboli E, Zelenika D, Boland A, Delepine M, Foglio M, Lechner D, Matsuda F, Blanche H, Gut I, Heath S, Lathrop M, Brennan P (2008) Lung cancer susceptibility locus at 5p15.33. Nat Genet 40: 1404–1406
Newcomb PA, Baron J, Cotterchio M, Gallinger S, Grove J, Haile R, Hall D, Hopper JL, Jass J, Le Marchand L, Limburg P, Lindor N, Potter JD, Templeton AS, Thibodeau S, Seminara D (2007) Colon Cancer Family Registry: an international resource for studies of the genetic epidemiology of colon cancer. Cancer Epidemiol Biomarkers Prev 16: 2331–2343
Penegar S, Wood W, Lubbe S, Chandler I, Broderick P, Papaemmanuil E, Sellick G, Gray R, Peto J, Houlston R (2007) National study of colorectal cancer genetics. Br J Cancer 97: 1305–1309
Peters U, Hutter CM, Hsu L, Schumacher FR, Conti DV, Carlson CS, Edlund CK, Haile RW, Gallinger S, Zanke BW, Lemire M, Rangrej J, Vijayaraghavan R, Chan AT, Hazra A, Hunter DJ, Ma J, Fuchs CS, Giovannucci EL, Kraft P, Liu Y, Chen L, Jiao S, Makar KW, Taverna D, Gruber SB, Rennert G, Moreno V, Ulrich CM, Woods MO, Green RC, Parfrey PS, Prentice RL, Kooperberg C, Jackson RD, Lacroix AZ, Caan BJ, Hayes RB, Berndt SI, Chanock SJ, Schoen RE, Chang-Claude J, Hoffmeister M, Brenner H, Frank B, Bezieau S, Kury S, Slattery ML, Hopper JL, Jenkins MA, Le Marchand L, Lindor NM, Newcomb PA, Seminara D, Hudson TJ, Duggan DJ, Potter JD, Casey G (2012) Meta-analysis of new genome-wide association studies of colorectal cancer risk. Hum Genet 131: 217–234
Power C, Elliott J (2006) Cohort profile: 1958 British birth cohort (National Child Development Study). Int J Epidemiol 35: 34–41
Rafnar T, Sulem P, Stacey SN, Geller F, Gudmundsson J, Sigurdsson A, Jakobsdottir M, Helgadottir H, Thorlacius S, Aben KK, Blondal T, Thorgeirsson TE, Thorleifsson G, Kristjansson K, Thorisdottir K, Ragnarsson R, Sigurgeirsson B, Skuladottir H, Gudbjartsson T, Isaksson HJ, Einarsson GV, Benediktsdottir KR, Agnarsson BA, Olafsson K, Salvarsdottir A, Bjarnason H, Asgeirsdottir M, Kristinsson KT, Matthiasdottir S, Sveinsdottir SG, Polidoro S, Hoiom V, Botella-Estrada R, Hemminki K, Rudnai P, Bishop DT, Campagna M, Kellen E, Zeegers MP, de Verdier P, Ferrer A, Isla D, Vidal MJ, Andres R, Saez B, Juberias P, Banzo J, Navarrete S, Tres A, Kan D, Lindblom A, Gurzau E, Koppova K, de Vegt F, Schalken JA, van der Heijden HF, Smit HJ, Termeer RA, Oosterwijk E, van Hooij O, Nagore E, Porru S, Steineck G, Hansson J, Buntinx F, Catalona WJ, Matullo G, Vineis P, Kiltie AE, Mayordomo JI, Kumar R, Kiemeney LA, Frigge ML, Jonsson T, Saemundsson H, Barkardottir RB, Jonsson E, Jonsson S, Olafsson JH, Gulcher JR, Masson G, Gudbjartsson DF, Kong A, Thorsteinsdottir U, Stefansson K (2009) Sequence variants at the TERT-CLPTM1L locus associate with many cancer types. Nat Genet 41: 221–227
Shete S, Hosking FJ, Robertson LB, Dobbins SE, Sanson M, Malmer B, Simon M, Marie Y, Boisselier B, Delattre JY, Hoang-Xuan K, El Hallani S, Idbaih A, Zelenika D, Andersson U, Henriksson R, Bergenheim AT, Feychting M, Lonn S, Ahlbom A, Schramm J, Linnebank M, Hemminki K, Kumar R, Hepworth SJ, Price A, Armstrong G, Liu Y, Gu X, Yu R, Lau C, Schoemaker M, Muir K, Swerdlow A, Lathrop M, Bondy M, Houlston RS (2009) Genome-wide association study identifies five susceptibility loci for glioma. Nat Genet 41: 899–904
Tenesa A, Farrington SM, Prendergast JGD, Porteous ME, Walker M, Haq N, Barnetson RA, Theodoratou E, Cetnarskyj R, Cartwright N, Semple C, Clark AJ, Reid FJL, Smith LA, Kavoussanakis K, Koessler T, Pharoah PDP, Buch S, Schafmayer C, Tepel J, Schreiber S, Volzke H, Schmidt CO, Hampe J, Chang-Claude J, Hoffmeister M, Brenner H, Wilkening S, Canzian F, Capella G, Moreno V, Deary IJ, Starr JM, Tomlinson IPM, Kemp Z, Howarth K, Carvajal-Carmona L, Webb E, Broderick P, Vijayakrishnan J, Houlston RS, Rennert G, Ballinger D, Rozek L, Gruber SB, Matsuda K, Kidokoro T, Nakamura Y, Zanke BW, Greenwood CMT, Rangrej J, Kustra R, Montpetit A, Hudson TJ, Gallinger S, Campbell H, Dunlop MG (2008) Genome-wide association scan identifies a colorectal cancer susceptibility locus on 11q23 and replicates risk loci at 8q24 and 18q21. Nat Genet 40: 631–637
Teo YY, Inouye M, Small KS, Gwilliam R, Deloukas P, Kwiatkowski DP, Clark TG (2007) A genotype calling algorithm for the Illumina BeadArray platform. Bioinformatics 23: 2741–2746
Tie J, Gibbs P, Lipton L, Christie M, Jorissen RN, Burgess AW, Croxford M, Jones I, Langland R, Kosmider S, McKay D, Bollag G, Nolop K, Sieber OM, Desai J (2010) Optimizing targeted therapeutic development: Analysis of a colorectal cancer patient population with the BRAF(V600E) mutation. Int J Cancer 128: 2075–2084
Tomlinson I, Webb E, Carvajal-Carmona L, Broderick P, Kemp Z, Spain S, Penegar S, Chandler I, Gorman M, Wood W, Barclay E, Lubbe S, Martin L, Sellick G, Jaeger E, Hubner R, Wild R, Rowan A, Fielding S, Howarth K, Silver A, Atkin W, Muir K, Logan R, Kerr D, Johnstone E, Sieber O, Gray R, Thomas H, Peto J, Cazier B, Houlston R, Consortium C (2007) A genome-wide association scan of tag SNPs identifies a susceptibility variant for colorectal cancer at 8q24.21. Nat Genet 39: 984–988
Tomlinson IP, Carvajal-Carmona LG, Dobbins SE, Tenesa A, Jones AM, Howarth K, Palles C, Broderick P, Jaeger EE, Farrington S, Lewis A, Prendergast JG, Pittman AM, Theodoratou E, Olver B, Walker M, Penegar S, Barclay E, Whiffin N, Martin L, Ballereau S, Lloyd A, Gorman M, Lubbe S, Howie B, Marchini J, Ruiz-Ponte C, Fernandez-Rozadilla C, Castells A, Carracedo A, Castellvi-Bel S, Duggan D, Conti D, Cazier JB, Campbell H, Sieber O, Lipton L, Gibbs P, Martin NG, Montgomery GW, Young J, Baird PN, Gallinger S, Newcomb P, Hopper J, Jenkins MA, Aaltonen LA, Kerr DJ, Cheadle J, Pharoah P, Casey G, Houlston RS, Dunlop MG (2011) Multiple common susceptibility variants near BMP pathway loci GREM1, BMP4, and BMP2 explain part of the missing heritability of colorectal cancer. PLoS Genet 7: e1002105
Tomlinson IP, Webb E, Carvajal-Carmona L, Broderick P, Howarth K, Pittman AM, Spain S, Lubbe S, Walther A, Sullivan K, Jaeger E, Fielding S, Rowan A, Vijayakrishnan J, Domingo E, Chandler I, Kemp Z, Qureshi M, Farrington SM, Tenesa A, Prendergast JG, Barnetson RA, Penegar S, Barclay E, Wood W, Martin L, Gorman M, Thomas H, Peto J, Bishop DT, Gray R, Maher ER, Lucassen A, Kerr D, Evans DG, Schafmayer C, Buch S, Volzke H, Hampe J, Schreiber S, John U, Koessler T, Pharoah P, van Wezel T, Morreau H, Wijnen JT, Hopper JL, Southey MC, Giles GG, Severi G, Castellvi-Bel S, Ruiz-Ponte C, Carracedo A, Castells A, Forsti A, Hemminki K, Vodicka P, Naccarati A, Lipton L, Ho JW, Cheng KK, Sham PC, Luk J, Agundez JA, Ladero JM, de la Hoya M, Caldes T, Niittymaki I, Tuupanen S, Karhu A, Aaltonen L, Cazier JB, Campbell H, Dunlop MG, Houlston RS (2008) A genome-wide association study identifies colorectal cancer susceptibility loci on chromosomes 10p14 and 8q23.3. Nat Genet 40: 623–630
Turnbull C, Rapley EA, Seal S, Pernet D, Renwick A, Hughes D, Ricketts M, Linger R, Nsengimana J, Deloukas P, Huddart RA, Bishop DT, Easton DF, Stratton MR, Rahman N, Collaboration, UKTC (2010) Variants near DMRT1, TERT and ATF7IP are associated with testicular germ cell cancer. Nat Genet 42: 604–607
Van Dyke AL, Cote ML, Wenzlaff AS, Abrams J, Land S, Iyer P, Schwartz AG (2009) Chromosome 5p region SNPs are associated with risk of NSCLC among women. J Cancer Epidemiol 2009: 242151
Wallis MC, Waters PD, Graves JA (2008) Sex determination in mammals-before and after the evolution of SRY. Cell Mol Life Sci 65: 3182–3195
Wang Y, Broderick P, Webb E, Wu X, Vijayakrishnan J, Matakidou A, Qureshi M, Dong Q, Gu X, Chen WV, Spitz MR, Eisen T, Amos CI, Houlston RS (2008) Common 5p15.33 and 6p21.33 variants influence lung cancer risk. Nat Genet 40: 1407–1409
Yeager M, Orr N, Hayes RB, Jacobs KB, Kraft P, Wacholder S, Minichiello MJ, Fearnhead P, Yu K, Chatterjee N, Wang Z, Welch R, Staats BJ, Calle EE, Feigelson HS, Thun MJ, Rodriguez C, Albanes D, Virtamo J, Weinstein S, Schumacher FR, Giovannucci E, Willett WC, Cancel-Tassin G, Cussenot O, Valeri A, Andriole GL, Gelmann EP, Tucker M, Gerhard DS, Fraumeni JF, Hoover R, Hunter DJ, Chanock SJ, Thomas G (2007) Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat Genet 39: 645–649
Acknowledgements
Cancer Research UK provided principal funding for this study individually to RSH. (C1298/A8362 – Bobby Moore Fund for Cancer Research UK), IPMT and MGD. At the Institute of Cancer Research additional funding was provided a Centre grant from CORE as part of the Digestive Cancer Campaign, the National Cancer Research Network and the NHS via the Biological Research Centre of the National Institute for Health Research at the Royal Marsden Hospital NHS Trust. BK and NW are in receipt of PhD Studentships from the Institute of Cancer Research, with BK receiving sponsorship from the Sir John Fisher Foundation. OS was supported by the Hilton Ludwig Cancer Metastasis Initiative. In Oxford, additional funding was provided by the Oxford Comprehensive Biomedical Research Centre. Core infrastructure support to the Wellcome Trust Centre for Human Genetics, Oxford, was provided by grant (090532/Z/09/Z). In Edinburgh funding was provided by a Cancer Research UK Programme Grant (C348/A12076) and a Centre Grant from the CORE Charity. Funding from the MRC and the Scottish Government Chief Scientist Office is also gratefully acknowledged. This work of the CFR was supported by the National Cancer Institute, National Institutes of Health under RFA #CA-95-011 and through cooperative agreements with members of the Colon CFR and Principal Investigators. Collaborating centres include the Australasian Colorectal Cancer Family Registry (U01 CA097735), Familial Colorectal Neoplasia Collaborative Group (U01 CA074799), Ontario Registry for Studies of Familial Colorectal Cancer (U01 CA074783) and the Seattle Colorectal Cancer Family Registry (U01 CA074794). The Colon CFR GWAS was supported by funding from the National Cancer Institute, National Institutes of Health (U01CA122839 to GC). This study made use of genotyping data from the 1958 Birth Cohort and NBS samples, kindly made available by the Wellcome Trust Case Control Consortium 2. A full list of the investigators who contributed to the generation of the data is available at http://www.wtccc.org.uk/. We would like to thank all individuals who participated in the study. We are grateful to many colleagues within UK Clinical Genetics Departments (for CORGI) and to many collaborators who participated in the VICTOR and QUASAR2 trials. We also thank colleagues from the UK National Cancer Research Network (for NSCCG). We are grateful to all the subjects for participating in this study.
Web-addresses
1000genomes: http://www.1000genomes.org/
IMPUTE: http://mathgen.stats.ox.ac.uk/impute/
SNPTEST: https://mathgen.stats.ox.ac.uk/genetics_software/snptest/
META: http://www.stats.ox.ac.uk/~jsliu/meta.html
SNAP http://www.broadinstitute.org/mpg/snap/
R project: http://www.r-project.org/
dbSNP: http://www.ncbi.nlm.nih.gov/projects/SNP/
TFSearch: http://www.cbrc.jp/research/db/TFSEARCH.html
deCode ECR Browser: http://ecrbrowser.dcode.org/
UCSC genome browser: http://genome.ucsc.edu/
Author information
Authors and Affiliations
Consortia
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
Supplementary Information accompanies the paper on British Journal of Cancer website
Supplementary information
Rights and permissions
From twelve months after its original publication, this work is licensed under the Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Kinnersley, B., Migliorini, G., Broderick, P. et al. The TERT variant rs2736100 is associated with colorectal cancer risk. Br J Cancer 107, 1001–1008 (2012). https://doi.org/10.1038/bjc.2012.329
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/bjc.2012.329
Keywords
This article is cited by
-
TERT Gene rs2736100 and rs2736098 Polymorphisms are Associated with Increased Cancer Risk: A Meta-Analysis
Biochemical Genetics (2022)
-
A genetic variant in telomerase reverse transcriptase (TERT) modifies cancer risk in Lynch syndrome patients harbouring pathogenic MSH2 variants
Scientific Reports (2021)
-
Genetic variant predictors of gene expression provide new insight into risk of colorectal cancer
Human Genetics (2019)
-
Telomere length and genetics are independent colorectal tumour risk factors in an evaluation of biomarkers in normal bowel
British Journal of Cancer (2018)
-
Meta-analysis of genome-wide association studies identifies common susceptibility polymorphisms for colorectal and endometrial cancer near SH2B3 and TSHZ1
Scientific Reports (2015)