Abstract
Background:
Brain metastasis from breast cancer is usually associated with a poor prognosis and early death. Alteration of p53 may contribute to malignant progression by abrogation of apoptosis induced by oncogene activation and by acquisition of gain-of-function properties, which promote tumour aggression. Mutation in TP53 occurs at high frequency in carcinomas of the lung and gastro-intestinal tract, but is much less frequent, at 25%, in primary breast cancer. The frequency of TP53 alteration in the central nervous system (CNS) metastatic breast cancer is not known.
Methods:
In all, 23 cases of histologically confirmed CNS metastatic breast cancer were identified and the coding sequence of TP53 determined. TP53 was also sequenced in two control series of primary breast carcinomas from independent clinical centres.
Results:
We demonstrate a strikingly high frequency of TP53 mutation in the CNS metastatic lesions with an over-representation of complex mutations (non-sense/deletions/insertions). Complex mutations occur in metastatic lesions in both triple-negative breast cancer and hormone receptor/HER2-positive cases. Analysis of paired primary carcinomas and brain metastatic lesions revealed evidence for both clonal selection and generation of new mutations (missense and complex) in progression from a primary breast carcinoma to brain metastasis.
Conclusion:
Mutation in TP53 is the most common genetic alteration reported during metastasis to the brain in breast cancer.
Similar content being viewed by others
Main
To all intents and purposes, development of metastatic disease implies that breast cancer is no longer curable. A survey of the literature shows that almost any distant organ or tissue can be the site of metastatic disease, but certain organs namely bone, lung, liver and brain are particularly common sites of such disease dissemination. Central nervous system (CNS) metastasis is the most common type of malignancy found in the brain and breast cancer is the second most common type of malignancy to cause CNS metastases (Sharma and Abraham, 2007). Central nervous system metastases are less common than bone or visceral metastases, but are less sensitive to systemic chemotherapy and are associated with significantly worse clinical outcomes. Recently, a trend towards increasing CNS recurrence has been noted, up to 25–34% as compared with historical rates of 1–16%. This may be due to increased use of sensitive detection methods such as contrast-enhanced magnetic resonance imaging, increased awareness by patients and clinicians, or an alteration of the natural history of breast cancer resulting from improvements in systemic therapies that are prolonging survival. Recent studies have revealed that breast cancer subtypes are associated with distinct patterns of metastatic spread, with luminal/HER2, HER2-enriched and basal-like tumours having a higher rate of brain metastasis. These subtypes are associated with a poorer prognosis (Kennecke et al, 2010). Two studies have sought to define changes in gene expression, which associate with brain metastasis in breast cancer and a number of candidate genes whose expression is up- or downregulated in the CNS metastatic lesions relative to the primary lesion have been identified (Bos et al, 2009; Klein et al, 2009).
Alteration of p53 by various mechanisms is a frequent finding in malignant disease, although the frequency of mutation in the gene varies considerably between tumour types, with a high proportion of mutations in lung cancer, lower gastrointestinal cancers and glial brain tumours, but low reported frequencies in melanoma and sarcomas (Soussi and Wiman, 2007). In breast cancer, an overall frequency of approximately 20% has been documented (Pharoah et al, 1999; Olivier et al, 2006). Many TP53 mutations identified in human cancer encode mutant proteins, which possess gain of function, such as the ability to cooperate with activated oncogenes to morphologically transform primary cells. The most common missense mutations in human cancer are termed ‘hotspot mutations’ (Walker et al, 1999). More complex mutations such as insertion/deletion/nonsense are also described. In breast cancer, the frequency of TP53 mutation is higher in carcinomas arising in a background of mutant BRCA1 and BRCA2 than in sporadic cases (Crook et al, 1997; Smith et al, 1999). Complex mutations appear more common in cancers arising in cancers of basal subtype, including but not restricted to those arising in a background of mutant BRCA1 (Holstege et al, 2009; Manié et al, 2009). Studies of TP53 mutations in metastatic breast cancer lesions have been limited by the lack of availability of tissue, perhaps because of the infrequency of re-biopsy of radiologically detected suspicious lesions in patients with a previous history of primary breast cancer. However, Ding et al (2010) used massively parallel sequencing to examine genetic changes in a primary breast carcinoma and subsequent CNS metastasis and identified a complex mutation in TP53. Here, we have analysed TP53 mutations in a series of CNS metastatic breast carcinomas.
Materials and methods
Tumours
Case ascertainment of surgically excised and histologically confirmed intra-cranial brain metastatic breast cancer identified 23 metastases subsequently retrieved from the neuropathology archives of the Charing Cross Hospital. As comparator primary breast cancers, we analysed using different sequencing techniques TP53 mutation in independent series of primary breast carcinomas from two different clinical centres. In each series, expression of the oestrogen receptor (ER), progesterone receptor (PR) and HER2 was performed according to standard protocols of clinical care (Purdie et al, 2010).
TP53 sequence analysis
Genomic DNA was isolated from archival cases by proteinase K digestion of 10 μ M formalin-fixed, paraffin-embedded (FFPE) sections using a standard xylene–phenol protocol. For the primary breast cancers in the Tayside Tissue Bank (Dundee, UK), genomic DNA was isolated from frozen tissues as described previously (Baker et al, 2010). Mutations in TP53 were detected using two methods. For the Tasyide cases, TP53 was analysed using the Roche Amplichip (Roche Molecular Systems, Pleasanton, CA, USA) as reported previously (Baker et al, 2010). For the CNS metastatic lesions and the FFPE cases, TP53 was analysed by direct sequencing of exons 4–10, which were individually amplified (Table 1). Polymerase chain reaction was carried out in a total volume of 10 μl consisting of 0.75 μ M of both primers, 1.5 mmol l−1 MgCl2, 200 μ M deoxynucleotide triphosphate, 0.25 U of AmpliTaq Gold 360 DNA Polymerase (Applied Biosystems, Foster City, CA, USA), 1 μl of 360 GC Enhancer, 50 ng of genomic DNA using a GeneAmp PCR System 9700 Thermal Cycler (Applied Biosystems). The PCR programme was an initial denaturation step of 10 min at 95 °C, followed by 40 cycles of 30 s at 95 °C, 45 s at specific annealing temperature for each fragment and 45 s at 72 °C, with a final step at 72 °c for 7 min. Amplified products of p53 exons (4–10) were directly sequenced in both directions on an ABI 3130 automated sequencer, using the BigDye terminator cycle sequencing reaction kit following the manufacturer's instructions (Applied Biosystem). The analysis was performed using the Variant Reporter Software v.1.0 (Applied Biosystems). Proposed mutations were confirmed on both strands on repeated, independent amplimers and validated by the IARC TP53 Mutation Database (http://www-p53.iarc.fr). The reference sequence is the genomic sequence NC_000017 version 9 (7512445–7531642) from GenBank.
Results
TP53 mutations in CNS metastatic breast cancer
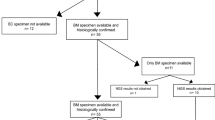
We analysed the genomic sequence of TP53 in 23 individual cases of surgically resected, histologically confirmed CNS metastatic breast cancer. As controls, we sequenced two independent series of primary breast carcinomas from two different oncology centres using two different techniques. TP53 mutations were present in 29 out of 91 (32%) cases from northern Italy (series 1) and in 55 out of 229 (24%) cases from the Tayside series (series 2). In the CNS metastatic lesions, mutations were present in 20 out of 23 (87%) cases (Figure 1 and Tables 2a and b). Two different TP53 mutations were detected in 2 out of 23 CNS metastatic lesions, 4 out of 29 primary carcinomas from series 1 and 0 out of 55 primary carcinomas from series 2 (Tables 2). In comparator series 1, 25 out of 87 ER-positive and 4 out of 4 ER-negative cases contained TP53 mutations. In series 2, TP53 mutations were present in 27 out of 177 ER-positive and 27 out of 52 ER-negative cases. The ER status was unavailable for one Her2-positive case (55P) with a TP53 mutation and for two ER-negative cases with TP53 mutations (37P and 38P). Mutation in TP53 was detected in 16 out of 34 triple-negative breast cancer (TNBC) in series 2. Frameshift, splice and nonsense mutations and in-frame insertions and deletions constitute complex TP53 mutations (Holstege et al, 2009). Out of 22 (41%) mutations, nine detected in the CNS metastases were complex and such mutations were present in 9 out of 20 (45%) such cases, compared with 0 out of 34 (0%) complex mutations in primary carcinomas from comparator series 1 and 7 out of 55 (13%) complex mutations in primary carcinomas from comparator series 2. In the CNS metastatic lesions, 13 mutations caused amino-acid substitutions and 9 out of 13 (69%) were hotspot mutations (Tables 2a and b). Hotspot mutations comprised 11 out of 34 (32%) substitution mutations in comparator series 1 (Tables 2c and d) and 30 out of 48 substitution mutations (62%) in comparator series 2 (Tables 2e and f). The frequency of hotspot mutations was not significantly different between primary cancers, which subsequently relapsed with metastasis and those which did not, in either series 1 or series 2. In the CNS metastases, complex mutations were present in both TNBC and hormone receptor cases: 6 out of 9 complex mutations occurred in hormone receptor-positive cancers compared to 3 out of 9 in TNBC. In comparator series 2, 6 out of 7 complex mutations were in ER-negative cases, of which four were TNBC.
Representative sequencing traces showing two examples of complex TP53 mutations in central nervous system (CNS) metastatic breast cancer lesions. Isolation of genomic DNA, sequencing and analysis were carried out as described in materials and methods. The top panel shows Case 1BM with a normal control sequencing trace immediately below. The second case is 16BM, again with a normal control immediately below. The deleted sequence is indicated by vertical lines.
Analysis of paired primary/metastatic lesions reveals clonal selection for loss of p53 function and generation of new mutations
The TP53 mutation frequency in comparator series 1 is 9 out of 30 (30%) in non-relapsed cases and 20 out of 61 (33%) in relapsed cases, and in comparator series 2, it is 29 out of 161 (18%) in non-relapsed cases and 26 out of 68 (38%) in relapsed cases. This implies that TP53 mutation frequency is higher in CNS metastatic lesions than in primary breast carcinomas, irrespective of whether the primary lesion ultimately relapsed. To further investigate this, we analysed four paired cases with tissue from both primary and CNS metastasis and we sequenced TP53 to determine whether mutations associated with subsequent CNS metastasis were detectable in primary cancers (Figure 2). In all four cases, TP53 mutations were present in the metastatic lesion (Table 2). In two of the four cases, the TP53 mutation present as the sole variant in the CNS metastasis was also detected in the primary cancer, in both cases in the presence of other mutations: in Case 19P, three mutations (Pro300Leu, Ser260Phe and the complex mutation Ser183Ter) were detected, but only Ser183Ter (homozygous) was detected in the CNS metastasis 19BM. In Case 20P, His178Tyr and Tyr163Cys were present, but only the hotspot mutation Tyr163Cys was detected (homozygous) in the CNS metastasis 20BM. In a third case (17P), no mutations were detected in the primary cancer, but hotspot mutation Pro151Ser was present as a homozygous mutation in the CNS metastasis (17BM). In Case 18, Ile232Phe and Tyr234His were detected in the primary (18P), but a new mutation Val216Met was detected in the CNS metastasis (18BM).
Representative sequencing traces showing TP53 mutations in matched primary and central nervous system (CNS) metastatic breast cancer lesions. Isolation of genomic DNA, sequencing and analysis were carried out as described in Materials and Methods. The upper panels show homozygous mutation Pro151Ser in CNS metastasis 17BM (arrowed), but only wild-type sequence in the primary carcinoma (17P). The lower panels show heterozygous Tyr163Cys and p.His178Tyr mutations (arrowed) in the primary carcinoma, 20P, but homozygous Tyr163Cys mutation (arrowed) in the CNS metastasis (20BM).
Discussion
Dissemination of breast cancer to the brain is a sinister event, associated with a poor prognosis. Treatment of such metastatic lesions is typically with radiotherapy, although solitary lesions can be resected, and in some cases an improved prognosis is observed. Nonetheless, the poor outlook and resistance to chemotherapy seen in most reported series strongly emphasises the need for improved understanding of the mechanisms of brain metastasis and for novel therapeutic approaches. Studies of the molecular basis of metastasis of breast carcinomas to the brain have been limited. Changes in gene expression in CNS metastatic lesions in breast cancer patients have been reported in two clinical studies (Nishizuka et al, 2002; Klein et al, 2009) and a pre-clinical study analysed changes in gene expression in MDA MB 231 cells with tropism for metastasis to the brain (Bos et al, 2009). Here, we have analysed the coding sequence of TP53 in a series of brain metastases, confirmed by histopathology to derive from primary breast carcinomas, and we report a high frequency of TP53 mutational alteration in breast cancer metastatic to the brain. This demonstrates that mutation of TP53 is the most common change (genetic or epigenetic) identified thus far in brain metastases from breast cancer.
As comparators for the metastatic brain lesions, we tested two independent series of primary breast carcinomas from geographically distinct clinical practices. One series comprised archival cases in which genomic DNA was prepared from FFPE tissue and the other series high-molecular-weight genomic DNA from fresh–frozen tissue. Cases were evaluated for tumour cell representation before analysis. The overall frequency of TP53 mutation detected in each series was 32 and 23%, with a low frequency of complex mutations, comparable to those reported in previous studies (Pharoah et al, 1999; Olivier et al, 2006). There have been very few previous studies of TP53 in CNS metastatic breast cancer lesions. However, our data are consistent with Ding et al (2010), who reported a frameshift mutation present in both primary and CNS metastasis of a basal phenotype breast cancer. Our results are also consistent with those of Piccirilli et al (1994), who showed 17p loss of heterozygosity and TP53 mutations in breast carcinomas metastatic to brain. Further, immunoreactivity for p53 in primary breast carcinomas, using antibodies pAb240 and pAb1801, is associated with increased risk for subsequent brain metastasis (Tham et al, 2006), consistent with a potential role for TP53 mutation in CNS metastasis. The frequency of complex mutations in the CNS metastatic lesions is higher than that in either of the series of primary carcinomas. The frequency of complex mutations is reported to be higher in basal subtype breast cancers, including cases arising in carriers of germ-line BRCA1 mutations (Holstege et al, 2009; Manié et al, 2009). In our series of CNS metastases, 7 out of 10 TNBC contained missense rather than complex mutations. Furthermore, 6 out of 9 complex mutations occurred in hormone receptor-positive cancers compared to 3 out of 9 in TNBC, implying that the high frequency of complex mutations in the CNS metastatic lesions cannot be explained on the basis of over-representation of TNBC. Analysis of matched primary and CNS metastatic lesions revealed selection of mutants during metastasis to the brain. For example, in one case, three TP53 mutations were detected in the primary lesion, but only the complex mutation resulting in premature termination at codon 183 was detected in the CNS metastasis from this patient (as a homozygous mutation), showing preferential selection of the complex mutation. A second primary cancer contained two detectable TP53 mutations, a non-hotspot His178Tyr and the hotspot Tyr163Cys. Only the hotspot mutation was detected (again homozygous) in the CNS metastasis.
In previous studies, conservation of TP53 mutation between primary and paired CNS metastasis was reported (Piccirilli et al, 1994). In another study, the same single mutation was detected in the primary breast carcinoma and paired metastatic lymph node in a subset of cases. In other cases, an additional mutation in the primary tumour only or a mutation in the metastasis only was observed (Peller et al, 1995). In our series, the final, clonal TP53 mutation was only detected in the primary lesion in two of four cases, and in both such cases, additional mutations were present in the primary lesion. We did not observe association between specific missense TP53 mutations and metastasis to CNS or other anatomical sites in the clinical series analysed. Rather, the common property in all such cases from series 2, which progressed to CNS metastatic disease was overexpression of Pin1 (data not shown), consistent with the critical role of this protein in promoting aggressive clinical phenotypes in breast cancers expressing mutant p53 (Girardini et al, 2011).
What are the implications of our data? First, the presence of TP53 mutations in the great majority of CNS metastatic lesions implies that therapeutic strategies aimed at exploiting loss of p53 function could be highly effective in brain metastasis, even if the primary breast cancer is wild type for TP53. Second, the results raise the possibility that patients at high risk for intra-cranial metastasis might be identifiable from detection of TP53 mutations in the primary cancer and interventions such as prophylactic cranial irradiation considered. However, our analysis of paired primary, metastatic cases, albeit with limited numbers, shows that TP53 mutations present in brain metastatic lesions are not always detectable in the primary. Also, despite the increased representation of complex mutations, many CNS metastases nonetheless occurred in cancers containing hotspot (substitution) mutations in TP53, reducing the likely sensitivity of detection of complex mutations as predictors of future CNS metastasis. As noted above, all primary cancers in series 2, which contained substitution mutations in TP53, and which subsequently progressed to CNS metastasis, overexpressed Pin1. Additional studies of the prognostic utility of Pin1 and mutant p53 are merited clearly. Third, our results further add to the evidence that changes in expression and structure of genes may occur in the progression of primary breast cancer to metastatic disease. In conclusion, our data reveal a strikingly high frequency of TP53 mutation in CNS metastatic breast cancer.
Change history
29 March 2012
This paper was modified 12 months after initial publication to switch to Creative Commons licence terms, as noted at publication
References
Baker L, Quinlan PR, Patten N, Ashfield A, Birse-Stewart-Bell LJ, McCowan C, Bourdon JC, Purdie CA, Jordan LB, Dewar JA, Wu L, Thompson AM (2010) p53 mutation, deprivation and poor prognosis in primary breast cancer. Br J Cancer 102: 719–726
Bos PD, Zhang XH, Nadal C, Shu W, Gomis RR, Nguyen DX, Minn AJ, van de Vijver MJ, Gerald WL, Foekens JA, Massagué J (2009) Genes that mediate breast cancer metastasis to the brain. Nature 459: 1005–1009
Crook T, Crossland S, Crompton MR, Osin P, Gusterson BA (1997) p53 mutations in BRCA1-associated familial breast cancer. Lancet 350: 638–639
Ding L, Ellis MJ, Li S, Larson DE, Chen K, Wallis JW, Harris CC, McLellan MD, Fulton RS, Fulton LL, Abbott RM, Hoog J, Dooling DJ, Koboldt DC, Schmidt H, Kalicki J, Zhang Q, Chen L, Lin L, Wendl MC, McMichael JF, Magrini VJ, Cook L, McGrath SD, Vickery TL, Appelbaum E, DeSchryver K, Davies S, Guintoli T, Lin L, Crowder R, Tao Y, Snider JE, Smith SM, Dukes AF, Sanderson GE, Pohl CS, Delahaunty KD, Fronick CC, Pape KA, Reed JS, Robinson JS, Hodges JS, Schierding W, Dees ND, Shen D, Locke DP, Wiechert ME, Eldred JM, Peck JB, Oberkfell BJ, Lolofie JT, Du F, Hawkins AE, O’Laughlin MD, Bernard KE, Cunningham M, Elliott G, Mason MD, Thompson Jr DM, Ivanovich JL, Goodfellow PJ, Perou CM, Weinstock GM, Aft R, Watson M, Ley TJ, Wilson RK, Mardis ER (2010) Genome remodeling in a basal-like breast cancer metastasis and xenograft. Nature 464: 999–1015
Girardini JE, Napoli M, Piazza S, Rustighi A, Marotta C, Radaelli E, Capaci V, Jordan L, Quinlan P, Thompson A, Mano M, Rosato A, Crook T, Scanziani E, Means AR, Lozano G, Schneider C, Del Sal G (2011) A Pin1/mutant p53 axis promotes aggressiveness in breast cancer. Cancer Cell 20: 79–91
Holstege H, Joosse SA, van Oostrom CT, Nederlof PM, de Vries A, Jonkers J (2009) High incidence of protein-truncating TP53 mutations in BRCA1-related breast cancer. Cancer Res 69: 3625–3633
Kennecke H, Yerushami R, Woods R, Cheang MC, Voduc D, Speers CH, Nielsen TO, Gelmon K (2010) Metastatic behavior of breast cancer subtypes. J Clin Oncol 28: 3271–3277
Klein A, Olendrowitz C, Schmutzler R, Hampl J, Schlag PM, Maass N, Arnold N, Wessel R, Ramser J, Meindl A, Scherneck S, Seitz S (2009) Identification of brain- and bone-specific breast cancer metastasis genes. Cancer Lett 276: 212–220
Manié E, Vincent-Salomon A, Lehmann-Che J, Pierron G, Turpin E, Warcoin M, Gruel N, Lebigot I, Sastre-Garau X, Lidereau R, Remenieras A, Feunteun J, Delattre O, de Thé H, Stoppa-Lyonnet D, Stern MH (2009) High frequency of TP53 mutation in BRCA1 and sporadic basal-like carcinomas but not in BRCA1 luminal breast tumors. Cancer Res 69: 663–671
Nishizuka I, Ishikawa T, Hamaguchi Y, Kamiyama M, Ichikawa Y, Kadota K, Miki R, Tomaru Y, Mizuno Y, Tominaga N, Yano R, Goto H, Nitanda H, Togo S, Okazaki Y, Hayashizaki Y, Shimada H (2002) Analysis of gene expression involved in brain metastasis from breast cancer using cDNA microarray. Breast Cancer 9: 26–32
Olivier M, Langerod A, Carrieri P, Bergh J, Klaar S, Eyford J, Theillet C, Rodriguez C, Lidereau R, Bieche I, Varley J, Bignon Y, Uhrhammer N, Winqvist R, Jukkola-Vuorinen A, Niederacher D, Kato S, Ishioka C, Hainaut P, Borresen-Dale AL (2006) The clinical value of somatic TP53 gene mutations in 1,794 patients with breast cancer. Clin Cancer Res 12: 1157–1167
Peller S, Halevy A, Slutzki S, Kopilova Y, Rotter V (1995) p53 mutations in matched primary and metastatic human tumors. Mol Carcinogen 13: 166–172
Pharoah PD, Day NE, Caldas C (1999) Somatic mutations in the p53 gene and prognosis in breast cancer: a meta-analysis. Br J Cancer 80: 1968–1973
Piccirilli C, Saxena A, Robertson J, Clark W, Ikejiri B, Oldfield E, Ali I (1994) Allelic deletions on chromosome-17 and mutations in the p53 gene in tumors metastatic to brain. Int J Oncol 1: 37–42
Purdie CA, Jordan LB, McCullough JB, Edwards SL, Cunningham J, Walsh M, Grant A, Pratt N, Thompson AM (2010) HER2 assessment on core biopsy specimens using monoclonal antibody CB11 accurately determines HER2 status in breast carcinoma. Histopathology 56: 702–707
Sharma M, Abraham J (2007) CNS metastasis in primary breast cancer. Expert Rev Anticancer Ther 7: 1561–1566
Smith PD, Crossland S, Parker G, Osin P, Brooks L, Waller J, Philp E, Crompton MR, Gusterson BA, Allday MJ, Crook T (1999) Novel p53 mutants selected in BRCA-associated tumours which dissociate transformation suppression from other wild-type functions. Oncogene 18: 2451–2459
Soussi T, Wiman KG (2007) Shaping genetic alterations in human cancer: the p53 mutation paradigm. Cancer Cell 12: 303–312
Tham YL, Sexton K, Kramer R, Hilsenbeck S, Elledge R (2006) Primary breast cancer phenotypes associated with propensity for central nervous system metastases. Cancer 107: 696–704
Walker DR, Bond JP, Tarone RE, Harris CC, Makalowski W, Boguski MS, Greenblatt MS (1999) Evolutionary conservation and somatic mutation hotspot maps of p53: correlation with p53 structural and functional features. Oncogene 18: 211–218
Acknowledgements
The work was supported by the Brain Tumour Research Charity (BTRC), Cancer Research-UK, Breast Cancer Research (Scotland) and the Tayside Tissue Bank. C Palmieri wishes to acknowledge the grant support from Cancer Research UK (Grant numbers: C20208/A8667). O Gojis wishes to acknowledge grant support from the European Society of Medical Oncology and the Ministry of Education of the Czech Republic (Project ‘Oncology’ MSM 0021620808). The Department of Oncology at Imperial College London/Imperial College HealthCare NHS Trust is an Experimental Cancer Medicine Centre (ECMC), which is supported by funds from Cancer Research UK and the Department of Health. We thank F Roncaroli for his contribution.
Author information
Authors and Affiliations
Corresponding author
Additional information
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
Rights and permissions
From twelve months after its original publication, this work is licensed under the Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Lo Nigro, C., Vivenza, D., Monteverde, M. et al. High frequency of complex TP53 mutations in CNS metastases from breast cancer. Br J Cancer 106, 397–404 (2012). https://doi.org/10.1038/bjc.2011.464
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/bjc.2011.464
Keywords
This article is cited by
-
Mutant p53 sustains serine-glycine synthesis and essential amino acids intake promoting breast cancer growth
Nature Communications (2023)
-
Brain metastasis in breast cancer: focus on genes and signaling pathways involved, blood–brain barrier and treatment strategies
Clinical and Translational Oncology (2023)
-
BTB protein family and human breast cancer: signaling pathways and clinical progress
Journal of Cancer Research and Clinical Oncology (2023)
-
Recent advances of therapeutic targets based on the molecular signature in breast cancer: genetic mutations and implications for current treatment paradigms
Journal of Hematology & Oncology (2019)
-
LCCC 1025: a phase II study of everolimus, trastuzumab, and vinorelbine to treat progressive HER2-positive breast cancer brain metastases
Breast Cancer Research and Treatment (2018)