Abstract
B-cell acute lymphoblastic leukemia (B-ALL) accounts for the most cancer incidences in children. We present here that autophagy is downregulated in pediatric B-ALL, suggesting a possible link between autophagy failure and pediatric B-ALL leukemogenesis. With a pediatric t(1;19) B-ALL xenograft mouse model, we show here that activation of autophagy by preventive administration of rapamycin improved the survival of leukemia animals by partial restoration of hematopoietic stem/progenitor cells, whereas treatment of the animals with rapamycin caused leukemia bone marrow cell-cycle arrest. Activation of autophagy in vitro or in vivo by rapamycin or starvation downregulated oncogenic fusion protein E2A/Pbx1. Furthermore, E2A/Pbx1 was found to be colocalized with autophagy marker LC3 in autolysosomes and with ubiquitin in response to autophagy stimuli, whereas autophagy or ubiquitination inhibitor blocked these colocalizations. Together, our data suggest a collaborative action between autophagy and ubiquitination in the degradation of E2A/Pbx1, thereby revealing a novel strategy for targeted preventive or treatment therapy on the pediatric ALL.
Similar content being viewed by others
Introduction
Autophagy is a catabolic pathway that involves lysosomal degradation and recycling of proteins and organelles,1 and is therefore considered as an important survival mechanism for both normal cells and cancer cells in response to metabolic stress or chemotherapy. In hematologic malignancies, autophagy either acts as a chemoresistance mechanism or has tumor suppressive functions, depending on the context.2 Intervening autophagy pathway is one of the current strategies in the treatment of leukemia. For instance, imatinib and its improved new compounds dasatinib and nilotinib are tyrosine kinase inhibitors, and are currently the frontline treatment for Ph+ acute lymphoblastic leukemia (Ph+ ALL) and chronic myeloid leukemia that carry the BCR-ABL1 fusion gene.3 Autophagy is downregulated in BCR-ABL-expressing leukemia cells.4 Activation of autophagy in these leukemias and chronic lymphoblastic leukemia frequently causes serious acute and late complications, resistance to chemotherapy and clinical relapse.4, 5, 6, 7, 8, 9, 10, 11 Therefore, suppression on autophagy improves the antileukemic effect of tyrosine kinase inhibitor. However, manipulation of autophagy may have an opposite effect on certain type of leukemias. In particular, autophagic targeting of oncogenic fusion proteins that block hematopoietic differentiation is one of the current strategies in targeted leukemia therapies.12, 13
B-cell acute lymphoblastic leukemia (B-ALL) accounts for the most cancer incidences in children. The t(1;19) translocation in pediatric B-ALL fuses the genes, which encode the transcriptional activator E2A and homeobox pre-B-cell leukemia transcription factor 1 (Pbx1), resulting in expression of the chimeric transcription factor E2A-Pbx1. E2A/Pbx1 has been proved to be an oncogene and could induce the malignant transformation of mice.14, 15, 16 The front line treatment for B-ALL involves an intense chemotherapy regimen with cure rate up to 80%.17 Nevertheless, about 20% of remission suffers a relapse with a very poor prognosis.17, 18 We have recently found that activation of autophagy by rapamycin inhibits in vitro pre-B ALL cells in part through downregulating DNA and RNA polymerases.19 But whether autophagy works alone or collaborates with other degradation mechanism in vivo in fighting against leukemia remains unknown. Using a pediatric B-ALL xenograft mouse model and in vitro pediatric B-ALL 697 cell line model, we show here that autophagy collaborates with ubiquitination in the degradation of E2A/Pbx1, thereby inhibiting the B-ALL cells.
Materials and methods
Patients bone marrow sample collection and Q-PCR
B-ALL patient bone marrow (BM) cells were collected from the affiliated Children’s Hospital of Soochow University. Seven patients confirmed of B-ALL were enrolled for this study. Normal BM cells from two healthy donors were used as a control. BM cells were collected and monocytes were separated by density gradient centrifugation using Ficoll (GE Healthcare, Pittsburgh, PA, USA). CD34, CD38, CD117, CD45, CD19 and CD10 were stained and analyzed with flow cytometry. CD34+38− and CD117+ were used as stem/progenitor cell markers. CD45+, CD19+ and CD10+ were used as mature B leukemia cell markers. Stem/progenitor cells (CD34+ CD38−) were sorted by staining of CD34 PE, CD38 FITC, leukemia B cells were obtained by staining of CD19 APC through FACS sorting (BD FACS Aria III, BD Bioscience, San Jose, CA, USA). The sorted cells were used for mRNA extraction and autophagy gene detection including Beclin1, Atg7, Atg5, LC3 and p62. GAPDH was used as an inner control. The Q-PCR was carried out in an ABI 7500 system (Applied Biosystems, Grand Island, NY, USA). The primers were as follows in Table 1.
Cell lines and drugs
ALL cell lines 697 were used. The cells were grown in RPMI-1640 medium (Hyclone, Thermo Fisher Scientific, Waltham, MA, USA) with 10% fetal bovine serum (Gibco, Life Technologies, Grand Island, NY, USA) in 37 °C, 5% CO2 incubator. For mice injection, rapamycin (Merck Calbiochem, Billerica, MA, USA) was dissolved in absolute ethanol at 10 mg/ml for stock solution and diluted in 5% Tween-80 and 5% PEG-400. For cell culture, rapamycin was dissolved in DMSO at 500 μg/ml for stock solution and 100 ng/ml for work concentration. 3-MA (Sigma-Aldrich, St Louis, MO, USA) was dissolved in phosphate-buffered saline (PBS) at 100 mM for stock solution and 1 mM for work concentration. Bafilomycin A1 (Sigma-Aldrich) was dissolved in DMSO at 10 μM for stock solution and 1 nM for work concentration. MG132 (Merck Calbiochem) was dissolved in DMSO at 1 mM for stock solution and 0.2 μM for work concentration.
Mice treatment
NOD-SCID male mice weighed 20–25 g were obtained from Shanghai SLAC laboratory animal company and divided into four groups randomly with 10 each for survival curve and 5 each for other experiments. The four groups were control (C), preventive group (P), model group (M) and treatment group (T), respectively. Mice in preventive group received 4 mg/kg rapamycin by intraperitoneal injection every other day for five times before the B-ALL 697 cell injection. Mice in treatment group received 4 mg/kg rapamycin by intraperitoneal injection every other day for five times after 1 week of the 697 cell injection. Mice in model group received the 697 cells only by intravenous injection. Each mouse received 1 × 107 pre-B ALL 697 cells for leukemogenesis. Control mice received PBS instead of the 697 cells.
Survival curve and mice killing
For survival curve, the mice were raised in SPF environment and the death time was recorded. For other experiments, the mice were killed at 3 weeks after the 697 cell injection due to the mice death time point. Peripheral blood, liver, spleen and BM cells were collected for experiments.
Peripheral blood were obtained from eye fundus vein of mice after anesthesia, 20 μl was counted with Blood cell counter Sysmex KX21N(Sysmex, Kobe, Japan) for white blood cell, red blood cell and platelet analysis. Then, the mice were killed.
After the mice being killed, liver and spleen were obtained. The weight was recorded and liver/spleen coefficients were calculated. Part of them was used for pathological observation with HE stain, and part was lysised for protein extraction. RIPA lysate from CST plus protease inhibitor (Roche, Basel, Switzerland) was used for protein extraction. Beclin1, LC3, GAPDH and E2A/Pbx1 expression of liver was detected by western blotting.
BM cells were isolated for cell-cycle and hematopoietic stem cell (HSC) analysis. After the mice being killed, BM cells were flushed by a 25-gauge needle from the long bones (tibias and femurs) with PBS without calcium or magnesium (Hyclone). For cell cycle, BM cells were fixed with 70% ethanol, and stained with Hoechst. For HSC analysis, lineage-negative cells were enriched with MACS lineage kit (Miltenyi Biotec, Bergisch Gladbach, Germany), and stained with CD34 FITC, Sca-1 PE and c-Kit APC. The CD34-LSK phenotype was used for long-term HSCs, and CD34+LSK phenotype was used for short-term HSCs. All the antibodies were obtained from BD company and analyzed by flow cytometry (BD Calibur or BD Aria III).
B-ALL 697 cell treatment and flow cytometry
The B-ALL 697 cells were treated with rapamycin (100 ng/ml), HBSS, 3-MA (1 mM), Baf A1 (1 nM) and MG132 (0.2 μM) for different time points. E2A/pbx1 expression in cells was measured by flow cytometry and western blotting. For flow cytometry, cells were collected and washed with PBS, fixed and performed with a kit according to the manufacturer’s protocol (ADG, Nordic Mubio, Susteren, The Netherlands). After fixation for 15 min, the cells were permeabilized and anti-mouse E2A/Pbx1 antibody (BD Bioscience) was stained for 30 min. PE anti-mouse second antibody was stained for 30 min, and after wash with PBS, the cells were measured for PE-E2A/Pbx1 expression by flow cytometry.
Western blotting
Cellular proteins were extracted by lysing cells in extraction buffer (RIPA lysate from CST plus protease inhibitor from Roche). The protein concentration was determined by BCA assay (Pierce, Thermo Fisher Scientific). Equal amounts of protein (30 μg) were fractionated by electrophoresis in SDS-polyacrylamide gel. The proteins were subsequently transferred onto PVDF membranes. Antibodies against E2A/Pbx1 (BD Bioscience), LC3 (Novus, Biologicals, Littleton, CO, USA), Beclin1, ubiquitin and GAPDH (Cell Signaling Technology, Danvers, MA, USA) were applied to probe the membranes, respectively. The secondary antibodies (anti-rabbit or anti-mouse, Cell Signaling Technology) were conjugated to horseradish peroxidase. Signals were detected using the ECL system (Biological Industries, Kibbutz Beit-Haemek, Israel).
Confocal microscopy
For confocal microscopy, the GFP-LC3 construct was transfected into the B-ALL 697 cells to generate a stable GFP-LC3 B-ALL 697 cell line. The GFP-LC3 697 cells were treated with rapamycin, HBSS or bafilomycin A1. At the time points indicated, cells were collected and pelleted by centrifugation for 5 min at 1000 r.p.m., washed in PBS, fixed in 4% paraformaldehyde at RT for 10 min, then aspirated fixative, rinsed three times in PBS for 5 min each time, and then blocked with 5% goat serum in PBS (with 0.3% Triton X-100) for 60 min. The cells were incubated with the primary antibody (E2A/Pbx1, diluted in PBS supplemented with 0.3% Triton X-100 and 1% bovine serum albumin, overnight in a humid chamber at 4 °C) and then with PE-conjugated secondary antibodies (diluted 1:1000, for 1 h at 37 °C in a humid chamber). Excess of unbound antibody was removed at each step by three washes with PBS. The nuclear was stained with 20 μg/ml Hoechst 33342 (Invitrogen, Life Technologies) at room temperature for 10 min. The images were obtained by using an Olympus confocal microscope (Olympus, Tokyo, Japan, FV1000MPE).
Image flow-cytometric analysis
Image flow cytometry was explored to quantify the co-localization of E2A/Pbx1 and LC3 or Ubiquitin (Ub). The 697 cells of different treatment groups were collected at the predicted time point. After fixation and permeabilization using the kit (ADG), cells were stained with primary antibody of anti-mouse E2A/Pbx1 and anti-rabbit LC3 or Ub for 30 min, and second antibody of PE anti-mouse and FITC anti-rabbit IgG for 30 min, then cells were explored to ImageStreamX Mark II (Amnis, Merck Millipore, Seattle, WA, USA) for image flow cytometry. Samples were visualized and analyzed for the expression of markers and proteins with IDEAS 6.0 software (Amnis, Merck Millipore). Cells were gated for single cells with the area and aspect ratio features, for focused cells, with the Gradient RMS feature. These subsets were then plotted for log intensities of channel with LC3 (autophagosome marker), Ub (Ubiquitination marker) and channel with E2A/Pbx1. Autophagy level was analyzed with LC3 level by spot count wizard. Co-localization of E2A/Pbx1 and LC3 or Ub was analyzed by the co-localization wizard.
Statistical analysis
The data were presented as mean values±s.d from three separate experiments. Statistical analysis was performed using GraphPad Prism 5 Software (downloaded from internet). Error bars represent s.e.m. and P-values calculated with a two-tailed Mann–Whitney test unless stated otherwise (ns means no significance, *P<0.05, **P<0.01, ***P<0.001).
Results
Occurrence of pediatric B-ALL is associated with reduced autophagy in patients
To explore the role of autophagy in clinical B-ALL leukemogenesis, BM cells from pediatric B-ALL patients and normal healthy donors were collected. Stem/progenitor cell markers CD34+38− and CD117+, and mature B leukemia cell markers CD45, CD19 and CD10 were stained for the BM cells. Flow-cytometric analysis showed significantly decreased percentage of CD34+38− and CD117+ cells, and significantly increased percentage of CD45, CD19 and CD10 cells in the B-ALL patients compared with the normal control (Figure 1a). The CD34+CD38− cells and CD19+ cells from the B-ALL patients were sorted and autophagy-essential genes of Beclin1, Atg7, Atg5, LC3 and p62 were detected by Q-PCR collectively representing autophagy level. Both the CD34+CD38− stem/progenitor cells and CD19+ leukemia cells from B-ALL patients showed low level of autophagy gene expression (Figures 1b and c). These data suggest that the occurrence of B-ALL is clinically associated with reduced autophagy in the BM cells of the leukemia patients.
Occurrence of pediatric B-cell ALL is associated with reduced autophagy. (a) BM cells of B-ALL patients and normal healthy control were collected and CD34, CD38, CD117, CD45, CD19 and CD10 were measured. It showed high percentage of hematological/ progenitor cells in normal control and high B lymphocytic cells in B-ALL patients. CD34+38− (b) and CD19+ B cells (c) were sorted separately and autophagy-essential genes of Beclin1, Atg5, Atg7, LC3 and p62 were detected by Q-PCR. It showed low autophagy level in both CD34+38- cells and CD19+ B cells of B-ALL patients. ***P<0.001, **P<0.01, *P<0.05, n=7.
Activation of autophagy by rapamycin improves the B-ALL animal survival and in vivo inhibits the transplanted ALL cells
To examine whether in vivo enhancement of autophagy is capable of fighting against B-ALL cells, we generated a human leukemia xenograft mouse model with B-ALL 697 cells. Ten mice each were used for survival curve in the four groups (control, model, preventative and treatment) administered with or without rapamycin as described in the Materials and methods. Western blotting analysis showed that LC3 lipidation, a typical autophagy activity indicator,20 was enhanced in the preventive or treatment groups, suggesting that rapamycin treatment ahead of, or right after B-ALL cell transplantation, activates or enhances autophagy response in the mouse models (Figure 2a).
Activation of autophagy before or after transplantation of B cell leukemia cells prolonged the survival of xenograft mice. The mice were divided into four groups: control (C), preventive group (P, rapamycin administrated before human B-ALL cell transplantation), model group (M, no rapamycin administrated before or after the B-ALL cell transplantation) and treatment group (T, rapamycin administrated 1 week after B-ALL cell transplantation). The survival curve (b) showed that activation of autophagy by rapamycin in the preventive and treatment group prolonged 5–9 days of the survival time compared with model group (with 10 mice in each group). The autophagy level (a, Beclin1 and LC3 expression), peripheral blood cell counting (c), the size and coefficient of liver and spleen (d), the pathological section of liver (e), B-ALL immune typing of human CD45, CD19 and CD10 expression in mice (f) were showed. It indicated that rapamycin slowed down the malignant transformation of leukemia cells in NOD-SCID mice. ***P<0.001, **P<0.01, *P<0.05.
The average survival time was 21.2±3.5, 26.7±5.4 and 30±2.8 days in the model group, preventive group and treatment group, respectively. The mice in the control group all survived. Mice began to die in 18 days after the B-ALL 697 cell injection in model group; activation of autophagy by rapamycin prolonged 5–9 days of the survival time compared with the model group, suggesting that in vivo enhanced autophagy may have a role in combating against leukemogenesis (Figure 2b).
Five mice were used for further experiments in each group and the killing time was set to 3 weeks due to the death time. Peripheral blood cell counting from the animal experiment showed that the white blood cell increased and the platelet decreased significantly in the model group, but rapamycin normalized the white blood cell number and platelet number, particularly in the treatment group (Figure 2c).
At the time of killing, liver and spleen were collected and weighed. At the appearance, the liver and spleen in the model group showed enlargement significantly (Figure 2d, left panel), but the enlargement was less significant in the preventive and treatment groups. The coefficient of liver and spleen (ratio of weight of liver or spleen to that of body weight) also showed that it was less tumid in the preventive and treatment groups than the model group (Figure 2d, right panel). The liver in the model group also showed significant invasion of cells, which was less significant in the preventive and treatment groups (Figure 2e).
To further confirm the role of autophagy in prevention or inhibition of B-ALL leukemogenesis, the expression of human CD45, CD19 and CD10 markers in the BM cells of the B-ALL mice administrated with or without rapamycin was measured. The results showed that the B-ALL markers expressed highly in the BM cells of model group, while reduced significantly in both rapamycin preventative and treatment groups, further suggesting that activation of autophagy in both prevention and treatment with rapamycin slows down or inhibits B-ALL leukemogenesis (Figure 2f).
Activation of autophagy induces cell-cycle arrest in the treatment animals and partially restores hematopoietic stem and progenitor cells in the preventive animals
To understand how activation of autophagy prevents or inhibits B-ALL leukemogenesis, cell cycle of the total BM cells of the four animal groups was first examined. The cell cycle in the preventive group did not show a difference from that in the model group, however, cell cycle of the treatment group showed that cell percentage of G0/G1 phase increased and S, G2/M phase decreased significantly compared with that of the model group (Figure 3a). This suggests that enhancement of autophagy by treatment with rapamycin after transplantation of the leukemia cells causes cell-cycle arrest of the total BM cells.
Manipulating autophagy affected the in vivo cell cycle of BM cells and LSK percentage in the xenograft mice. (a) The cell cycle of BM cells showed that the cells in G0/G1 phase decreased significantly and the cells in S and G2/M increased in model group, but the cells in G0/G1 phase increased and the cells in S and G2/M phase decreased significantly in rapamycin treatment group, whereas the rapamycin preventative group did not show a significant change in cell cycle as compared with model group. (b, c) The LSK (both CD34−LSK and CD34+LSK) percentage decreased significantly in model group, but the LSK number (both CD34−LSK and CD34+LSK) restored significantly in the preventive group, not in the treatment group. **P<0.01, *P<0.05.
We purified bone morrow LSK cells from the mice with Ficoll and MACS lineage kit. CD34−LSK markers were used as long-term HSC phenotype, and CD34+LSK markers were used as short-term HSC phenotype, shown by the representative flow gating for hematopoietic stem/progenitor cells (Figure 3b). Flow-cytometric analysis discloses a reduced cellurality of LSK cell population in the model group. In the preventive group, but not the treatment group, LSK (including CD34− LSK, CD34+ LSK) cell percentage increased significantly compared with that of model group (Figure 3c), suggesting that enhancement of autophagy by preventive administration of rapamycin restores, at least in part, the pool of hematopoietic stem and progenitor cells.
Activation of autophagy downregulates E2A/Pbx1 level in B-ALL 697 cells
To further explore the mechanism by which activation of autophagy combating against B-ALL, autophagy of the B-ALL 697 cells was activated with rapamycin or starvation (amino-acid depletion with HBSS medium). Autophagy level was analyzed by the LC3 Spot count wizard through image flow cytometry, showing representative and statistical accumulated LC3 puncta upon treatment with rapamycin or starvation (Figure 4a). Flow-cytometry analysis showed that E2A/Pbx1 protein level downregulated significantly after activation of autophagy by rapamycin or nutrient depletion by HBSS culture medium (Figure 4b). Western blotting analysis of total cellular protein, cytoplasmic protein and nucleus protein also showed downregulated E2A/Pbx1 protein level through activation of autophagy by rapamycin and in particular starvation. The downregulation of E2A/Pbx1 not only occurs mostly in the cytoplasm, nuclear E2A/Pbx1 level was also reduced by activation of autophagy by rapamycin or starvation, suggesting that autophagy activation might cause relocalization of the oncogenic nuclear E2A/Pbx1 to the cytoplasm for degradation (Figure 4c). The western blotting results were confirmed by confocal microscopic analysis, which discloses that E2A/Pbx1-PE was merged with GFP-LC3 in the HBSS and rapamycin treatment groups in the cytoplasm, while nuclear E2A/Pbx1 was relocalized to the cytoplasm upon treatment with HBSS or rapamycin and this relocalization was blocked by bafilomycin A1, an autophagy inhibitor (Figure 4d).
Activation of autophagy by rapamycin or nutrient depletion degraded E2A/Pbx1 protein in in vitro B-ALL 697 cells and in vivo NOD-SCID leukemia mice. Image flow cytometry (a) showed that autophagy was activated by rapamycin or nutrient depletion with HBSS medium (high LC3 spot count), and E2A/Pbx1 protein decreased in Rap and HBSS groups (b). Western blot (c) of total protein, cytoplasm protein and nucleus protein also showed that activation of autophagy could degrade E2A/Pbx1 protein and the degradation mostly happen in cytoplasm protein, which is confirmed by confocal microscopy (d), E2A/Pbx1-PE was merged with GFP-LC3 in HBSS and rapamycin groups in cytoplasm, indicating that E2A/Pbx1 was degraded by autophagy. E2A/Pbx1 was also degraded in the rapamycin treatment group of leukemia NOD-SCID mice liver protein (e). ***P<0.001.
Furthermore, western blotting analysis of liver proteins from the B-ALL xenograft mouse model indicates that preventive administration with rapamycin partially downregulated E2A/Pbx1 protein level, whereas treatment with rapamycin significantly minimized the E2A/Pbx1 protein level (Figure 4e), further suggesting that E2A/Pbx1 might be degraded at least in part, by autophagy.
Inhibition of autophagy or ubiquitination accumulates E2A/Pbx1
To explore whether other cellular mechanism might be implicated in the downregulation of the oncogenic E2A/Pbx1 protein, the B-ALL 697 cells were treated with PI3K inhibitor 3MA to inhibit autophagy, and ubiquitin-proteasome inhibitor MG132 to inhibit ubiquitination at different time points. While at basal autophagy level where autophagy was not manipulated with HBSS or rapamycin, treatment with these inhibitors did not cause an obvious change at E2A/Pbx1 level, detected by flow cytometry (Figure 5a). In contrast, the results by western blotting analysis showed that similar to the inhibition of basal autophagy with 3-MA that caused accumulation of E2A/Pbx1 protein, inhibition of ubiquitination with MG132 alone or in combination with 3-MA also caused an accumulation of E2A/Pbx1 protein (Figure 5b).
Inhibition of either autophagy or ubiquitination fails to degrade E2A/Pbx1. Flow cytometry (a) showed that inhibition of autophagy by 3-MA and inhibition of ubiquitination by ubiquitin-proteasome inhibitor MG132 fails to degrade E2A/Pbx1, and the oncoprotein accumulated in the combined group in western botting result (b), indicating that both autophagy and ubiquitination contribute to the degradation of E2A/Pbx1. Nutrient depletion with HBSS medium also induced ubiquitination of 697 cells (c), and MG132 restored the degradation of E2A/Pbx1 induced by nutrient depletion with HBSS (d).
Furthermore, western blotting analysis with pan-ubiquitin indicates that pan ubiquitination in the B-ALL 697 cells was increased in HBSS starvation (Figure 5c), suggesting that autophagy might promote ubiquitinational degradation of the E2A/Pbx1 protein. While MG132 alone effectively blocks the degradation of E2A/Pbx1, it also partially blocked the degradation of the oncogenic protein caused by HBSS starvation (Figure 5d). Interestingly, Amnis image flow-cytometric analysis showed that MG132 blocked HBSS-caused E2A/Pbx1 cytoplasmic relocalization (Figure 6a) and statistically, MG132 blocked HBSS starvation-caused degradation of E2A/Pbx1 (Figure 6d). These data suggest that autophagy and ubiquitination might be collaborated to downregulate E2A/Pbx1 level.
Colocalization of E2A/Pbx1 with LC3 or Ub in pediatric B-ALL 697 cells. (a,b) The images of fluorescence intensity of LC3-FITC, Ubiquitin-FITC, E2A/Pbx1-PE and merged images of E2A/Pbx1 with LC3 or Ub are shown. Simultaneously, autophagy level presented by LC3 spot count (c) and E2A/Pbx1 expression (d) was also analyzed. The colocalization statistical percentage of E2A/Pbx1 with LC3 and E2A/Pbx1 with Ub was showed in (e) and (f), in rapamycin treatment, colocalization of E2A/Pbx1 with LC3 increased significantly, but in HBSS treatment, both colocalization of E2A/Pbx1 with LC3 and Ub increased, especially with Ub, and MG132 reversed the colocalization of E2A/Pbx1 with Ub in HBSS treatment ***P<0.001, **P<0.01, *P<0.05.
Activation of autophagy causes colocalization of E2A/Pbx1 with LC3 or Ubiquitin
Experiment with ubiquitination inhibitor indicates that ubiquitination might be involved in the degradation of E2A/Pbx1 as autophagy does. One would expect that the oncogenic protein E2A/Pbx1 and ubiquitin should be colocalized for ubiquitinational degradation. To this end, Amnis image flow cytometer was used to visually and statistically examine whether this colocalization occurs. Similar to the observation that autophagy activation by amino-acid depletion or rapamycin causes more LC3 or E2A/Pbx1 moving out of the nucleus, and subsequently colocalization in the cytoplasm (Figure 6a), starvation by amino-acid depletion with HBSS medium caused colocalization between ubiquitin and E2A/Pbx1, and treatment with MG132 significantly blocked E2A-Pbx1/Ub colocalization (Figures 6b and f). These data further suggest that ubiquitination is also involved in the degradation of the E2A/Pbx1 protein.
Discussion
Pharmacological manipulation of autophagy has been reported to enhance the activity of anticancer agents. We previously found that autophagy is essential for vitamin D3-induced differentiation of myeloid leukemia cells.12 Other groups also reported that activation of autophagy causes autophagic cell death in pediatric AML, chronic myeloid leukemia, ALL and chronic lymphoblastic leukemia.21, 22, 23, 24, 25, 26, 27, 28 Of particular interest, autophagy was shown to degrade oncogenic fusion proteins such as PML-RARα in acute promyelocytic leukemia13, 29, 30, 31, 32 and in the human T-cell lymphocytic leukemia cell line Molt4.33 The downregulation of the PML/RARα oncoprotein is mainly affected by autophagy since its degradation in cells treated with arsenic trioxide or all-trans retinoic acid depends upon and is related to increased autophagic activity.13 Indeed, the above antileukemic effects were largely attributable to the activation of autophagy, which either non-specifically overdigests intracellular components, leading to cellular demise or specifically digests oncogenic fusion proteins to promote terminal differentiation of leukemia cells.
Our recent study indicates that autophagy activated by rapamycin inhibits proliferation and induces cell-cycle arrest in in vitro childhood pre-B ALL cells by specifically downregulating DNA pol δ1 and RNA pol II, at both the transcriptional and post-translational level. The post-translational mechanism possibly involves autophagic degradation of the polymerases. The downregulation of two key polymerases in DNA and RNA synthesis causes the reduced expression of multiple positive cell-cycle regulators, ultimately leading to a proliferation blockade and growth arrest.19 However, whether enhancement of autophagy can inhibit in vivo pre-B ALL cells and whether autophagy collaborates with other degradation mechanism to prevent or cure this B-ALL remains unclear.
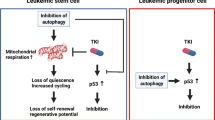
In the present study, we show that autophagy is downregulated in childhood B-ALL (Figure 1), suggesting a possible link between autophagy failure and pediatric B-ALL leukemogenesis. We thus generated a pediatric t(1;19) B-ALL xenograft mouse model and administrated with rapamycin before or after transplanting the leukemia cells to mimic a preventive or treatment regimen on the B-ALL animal model. With this ALL xenograft mouse model, we show here that activation of autophagy by preventive administration of rapamycin improves the survival of leukemia animals by partial restoration of hematopoietic stem/progenitor cells, whereas treatment of the animal with rapamycin caused leukemia BM cell-cycle arrest, suggesting that in addition to treatment of leukemia, enhancement of autophagy might be a possible strategy to prevent or delay leukemia occurrence. We observed that activation of autophagy in vitro or in vivo by administration with rapamycin or starvation with amino-acid depletion downregulated oncogenic fusion protein E2A/Pbx1. Furthermore, E2A/Pbx1 was found to be colocalized with autophagy marker LC3 in autolysosomes, and this oncogenic protein was also colocalized with ubiquitin in response to autophagy stimuli, whereas autophagy inhibitor or ubiquitination inhibitor blocked these colocalizations, supporting our notion that both autophagy and ubiquitination are involved in the degradation of E2A/Pbx1. Interestingly, ubiquitination inhibitor MG132 blocked starvation-caused degradation of E2A/Pbx1 (Figure 6d), confirming that autophagic degradation of E2A/Pbx1 depends on ubiquitination. Hence, autophagy and ubiquitination could collaborate with each other to downregulate oncogenic E2A/Pbx1 level. To our knowledge, this is the first evidence that autophagy collaborates with ubiquitination to fight against cancer cells via degrading an oncogenic protein. Our findings thus reveal a novel strategy for targeted prevention or targeted therapy on the pediatric B-ALL.
References
Levine B, Klionsky DJ . Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell 2004; 6: 463–477.
Nencioni A, Cea M, Montecucco F, Longo VD, Patrone F, Carella AM et al. Autophagy in blood cancers: biological role and therapeutic implications. Haematologica 2013; 98: 1335–1343.
Helgason GV, Karvela M, Holyoake TL . Kill one bird with two stones: potential efficacy of BCR-ABL and autophagy inhibition in CML. Blood 2011; 118: 2035–2043.
Altman BJ, Jacobs SR, Mason EF, Michalek RD, Maclntyre AN, Coloff JL et al. Autophagy is essential to suppress cell stress and to allow BCR-Abl–mediated leukemogenesis. Oncogene 2011; 30: 1855–1867.
Ertmer A, Huber V, Gilch S, Yoshimori T, Erfle V, Duyster J et al. The anticancer drug imatinib induces cellular autophagy. Leukemia 2007; 21: 936–942.
Bellodi C, Lidonnici MR, Hamilton A, Helgason GV, Soliera AR, Ronchetti M et al. Targeting autophagy potentiates tyrosine kinase inhibitor-induced cell death in Philadelphia chromosome-positive cells, including primary CML stem cells. J Clin Invest 2009; 119: 1109–1123.
Carew JS, Nawrocki ST, Kahue CN, Zhang H, Yang C, Chung L et al. Targeting autophagy augments the anticancer activity of the histone deacetylase inhibitor SAHA to overcome Bcr-Abl-mediated drug resistance. Blood 2007; 110: 313–322.
Kamitsuji Y, Kuroda J, Kimura S, Toyokuni S, Watanabe K, Ashihara E et al. The Bcr-Abl kinase inhibitor INNO-406 induces autophagy and different modes of cell death execution in Bcr-Abl-positive leukemias. Cell Death Differ 2008; 15: 1712–1722.
Crowley LC, Elzinga BM, O'Sullivan GC, McKenna SL . Autophagy induction by Bcr-Abl-expressing cells facilitates their recovery from a targeted or nontargeted treatment. Am J Hematol 2011; 86: 38–47.
Amrein L, Soulières D, Johnston JB, Aloyz R . p53 and autophagy contribute to dasatinib resistance in primary CLL lymphocytes. Leuk Res 2011; 35: 99–102.
Liu L, Yang M, Kang R, Wang Z, Zhao Y, Yu Y et al. HMGB1-induced autophagy promotes chemotherapy resistance in leukemia cells. Leukemia 2011; 25: 23–31.
Wang J, Lian H, Zhao Y, Kauss MA, Spindel S . Vitamin D3 induces autophagy of human myeloid leukemia cells. J Biol Chem 2008; 283: 25596–25605.
Isakson P, Bjoras M, Boe SO, Simonsen A . Autophagy contributes to therapy-induced degradation of the PML/PARA oncoprotein. Blood 2010; 116: 2324–2331.
Crist WM, Carroll AJ, Shuster JJ, Behm FG, Whitehead M, Vietti TJ et al. Poor prognosis of children with pre-B acute lymphoblastic leukemia is associated with the t(1;19)(q23;p13): A Pediatric Oncology Group study. Blood 1990; 76: 117–122.
Kamps MP, Look AT, Baltomore D . The human t(1;19) translocation in pre-B ALL produces multiple nuclear E2A-Pbx1 fusion proteins with differing transforming potentials. Gene Dev 1991; 5: 358–368.
Raimondi SC, Behm FG, Roberson PK, Williams DL, Pui CH, Crist WM et al. Cytogenetics of pre-B-cell acute lymphoblastic leukemia with emphasis on prognostic implications of the t(1;19). J Clin Oncol 1990; 8: 1380–1388.
Pui CH, Jeha S . New therapeutic strategies for the treatment of acute lymphoblastic leukaemia. Nat Rev Drug Discov 2007; 6: 149–165.
Jeha S . New therapeutic strategies in acute lymphoblastic leukemia. Semin Hematol 2009; 46: 76–88.
Wang Z, Xu F, Yuan N, Niu Y, Lin W, Cao Y et al. Rapamycin inhibits pre-B acute lymphoblastic leukemia cells by downregulating DNA and RNA polymerases. Leuk Res 2014; 38: 940–947.
Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K et al. Guideline for the use and interpretation of assays for monitoring autophagy. Autophagy 2012; 8: 445–544.
Crazzolara R, Cisterne A, Thien M, Hewson J, Baraz R, Bradstock KF et al. Potentiating effects of RAD001 (Everolimus) on vincristine therapy in childhood acute lymphoblastic leukemia. Blood 2009; 113: 3297–3306.
Bonapace L, Bornhauser BC, Schmitz M, Cario G, Ziegier U, Niggli FK et al. Induction of autophagy-dependent necroptosis is required for childhood acute lymphoblastic leukemia cells to overcome glucocorticoid resistance. J Clin Invest 2010; 120: 1310–1323.
Laane E, Tamm KP, Buentke E, Ito K, Kharaziha P, Oscarsson J et al. Cell death induced by dexamethasone in lymphoid leukemia is mediated through initiation of autophagy. Cell Death Differ 2009; 16: 1018–1029.
Ivanov A, Beers SA, Walshe CA, Honeychurch J, Alduaij W, Cox KL et al. Monoclonal antibodies directed to CD20 and HLA-DR can elicit homotypic adhesion followed by lysosome-mediated cell death in human lymphoma and leukemia cells. J Clin Invest 2009; 119: 2143–2159.
Samudio I, Kurinna S, Ruvolo P, Korchin B, Kantarjian H, Beran M et al. Inhibition of mitochondrial metabolism by methyl-2-cyano-3,12-dioxooleana-1,9-diene-28-oate induces apoptotic or autophagic cell death in chronic myeloid leukemia cells. Mol Cancer Ther 2008; 7: 1130–1139.
Robert G, Ben Sahra I, Puissant A, Colosetti P, Belhacene N, Gounon P et al. Acadesine kills chronic myelogenous leukemia (CML) cells through PKC-dependent induction of autophagic cell death. PLoS One 2009; 4: e7889.
Chen YJ, Huang WP, Yang YC, Lin CP, Chem SH, Hsu ML et al. Platonin induces autophagy-associated cell death in human leukemia cells. Autophagy 2009; 5: 173–183.
Grimaldi C, Chiarini F, Tabellini G, Ricci F, Tazzari PL, Battistelli M et al. AMP-dependent kinase/mammalian target of rapamycin complex 1 signaling in T-cell acute lymphoblastic leukemia: therapeutic implications. Leukemia 2012; 26: 91–100.
Qian W, Liu J, Jin J, Ni W, Xu W . Arsenic trioxide induces not only apoptosis but also autophagic cell death in leukemia cell lines via up regulation of Beclin1. Leuk Res 2007; 31: 329–339.
Yang C, Tong Y, Ni W, Liu J, Xu W, Li L et al. Inhibition of autophagy induced by overexpression of mda-7/interleukin-24 strongly augments the antileukemia activity in vitro and in vivo. Cancer Gene Ther 2010; 17: 109–119.
Ren Y, Xie Y, Chai L, Wang S, Cheng M . Autophagy modification augmented the treatment effects initiated by arsenic trioxide in NB4 cells. Med Oncol 2011; 28: 231–236.
Goussetis DJ, Altman JK, Glaser H, McNeer JL, Tailman MS, Platanias LC et al. Autophagy is a critical mechanism for the induction of the antileukemic effects of arsenic trioxide. J Biol Chem 2010; 285: 29989–29997.
Song J, Qu Z, Guo X, Zhao Q, Zhao X, Gao L et al. Hypoxia in duced autophagycontributes to the chemoresistance of hepatocellular carcinoma cells. Autophagy 2009; 5: 1131–1144.
Acknowledgements
This work was supported by grants from National Science Foundation of China (No. 31071258, No. 81272336 and No. 31201073), National Basic Research Program of China, The Ministry of Science and Technology of China (No. 2011CB512101), Basic Research Program of Jiangsu Province (No. BK20130333) and a project funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/4.0/
About this article
Cite this article
Yuan, N., Song, L., Lin, W. et al. Autophagy collaborates with ubiquitination to downregulate oncoprotein E2A/Pbx1 in B-cell acute lymphoblastic leukemia. Blood Cancer Journal 5, e274 (2015). https://doi.org/10.1038/bcj.2014.96
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/bcj.2014.96
This article is cited by
-
The advances of E2A-PBX1 fusion in B-cell acute lymphoblastic Leukaemia
Annals of Hematology (2023)
-
Molecular processes involved in B cell acute lymphoblastic leukaemia
Cellular and Molecular Life Sciences (2018)
-
Autophagy therapeutics: preclinical basis and initial clinical studies
Cancer Chemotherapy and Pharmacology (2018)
-
Abrogation of Nrf2 impairs antioxidant signaling and promotes atrial hypertrophy in response to high-intensity exercise stress
Journal of Translational Medicine (2016)