Abstract
Background:
The CCAAT/enhancer binding protein delta (CEBPδ) is a member of a highly conserved family of basic region leucine zipper transcription factors. It has properties consistent with a tumour suppressor; however, other data suggest that CEBPδ may be involved in the metastatic process.
Methods:
We analysed the expression of CEBPδ and the methylation status of the CpG island in human breast cancer cell lines, in 107 archival cases of primary breast cancer and in two series of metastatic breast cancers using qPCR and pyrosequencing.
Results:
Expression of CEBPδ is downregulated in primary breast cancer by site-specific methylation in the CEBPδ CpG island. Expression is also downregulated in 50% of cases during progression from primary carcinoma to metastatic lesions. The CEBPδ CpG island is methylated in 81% metastatic breast cancer lesions, while methylation in the CEBPδ CpG island in primary cancers is associated with increased risk of relapse and metastasis.
Conclusion:
CCAAT/enhancer binding protein delta CpG island methylation is associated with metastasis in breast cancer. Detection of methylated CEBPδ genomic DNA may have utility as an epigenetic biomarker of primary breast carcinomas at increased risk of relapse and metastasis.
Similar content being viewed by others
Main
The CCAAT/enhancer binding proteins (CEBPs) are a highly conserved family of basic region leucine zipper (bZip) transcription factors, and comprises six family members (CEBPα to CEBPζ) (Ramji and Foka, 2002). The CEBP proteins exhibit significant amino acid homology (>90%) in the bZip (C-terminal) domain, while the N-terminal regions are quite divergent exhibiting <20% sequence homology (Ramji and Foka, 2002). The CEBP form homo- or heterodimers with each other as well as other bZip-containing proteins such as Jun and Fos (Vinson et al, 2002), with the dyad symmetrical repeat RTTGCGYAAY (where R is A or G, and Y is C or T) considered to be the optimal CEBP binding site (Osada et al, 1997). The CEBPδ unlike other family members lacks an activation domain and, therefore, represses gene transcription by forming inactive heterodimers with other members (Cooper et al, 1995). The CEBP family is involved in a number of key cellular processes including differentiation, metabolism, inflammation, apoptosis and proliferation (Wang et al, 1995; Yamanaka et al, 1997; Zinszner et al, 1998; Robinson et al, 1998).
CCAAT/enhancer binding protein delta has been proposed to have tumour suppressor function given its ability to decrease levels of cyclin D1 and cyclin E, while increasing p27 (Gery et al, 2005; Ikezoe et al, 2005; Pawar et al, 2010), as well as regulating proapoptotic gene expression during mammary gland involution (Thangaraju et al, 2005; Stein et al, 2009). Treatments in vitro, which induce growth arrest such as serum and growth factor withdrawal, increase CEBPδ expression and induce growth arrest in breast cancer cell lines as well as in human mammary epithelial cells (O’Rourke et al, 1997; Sivko and DeWille, 2004). However, in vivo loss of CEBPδ results in increased mammary epithelial cell proliferation and ductal hyperplasia, supporting the importance of CEBPδ in regulating mammary epithelial growth in vivo (Gigliotti et al, 2003). These data are supported by the reduction observed in CEBPδ expression in mammary tumour-prone MMTV-c-neu transgenic mice and in carcinogen-induced rodent mammary tumours (Porter et al, 2001; Kuramoto et al, 2002). Further evidence that CEBPδ is a tumour suppressor comes from animal data using mice with a germ-line deletion of CEBPδ (on a MMTV-c-neu background), with these animals developing significantly more breast tumours compared with controls (Balamurugan et al, 2010). Interestingly, in the context of this mouse knockout model absence of CEBPδ resulted in less efficient metastasis under hypoxia, implying that the protein may be required for metastasis at least under these conditions (Balamurugan et al, 2010).
CCAAT/enhancer binding protein delta is downregulated via methylation in cervical cancer, hepatocellular carcinoma and AML (Agrawal et al, 2007; Ko et al, 2008). CCAAT/enhancer binding protein delta protein expression also correlates with low-grade histology and disease-free survival in meningiomas (Barresi et al, 2009). CCAAT/enhancer binding protein delta has also been shown to be downregulated in ductal carcinoma in situ as compared to normal breast tissue (Porter et al, 2003). In a series of primary human breast cancers, CEBPδ mRNA levels were very low in 32% (18 out of 57) of cases, and in those cases with low mRNA levels, and this was associated with CpG methylation in the CEBPδ gene promoter and 5′ coding region (Tang et al, 2006). CEBPδ also formed part of 70-gene signature, which predicted better survival of breast cancer patients (Naderi et al, 2007).
To date, there have been no reports regarding the involvement of CEBPδ in metastasis in human cancer, nor of the utility of CEBPδ as a prognostic biomarker in breast cancer. Here we have performed quantitative analysis of CEBPδ CpG island methylation to test these possibilities.
Materials and Methods
Breast cancer cell lines
Breast carcinoma cell lines SKBR3, MDA-MB231, MDA-MB 453, MDA-MB468, MDA-MB 435, MCF7, T47D, ZR75.1, HCC1937, HS578 were grown as described previously (Shah et al, 2009).
Two series of cases were analysed in the study. The first was 107 primary breast carcinomas. The characteristics of this patient population are shown in Table 1. These cases were randomly selected from the tissue archives of S. Croce General Hospital, Cuneo, Italy. For all 107 cases, genomic DNA was available and was analysed by pyrosequencing for CEBPδ CpG island methylation. For 26 of the 107 cases, mRNA was available and was used to analyse CEBPδ expression by qPCR. At the time of the study, metastatic relapse had occurred in 31 of the 107 patients. For 14 of 31 relapsed cases, tissue from the metastasis was available and was analysed in parallel with matched tissue from the primary cancer for CEBPδ expression. The second series comprised 21 central nervous system (CNS) metastatic lesions from Imperial Healthcare NHS Trust. These cases were identified from the neuropathology archives at Charing Cross Hospital, London. Tissue was originally obtained at neurosurgical resection of intracranial disease in patients with a pre-existing diagnosis of breast cancer and was confirmed by histopathology to be metastatic breast cancer. Genomic DNA from this series was analysed by pyrosequencing for CEBPδ CpG island methylation. The study received ethical committee approval in both centres. In all cases, the original diagnosis and adequate representation of neoplastic tissue was confirmed by histopathological review prior to inclusion in the study. Expression of the oestrogen receptor (ER), the progesterone receptor and HER2 was determined according to local protocols.
Analysis of CEBPδ expression
Total RNA was extracted from formalin-fixed paraffin-embedded (FFPE) tissue using Recover All kit (Ambion, Carlsbad, CA, USA). cDNA was synthesised from 1 μg total RNA using the High Capacity cDNA Reverse Transcription kit (Applied Biosystems, Foster City, CA, USA). For demethylation, cells were treated with 5 μ M 5′azacytidine (Sigma, Gillingham, UK) for 7 days. Cells were split every 2–3 days with the addition of fresh drug. After drug treatment, cells were harvested for qPCR. For qPCR analysis, 25-μl PCRs were performed using 50 ng of cDNA obtained by reverse transcription. Amplification and analysis were done according to the manufacturer’s protocol in 96-well plates in an ABI PRISM 7000 Sequence Detection System (Applied Biosystems) and using the pre-cast ‘TaqMan Gene Expression Assays’ (Applera, https://products.appliedbiosystems.com/) for CEBPδ (Hs00270931_s1). Quantification of target transcripts was performed in comparison to the reference transcript β2-microglobulin (Hs99999907_m1) using the ‘ΔΔCt method for comparing relative expression results in real-time PCR’ as outlined by PE Applied Biosystems (Perkin Elmer, Forster City, CA, USA).
Pyrosequencing
Genomic DNA was extracted from cellular pellets and FFPE sections using the DNeasy Mini kit (Qiagen, Crawley, UK) according to the manufacturer’s instructions, and from 10 micron sections of FFPE using phenol with the traditional protocol. Methylation in the CpG island of CEBPδ was analysed by pyrosequencing technology, which allows the quantification of the degree of methylation at each CG site through the calculation of the ratio between T and C. PCR primers were as follows:
Forward: BIOT-5′-GGAGTGTTGGTAGAGGGAG-3′
Reverse: 5′-CCCTAAAAACCCCCAACCC-3′.
The PCR conditions were as follows: 95°C for 10 min, 95°C for 30 s, 58°C for 30 s, 72°C for 40 s for 40 cycles, 72°C for 7 min. The PCR products were then analysed by pyrosequencing using the Sample Prep kit (Diatech, Jesi, Italy).
After pyrosequencing, analysis of percentage methylation at each CG was determined using Pyromark Q CpG Software (Qiagen, Venlo, The Netherland). DNA from five normal breast samples and placental DNA were used as a negative control for methylation (0% average methylation), and a commercial methylated DNA (Millipore, Billerica, MA, USA) was used as positive control (98% average methylation).
Statistical analysis
The CEBPδ CpG island methylation status and presence of metastatic profile were assessed for associations using the χ2-test, with Yates correction or Fisher exact test when appropriate. All the comparisons are two-tailed.
Results
Site-specific CpG methylation correlates with silencing of CEBPδ in breast cancer cell lines
We analysed expression and epigenetic regulation of CEBPδ in a panel of breast carcinoma cell lines. Because a previous report has identified methylation-associated downregulation of CEBPδ in the SUM-52PE breast carcinoma cell line (Tang et al, 2006), we were interested to further test and characterise the relationship between expression of CEBPδ mRNA and methylation in breast carcinoma cell lines. We wished to use a fully quantitative analytical technique rather than methylation-specific PCR (MSP) and we therefore used pyrosequencing to analyse a section of the CEBPδ CpG island (Figure 1). In breast cancer cell lines, methylation was predominantly but not exclusively seen at CG 5 in the fragment analysed by pyrosequencing (Figure 1). CCAAT/enhancer binding protein delta mRNA was detectable in many cell lines. Expression was highest in HCC1937 and ZR75.1, but was downregulated relative to normal breast cells in several cell lines (Figure 1). There was a good correlation between methylation at CG 5 of the analysed fragment and downregulation of the mRNA expression (Figure 1).
Downregulation of CEBPδ mRNA in breast carcinoma cell lines correlates with site-specific CpG methylation in the CEBPδ CpG island. (A) Expression of CEBPδ in breast carcinoma cell lines. qPCR was performed as described in Materials and methods. (B) Site-specific CpG island methylation in breast carcinoma cell lines. The percentage methylation at CG 5, as determined by pyrosequencing, is indicated. (C) Map of CpG methylation in the CEBPδ CpG island in breast cancer cell lines. Pyrosequencing was performed as described in Materials and methods. The level of methylation is represented by the intensity of shading in the circles, each of which represents an individual CG dinucleotide in the amplified fragment.
CEBPδ is downregulated in primary breast carcinomas compared with normal breast tissue
Next we sought to investigate whether there is downregulation of CEBPδ mRNA in clinical cases of breast cancer, and we performed qPCR in 26 primary breast carcinomas. CCAAT/enhancer binding protein delta mRNA was reduced relative to normal breast tissue in many cases (Figure 2A). Expression was reduced most strikingly in the series of primary cancers, which later relapsed in comparison to cases which did not relapse: downregulation by at least 50% compared with normal breast epithelium was observed in 1 out of 7 non-relapsing cases and 11 out of 19 relapsing cases (compare upper and lower panels in Figure 2A).
Expression of CEBPδ is downregulated in clinical cases of breast cancer. (A) Expression of CEBPδ in primary breast carcinomas. The figure shows mRNA levels determined by qPCR as described in Materials and Methods. was calculated as described in Materials and Methods relative to CEBPδ expression in control normal breast tissue (arrowed). The upper panel shows expression in cases which had not relapsed at the time of censor, the lower panel shows cases which had relapsed at the time of censor. (B) Expression of CEBPδ is frequently downregulated in metastatic breast cancer. The figure shows 14 paired primary/metastasis cases. In each case, expression in the primary breast cancer (open box) is set at 1 and expression in the metastasis (black box) is relative to this. Expression is downregulated in cases 3, 6, 7, 8, 10, 12 and 13.
Downregulation of CEBPδ in metastatic breast cancer lesions
We then analysed expression of CEBPδ in metastatic breast cancer lesions. We examined a series of 14 cases comprising the primary breast carcinoma together with the paired metastasis, which had been confirmed by histopathology. Clinico-pathological details and sites of metastasis for each pair are shown in Table 2. Using qPCR, we analysed expression of CEBPδ. In 7 out of 14 (50%) cases, we observed a significant reduction in CEBPδ mRNA in the metastasis relative to the primary cancer, consistent with selective pressure for loss of CEBPδ expression with acquisition of a metastatic phenotype in breast cancer (Figure 2B).
CEBPδ CpG island methylation predicts breast cancer relapse
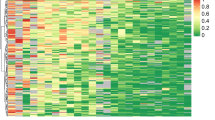
We next analysed a series of 107 cases of primary breast cancer from the same patient population to determine whether analysis of CpG island methylation in CEBPδ has utility as a biomarker predictive of clinical relapse. Clinico-pathological details of the study population are shown in Table 1. Representative analyses showing the distribution of methylation in the amplified area of the CpG island are shown in Figure 3. Consistent with breast carcinoma cell lines, methylation at individual CG dinucleotides was variable. In the clinical cases, methylation was most dense in CG 4–7 in the analysed fragment (Figure 3A), CGs 2 and 3 being almost entirely unmethylated in all cases. Methylation correlated well with reduced expression of CEBPδ mRNA (Figure 3B). The distribution of CEBPδ CpG island methylation between cases relapsing with metastatic disease and non-relapsing cases is shown in Figure 4. At the time of censor, 29% (31 of 107) of cases had relapsed. By using a mean percentage CpG methylation cutoff of 8%, as determined by pyrosequencing, relapse was significantly more frequent in cases in which the CEBPδ CpG island was positive for methylation (P=0.0006 by Fisher’s Exact test; P=0.001 with Yates correction).
Methylation-associated transcriptional silencing of CEBPδ in primary breast carcinomas. (A) Representative pyrosequencing analyses of the CEBPδ CpG island in primary breast carcinomas. The upper 12 cases were non-relapsing, the lower 12 cases later relapsed at distant metastatic sites. The level of methylation is represented by the intensity of shading in the circles, each of which represents an individual CG dinucleotide in the amplified fragment as indicated in the figure. (B) Association of CEBPδ CpG island methylation with downregulation of CEBPδ mRNA levels. Expression of CEBPδ was determined by qPCR and CpG methylation by pyrosequencing as described in Materials and Methods. Cases are divided into those with mean % CG methylation below (<) or above (>) 8. Also shown is expression in normal breast epithelium (arrowed).
CEBPδ CpG island methylation is increased in primary breast carcinomas which subsequently relapse. The figure shows distribution of CEBPδ CpG island methylation in cases which at the time of censor had either relapsed or not relapsed. Mean percentage CpG methylation, determined by pyrosequencing, is shown in relapsing and non-relapsing cases.
CEBPδ CpG island methylation is associated with metastatic breast cancer
We had previously shown that expression of CEBPδ is downregulated in metastatic breast cancer lesions (Figure 2B). We wished to test whether CEBPδ CpG island methylation is associated with increased risk of distant organ metastasis in breast cancer and we asked whether there was an association between CEBPδ CpG island methylation and metastasis at specific organ sites. Metastases in liver (P=0.01), lymph node (P=0.02) and skin (P=0.02) were more common in cases in which the primary cancer was positive for methylation (using a mean percentage CpG methylation cutoff of 8% as determined by pyrosequencing). In contrast, metastases in bone and lung were not significantly affected by the methylation status of the CEBPδ CpG island (P=0.13 and P=0.24, respectively). The frequency of brain metastases was higher in cases in which the primary cancer was positive for CEBPδ CpG island methylation, but this just failed to reach significance (P=0.06) due to the small number of cases with brain metastases. To extend these observations, we analysed CEBPδ CpG island methylation by pyrosequencing in a series of 21 CNS metastases, confirmed by histopathology to be derived from primary breast carcinomas (Figure 5). As observed previously, methylation was most dense at CG 4 and CG 5 and was detected in 81% (17 out of 21) of cases (Figure 5). For three of the cases, the paired breast cancer primary was also available to us. In one of the three cases, there was a change in methylation in the CNS metastasis in comparison with the primary, with acquisition of methylation at CGs 4, 5 and 6 in the metastasis (Figure 5).
CEBPδ CpG island methylation in breast cancer central nervous system (CNS) metastases. The figure shows pyrosequencing analysis of the CEBPδ CpG island in CNS metastatic lesions, confirmed by histopathology to be metastatic breast cancer. Also shown (top) are three paired primary/CNS metastatic breast cancers. Pyrosequencing was done as described in Materials and methods. The level of methylation is represented by the intensity of shading in the circles, each of which represents an individual CG dinucleotide in the amplified fragment.
Discussion
We have investigated the expression and regulation of CEBPδ in breast cancer. We show that the gene is a frequent target for downregulation in primary breast carcinomas, as a result of methylation in the CpG island. Furthermore, we demonstrate that methylation in CEBPδ is associated with metastasis and that methylation, when analysed with high-resolution, quantitative methodology may have utility as a biomarker predictive of future metastatic relapse. We were initially interested to investigate this gene because there is experimental evidence that CEBPδ has tumour suppressor properties (Porter et al, 2001; Kuramoto et al, 2002) and yet, at least in some animal models, may be involved in metastasis (Balamurugan et al, 2010). The data we present are consistent with and supportive of a tumour suppressor and metastasis suppressor function in human breast cancer.
We initially studied expression in a panel of established breast carcinoma cell lines and demonstrated that expression was reduced in several of the cell lines. Some studies of candidate epigenetically regulated biomarker genes use non-quantitative, low-resolution techniques such as methylation-dependent PCR (MSP) to analyse methylation. Here we have used pyrosequencing that allows high-resolution quantification of percentage methylation at individual CG dinucleotides within a defined section of the CpG island. We observed using pyrosequenicng that methylation at CG5 in the analysed region of the CpG island showed a good correlation with transcriptional silencing in a panel of breast cancer cell lines. Our data showing correlation between methylation and downregulation of expression in both breast cancer cell lines and primary carcinomas are supported by other studies in hepatocellular carcinoma (Ko et al, 2008).
We then extended these initial studies to investigate the possible role of downregulation of CEBPδ in breast cancer metastasis. Several lines of evidence from our studies support loss of CEBPδ expression as a determinant of metastasis. First, analysis of paired primary/metastatic lesions showed clear downregulation in the metastases in 50% of cases. Very few studies have specifically examined changes in expression of individual genes during metastasis in breast cancer. Our data are clearly consistent with selective pressure for loss of CEBPδ during acquisition of a metastatic phenotype. Second, we have shown that the presence of methylation in the CEBPδ CpG island in primary breast carcinomas is associated with an increased risk of relapse and of distant organ metastasis. Third, we show that the CEBPδ CpG island is frequently methylated in CNS metastases shown to originate in primary breast carcinomas. Further we have also shown herein, albeit in limited numbers of cases, that methylation in CEBPδ may be acquired during metastasis to the CNS, consistent with epigenetic evolution as cells acquire metastatic properties. Methylation of the CEBPδ CpG island as an important event in breast cancer metastasis is consistent with a previous report, implicating downregulation of CEBPδ (among other genes) in breast carcinoma cell lines with increased propensity for CNS metastasis (Bos et al, 2009).
Our current data in early breast cancer are consistent with CEBPδ being a tumour suppressor (Gery et al, 2005; Ikezoe et al, 2005; Balamurugan et al, 2010; Pawar et al, 2010). However, our data reporting fewer metastasis when CEBPδ is not methylated are at face value at odds with the in vivo data, where loss of CEBPδ is associated with fewer metastasis (Balamurugan et al, 2010). Possible explanations for the difference include the fact that the study of Balamurugan et al (2010) is from an animal experimental system, whereas the present data are derived from human breast cancer samples. Furthermore, the model used was in a HER2 overexpressing background (Guy et al, 1992), while in the current series only 13% of cases were HER2 positive. In addition, only data relating to lung metastasis were reported, with no data with regard to the effect of CEBPδ on involvement of other common sites for metastasis or overall metastatic tumour burden. It is known within the context of human breast cancer that HER2-positive breast cancer not only has a predilection to metastasis to the lung but also to the brain and liver (Kennecke et al, 2010). Therefore, the phenotype seen may be specific to the animal model in question. Furthermore, the underlying mechanism proposed for the effect observed in vivo was related to hypoxic HIF-1α accumulation and hypoxia adaptation. As such, these conditions may therefore be prerequisite for the effect observed and may in be part dependent on HER2 (Balamurugan et al, 2010).
It should be noted, of course, that CEBPδ is not the only gene contributing to a metastatic profile. The current data show that methylation of CEBPδ in the primary tumour (using a mean percentage CpG methylation cutoff of 8% as determined by pyrosequencing) is associated with metastasis in the liver, lymph node and skin, with metastases in bone and lung not being significantly influenced by the methylation status of CEBPδ. Multiple additional genes must be at play and we have previously shown the importance of one such candidate CACNA2D3 in the metastatic process (Palmieri et al, 2012).
The patient population analysed in our study consisted predominantly of ER-positive cases treated with adjuvant endocrine therapy. A key question in the management of early breast cancer continues to be risk stratification to identify patients likely to relapse despite being deemed to be at low risk by clinico-pathological parameters such as those in the St Gallen criteria. Such patients could then be offered appropriate systemic therapy such as adjuvant chemotherapy. While others could be spared such treatment if their risk could objectively be determined to be low. We have shown herein that methylation in the CEBPδ CpG island correlates with a significantly increased risk of metastatic relapse at distant organ sites including brain and liver. Our data imply that CEBPδ may have utility as a biomarker predictive of relapse and metastasis and thus in the identification of patients who may derive greater benefit from adjuvant treatment.
In conclusion, we show that transcriptional silencing of CEBPδ is associated with metastasis in breast cancer. Validation of these results in larger, independent cohorts of patients is required and if positive would encourage the inclusion of CEBPδ in molecular risk assessment algorithms to inform the use of adjuvant therapy in breast cancer.
Change history
30 July 2012
This paper was modified 12 months after initial publication to switch to Creative Commons licence terms, as noted at publication
References
Agrawal S, Hofmann WK, Tidow N, Ehrich M, van den Boom D, Koschmieder S, Berdel WE, Serve H, Müller-Tidow C (2007) The C/EBPdelta tumor suppressor is silenced by hypermethylation in acute myeloid leukemia. Blood 109: 3895–3905
Balamurugan K, Wang JM, Tsai HH, Sharan S, Anver M, Leighty R, Sterneck E (2010) The tumour suppressor C/EBPδ inhibits FBXW7 expression and promotes mammary tumour metastasis. EMBO J 29: 4106–4117
Barresi V, Vitarelli E, Cerasoli S, Barresi G (2009) The cell growth inhibitory transcription factor C/EBPdelta is expressed in human meningiomas in association with low histological grade and proliferation index. J Neurooncol 97: 233–240
Bos PD, Zhang XH, Nadal C, Shu W, Gomis RR, Nguyen DX, Minn AJ, van der Vijver MJ, Gerald WL, Foekens JA, Massague J (2009) Genes that mediate breast cancer metastasis to the brain. Nature 459: 1005–1009
Cooper C, Henderson A, Artandi S, Avitahl N, Calame K (1995) Ig/EBP(C/EBPδ) is a transdominant negative inhibitor of C/EBP family of transcriptionalactivators. Nucleic Acids Res 23: 4371–4377
Gery S, Tanosaki S, Hofmann WK, Koppel A, Koeffler HP (2005) C/EBPdelta expression in a BCR-ABL-positive cell line induces growth arrest and myeloid differentiation. Oncogene 24: 1589–1597
Gigliotti AP, Johnson PF, Sterneck E, DeWille JW (2003) Nulliparous CCAAT/ enhancer binding proteindelta (C/EBPdelta) knockout mice exhibit mammary gland ductal hyperlasia. Exp Biol Med 228: 278–285
Guy CT, Webster MA, Schaller M, Parsons TJ, Cardiff RD, Muller WJ (1992) Expression of the neu protooncogene in the mammary epithelium of transgenic mice induces metastatic disease. Proc Natl Acad Sci USA 89: 10578–10582
Ikezoe T, Gery S, Yin D, O’Kelly J, Binderup L, Lemp N, Taguchi H, Koeffler HP (2005) CCAAT/enhancer-binding protein delta: a molecular target of 1,25-dihydroxyvitaminD3 in androgen-responsive prostate cancer LNCaP cells. Cancer Res 2005 65 4762–4768
Kennecke H, Yerushalmi R, Woods R, Cheang MC, Voduc D, Speers CH, Nielsen TO, Gelmon K (2010) Metastatic behavior of breast cancer subtypes. J Clin Oncol 28: 3271–3277
Ko C-Y, Hsu H-C, Shen M-R, Chang W-C, Wang J-M (2008) Epigenetic silencing of CCAAT Enhancer-binding Protein d activity by YY1/Polycomb Group/DNA methyltransferase complex. J Biol Chem 283: 30919–30932
Kuramoto T, Morimura K, Yamashita S, Okochi E, Watanabe N, Ohta T, Ohki M, Fukushima S, Sugimura T, Ushijima T (2002) Etiology-specific gene expression profiles in rat mammary carcinomas. Cancer Res 62: 3592–3597
Naderi A, Teschendorff AE, Barbosa-Morais NL, Pinder SE, Green AR, Powe DG, Robertson JF, Aparicio S, Ellis IO, Brenton JD, Caldas C (2007) A gene-expression signature to predict survival in breast cancer across independent data sets. Oncogene 26: 1507–1516
O’Rourke J, Yuan R, DeWille J (1997) CCAAT/enhancer-binding protein delta (C/EBP-delta) is induced in growth-arrested mouse mammary epithelial cells. J Biol Chem 272: 6291–6296
Osada S, Yamamoto H, Nishihara T, Imagwa M (1997) DNA binding specifcity of the CCAAT/enhancer-binding protein transcription factor family. J Biol Chem 271: 3891–3896
Palmieri C, Rudraraju B, Monteverde M, Lattanzio L, Gojis O, Brizio R, Garrone O, Merlano M, Syed N, Lo Nigro C, Crook T (2012) Methylation of the calcium channel regulatory subunit α2δ-3 (CACNA2D3) predicts site-specific relapse in oestrogen receptor-positive primary breast carcinomas. Br J Cancer e-pub ahead of print 29 May doi:10.1038/bjc.2012.231
Pawar SA, Roy Sarkar T, Balamurugan K, Sharan S, Wang J, Zhang Y, Dowdy SF, Huang AM, Sterneck E (2010) C/EBP{delta} targets cyclin D1 for proteasome-mediated degradation via induction of CDC27/APC3 expression. Proc Natl Acad Sci USA 107: 9210–9215
Porter D, Lahti-Domenici J, Keshaviah A, Bae YK, Argani P, Marks J, Richardson A, Cooper A, Strausberg R, Riggins GJ, Schnitt S, Gabrielson E, Gelman R, Polyak K (2003) Molecular markers in ductal carcinoma in situ of the breast. Mol Cancer Res 1: 362–375
Porter DA, Krop IE, Nasser S, Sgroi D, Kaelin CM, Marks JR, Riggins G, Polyak K (2001) A SAGE (serial analysis of gene expression) view of breast tumor progression. Cancer Res 61: 5697–5702
Ramji DP, Foka P (2002) CCAAT/enhancer-binding proteins: structure, function and regulation. Biochem J 365: 561–575
Shah R, Smith P, Purdie C, Quinlan P, Baker L, Aman P, Thompson AM, Crook T (2009) The Prolyl 3-Hydroxylases P3H2 and P3H3 are novel targets for epigenetic silencing in breast cancer. Br J Cancer 100: 1687–1696
Stein T, Salomonis N, Nuyten DS, van de Vijver MJ, Gusterson BA (2009) A mouse mammary gland involution mRNA signature identifies biological pathways potentially associated with breast cancer metastasis. J Mammary Gland Biol Neoplasia 14: 99–116
Sivko GS, DeWille JW (2004) CCAAT/Enhancer binding protein delta (c/EBPdelta) regulation and expression in human mammary epithelial cells I. ‘Loss of function’ alterations in the c/EBPdelta growth inhibitory pathway in breast cancer cell lines. J Cell Biochem 93: 830–843
Tang D, Sivko GS, DeWille JW (2006) Promoter methylation reduces C/EBPdelta (CEBPD) gene expression in the SUM-52PE human breast cancer cell line and in primary breast tumors. Breast Cancer Res Treat 95: 161–170
Thangaraju M, Rudelius M, Bierie B, Raffeld M, Sharan S, Hennighausen L, Huang A-M, Sterneck E (2005) C/EBPd is a crucial regulator of pro-apoptotic gene expression during mammary gland involution. Development 132: 4675–4685
Robinson GW, Johnson PF, Hennighausen L, Sterneck E (1998) The C/EBPb transcription factor regulates epithelial cell proliferation and differentiation in the mammary gland. Genes Dev 12: 1907–1916
Vinson C, Myakishev M, Acharya A, Mir AA, Moll JR, Bonovich M (2002) Classification of human B-ZIP proteins based on dimerization properties. Mol Cell Biol 22: 6321–6335
Wang ND, Finegold MJ, Bradley A, Ou CN, Abdelsayed SV, Wilde MD, Taylor LR, Wilson DR, Darlington GJ (1995) Impaired energy homeostasis in C/EBPa knockout mice. Science 269: 1108–1112
Yamanaka R, Kim GD, Radomska HS, Lekstrom-Himes J, Smith LT, Antonson P, Tenen DG, Xanthopoulos KG (1997) CCAAT/enhancer binding protein e is preferentially up-regulated during granulocyte differentiation and its functional versatility is determined by alternative use of promoters and differential splicing. Proc Natl Acad Sci USA 94: 6462–6467
Zinszner H, Kuroda M, Wang X-Z, Batchvarova N, Lightfoot RT, Remotti H, Stevens JL, Ron D (1998) CHOP is implicated in programmed cell death in response to impaired function of the endoplasmic reticulum. Genes Dev 12: 982–995
Acknowledgements
Carlo Palmieri is the recipient of a Cancer Research UK Clinician Scientist award and also acknowledges support from Imperial College Healthcare Charity. Ondrej Gojis is in part funded by a grant from the Ministry of Education of the Czech Republic (Project ‘Oncology’ MSM 0021620808) and is also a recipient of the Translational Research Fellowship from the European Society of Medical Oncology and a fellowship from the European Society for Surgical Oncology. The Division of Cancer at Imperial College London, Imperial College Healthcare NHS Trust is an Experimental Cancer Medicine Centre that is supported by funds from Cancer Research UK and the Department of Health (C37/A7283), and also forms part of the Imperial Cancer Research UK Centre (C42671/A12196). We would like to thank Prof Gerry Thomas, Sarah Chilcott-Burns and the Imperial College Healthcare NHS Trust, Human Biomaterials Resource Centre (Tissue Bank). Nelofer Syed acknowledges support from the Brain Tumour Research Campaign. Tim Crook is a Scottish Senior Clinical Fellow in Medical Oncology. We thank De Federico Roncaroli for his contribution to the work.
Author information
Authors and Affiliations
Corresponding author
Additional information
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
Rights and permissions
From twelve months after its original publication, this work is licensed under the Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Palmieri, C., Monteverde, M., Lattanzio, L. et al. Site-specific CpG methylation in the CCAAT/enhancer binding protein delta (CEBPδ) CpG island in breast cancer is associated with metastatic relapse. Br J Cancer 107, 732–738 (2012). https://doi.org/10.1038/bjc.2012.308
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/bjc.2012.308
Keywords
This article is cited by
-
DNA methylation in ductal carcinoma in situof the breast
Breast Cancer Research (2013)