Abstract
CCAAT/enhancer-binding protein alpha (CEBPA) mutations are a favorable prognostic factor in adult acute myeloid leukemia (AML) patients; however, few studies have examined their significance in pediatric AML patients. Here we examined the CEBPA mutation status and clinical outcomes of pediatric AML patients treated in the AML-05 study. We found that 47 (14.9%) of the 315 evaluable patients harbored mutations in CEBPA; 26 cases (8.3%) harbored a single mutation (CEBPA-single) and 21 (6.7%) harbored double or triple mutations (CEBPA-double). After excluding core-binding factor-AML cases, patients harboring CEBPA mutations showed better overall survival (OS; P=0.048), but not event-free survival (EFS; P=0.051), than wild-type patients. Multivariate analysis identified CEBPA-single and CEBPA-double as independent favorable prognostic factors for EFS in the total cohort (hazard ratio (HR): 0.47 and 0.33; P=0.02 and 0.01, respectively). CEBPA-double was also an independent favorable prognostic factor for OS (HR: 0.30; P=0.04). CEBPA-double remained an independent favorable factor for EFS (HR: 0.28; P=0.04) in the normal karyotype cohort. These results suggest that CEBPA mutations, particularly CEBPA-double, are an independent favorable prognostic factor in pediatric AML patients, which will have important implications for risk-stratified therapy.
Similar content being viewed by others
Introduction
CCAAT/enhancer-binding protein alpha (CEBPA) is a transcription factor that co-ordinates cellular differentiation. CEBPA is expressed in myeloid precursors during hematopoiesis, where it regulates the expression of several granulocyte-specific genes.1 CEBPA inhibits E2F pathways, thereby downregulating c-Myc and allowing myeloid precursors to enter the granulocytic differentiation pathway.2, 3 The CEBPA gene is located on chromosome 19 band q13.1. Approximately 10% of acute myeloid leukemia (AML) patients harbor mutations in CEBPA genes, and these mutations can occur across the whole gene, but there are two main hotspots.4, 5 N-terminal out-of-frame mutations are located between the major translational start site and a second ATG further downstream. They abolish translation of the full-length p42 isoform of CEBPA, leading to overexpression of a shorter dominant-negative p30 isoform.6 C-terminal mutations are generally in-frame insertions/deletions located in the basic leucine zipper (bZIP) domain; these mutations disrupt binding to DNA or dimerization.7 Most AML patients with double CEBPA mutations harbor both N- and C-terminal mutations, which are typically present on different alleles; however, homozygous mutations have also been described.8
CEBPA mutations are a favorable prognostic factor for AML, particularly in patients harboring double CEBPA mutations and a normal karyotype.8, 9, 10, 11, 12, 13 However, the prognostic value of CEBPA mutations has been studied mostly in adult AML patients, with few studies examining mutations in pediatric AML patients. The first set of pediatric data was presented by the Taiwan Pediatric Oncology Group, but the report lacked data regarding clinical outcome.14 The prognostic impact of CEBPA in pediatric AML was reported by two other groups, namely, the Children’s Oncology Group and the Dutch Childhood Oncology Group/the Berlin–Frankfurt–Münster Study Group,15, 16 which both reported that, after excluding core-binding factor (CBF)-AML cases, patients harboring CEBPA mutations had a significantly better clinical outcome than those harboring the wild-type (WT) gene; however, the clinical implications of single vs double mutations were unclear. A more recent study conducted by the Nordic Society of Pediatric Hematology and Oncology suggests that CEBPA mutations in pediatric AML patients are not associated with improved survival;17 thus the clinical significance of CEBPA mutations in pediatric AML patients is unclear. Although we previously reported the characteristics of CEBPA mutations in Japanese children with AML, the small sample size meant that further study was required.18
Here we examined the CEBPA mutation status and clinical outcomes of pediatric AML patients treated in the Japanese Pediatric Leukemia/Lymphoma Study Group (JPLSG) AML-05 study.
Subjects and methods
Patients and study protocol
The AML-05 study is a Japanese nationwide multi-institutional study of children (age <18 years) with de novo AML, all of whom were enrolled between 1 November 2006 and 31 December 2010. The trial is registered with the UMIN Clinical Trials Registry (UMIN–CTR; http://www.umin.ac.jp/ctr/index.htm; number UMIN000000511).
In all, 485 patients with suspected AML (diagnosed at 118 centers and hospitals in Japan) were registered in the AML-05 study. Patients with acute promyelocytic leukemia, Down’s syndrome, secondary AML, myeloid/natural killer cell leukemia and myeloid sarcoma, were not eligible. Overall, 38 patients were excluded, mainly because of misdiagnosis, while four additional patients were excluded for the following reasons: the patient’s guardian refused permission to participate (n=1); there was a significant protocol violation during the initial induction course (n=1); the hospital withdrew from the JPLSG (n=1); and the patient was transferred to a non-JPLSG member hospital (n=1). Patients were stratified into three risk groups according to specific cytogenetic characteristics and morphological responses to treatment. CBF-AML patients were assigned to the low-risk group; those with unfavorable cytogenetics (−7, 5q-, t(16;21)(p11;q22), Ph1, Fms-like tyrosine kinase 3 internal tandem duplications (FLT3-ITD)) and poor induction responders were assigned to the high-risk group; and the rest were assigned to the intermediate-risk group. Details of the patient disposition, treatment schedules and risk stratification have been described previously.19, 20 In the present study, morphology was diagnosed prospectively using a central review system. Cytogenetic tests were performed in regional laboratories, but the reports were reviewed centrally. The study was conducted in accordance with the principles set down in the Declaration of Helsinki and was approved by the Ethics Committees of all participating institutions. All patients, or the patients’ parents/guardians, provided written informed consent.
Mutation analysis
cDNA was synthesized from RNA obtained from diagnostic bone marrow samples using the Omniscript Reverse Transcription Kit (Qiagen, Chatsworth, CA, USA), according to the manufacturer’s recommendations. The entire coding region of the CEBPA gene was amplified using the overlapping PCR primer pairs followed by direct sequencing, as previously described.6, 18
Statistical analysis
Patient characteristics were analyzed using Fisher’s exact test (categorical variables) and the Kruskal–Wallis test (continuous variables). Event-free survival (EFS) was defined as the time from the diagnosis of AML to the last follow-up or the first event (failure to achieve remission, relapse, secondary malignancy or any-cause death). Overall survival (OS) was defined as the time from the diagnosis of AML to any-cause death. The Kaplan–Meier method was used to estimate EFS and OS, and data were compared using the log-rank test. To determine the prognostic value of CEBPA mutation, Cox regression analysis was performed using SAS version 9.2 (SAS Institute Inc., Cary, NC, USA). All tests were two-tailed, and a P-value <0.05 was considered statistically significant.
Results
Mutation analysis
Diagnostic samples from 315/443 (71.1%) eligible AML patients were analyzed for CEBPA mutations; CEBPA data were unavailable for 128 patients. There were no significant differences in the major characteristics or clinical outcomes of the 315 patients and the 128 patients for whom no data were available (EFS P=0.78, OS P=0.30). We found that 47/315 patients (14.9%) harbored a mutation in CEBPA, 26 (8.3%) harbored a single mutation and 21 (6.7%) harbored double or triple mutations. The location and combination pattern of all the detected mutations are shown in Figure 1 and Table 1. Single mutations were distributed across the entire gene, but most in-frame insertions/deletions were located in the bZIP domain. By contrast, double or triple mutations were clustered in the N- and C-terminal hotspots. Thirteen out of the 21 cases (61.9%) harbored both an N-terminal out-of-frame mutation and an in-frame mutation in the bZIP domain, which were predicted to result in a lack of WT CEBPA p42 expression. We identified five patients with triple mutations but could not exclude the possibility that these mutations occurred in different cells. Moreover, the method we used cannot identify whether mutations are located on different alleles. Further study is required to overcome these limitations.
Polymorphisms in the CEBPA mutations
Overall, 131 patients (41.6%) harbored an in-frame 6-bp insertion (ACCCGC) in the transactivation domain 2 (TAD2), resulting in a His-Pro duplication (HP196–197 insertion). This mutation is observed in approximately 10% of healthy controls and AML patients and is reported as a germline polymorphism.21, 22 We did not identify any differences in characteristics between the HP196–197 insertion-positive and -negative groups, and the clinical outcomes of both groups were similar (data not shown). Therefore, we ignored this mutation during our analysis of clinical outcome, along with other mutations that did not result in amino-acid changes.
Patient characteristics
Patient characteristics according to CEBPA mutation status are shown in Table 2. Patients harboring a single CEBPA mutation were described as ‘CEBPA-single’ and those harboring double or triple CEBPA mutations were described as ‘CEBPA-double’. CEBPA-double patients showed a significantly higher percentage of M1 or M2 French–American–British subtypes (P< 0.001). Compared with WT patients, patients with CEBPA mutations were older (P=0.03) at the time of diagnosis. CEBPA mutations were predominant in those with an intermediate risk (P=0.002) and a normal karyotype (P<0.001). There was a well-balanced gender distribution (P=0.84), and there were no significant differences in the number of patients with FLT3-ITD and NPM1 mutations among the three CEBPA subgroups.
Prognostic impact of CEBPA in the total cohort
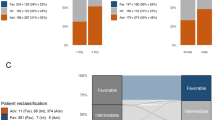
We first analyzed the clinical outcomes of patients harboring CEBPA mutations and then compared them with the outcomes of CBF-AML patients and patients without CBF or CEBPA mutations (denoted ‘WT non-CBF’) (Figure 2). The CBF-AML group included AML patients harboring t(8;21)(q22;q22) along with inv(16)(p13.1q22) or its variant t(16;16)(p13.1;q22). Seven CBF-AML patients harboring CEBPA mutations were categorized as ‘CEBPA-mutant’. Patients harboring CEBPA mutations showed better OS (P=0.048), but not EFS (P=0.051), than WT non-CBF patients (Figures 2a and b). However, patients with CEBPA mutations showed poorer OS (P=0.0006) than patients with CBF-AML. Furthermore, we examined whether the number of CEBPA mutations had an impact on prognosis (Figures 2c and d). CEBPA-double patients did not show significantly better EFS and OS than CEBPA-single patients (P=0.33 each). There was also no significant difference in EFS and OS between CEBPA-double patients and WT non-CBF patients (P=0.055 and P=0.057, respectively).
Kaplan–Meier survival curves showing EFS and OS from the time of diagnosis according to CEBPA mutation status. (a) EFS and (b) OS of patients harboring CEBPA mutations, patients harboring WT CEBPA (excluding core-binding factor-acute myeloid leukemia (CBF-AML) cases (WT non-CBF)) and patients with CBF-AML. (c) EFS and (d) OS of patients harboring a single CEBPA mutation (CEBPA-single), patients harboring double or triple CEBPA mutations (CEBPA-double), WT patients (excluding CBF-AML cases (WT non-CBF)) and patients with CBF-AML. P-values were determined using the log-rank test.
Prognostic impact of CEBPA in the normal karyotype cohort
We next examined prognosis in the normal karyotype cohort, because CEBPA mutations have been described as a favorable prognostic factor, particularly in cytogenetically normal AML (Figure 3). There was no significant difference in EFS and OS between CEBPA-double patients and WT or CEBPA-single patients (EFS: CEBPA-double vs WT, P=0.15; CEBPA-double vs CEBPA-single, P=0.21; OS: CEBPA-double vs WT, P=0.28; CEBPA-double vs CEBPA-single; P=0.44). Patients with CEBPA-single showed almost identical EFS (P=0.97) and OS (P=0.77) to those of WT patients.
Kaplan–Meier survival curves showing EFS and OS in acute myeloid leukemia patients with a normal karyotype. (a) EFS and (b) OS of patients harboring a single CEBPA mutation (CEBPA-single), patients harboring double or triple CEBPA mutations (CEBPA-double) and WT patients. P-values were determined using the log-rank test.
Prognostic impact of CEBPA mutation type
We also examined the prognostic impact of the location of the CEBPA mutations, which has never been examined in pediatric AML patients. Only patients with hotspot mutations predicted to cause translation of the p30 isoform and/or disruption or loss of the C-terminal bZIP domain were included in the analysis. In the total cohort, patients with an N-terminal out-of-frame mutation and a C-terminal in-frame mutation (n=13, denoted as CEBPA-double N+C-term) had significantly better EFS (P=0.01), but not OS (P=0.06), than WT non-CBF patients (Figures 4a and b). This patient group also had significantly better EFS, but not OS, than other CEBPA-double patients (n=8), suggesting that a combination of N-terminal and C-terminal mutations results in a better prognosis for CEBPA-double patients (data not shown). We also investigated differences in outcome between CEBPA-single patients with an N-terminal mutation and those with a C-terminal mutation and found that the clinical outcomes were nearly identical. In the normal karyotype cohort, we found no significant difference in the outcome of four groups: patients with an N-terminal out-of-frame mutation, patients with a C-terminal in-frame mutation, patients with an N-terminal out-of-frame mutation and a C-terminal in-frame mutation, and WT patients, which may be due to the small sample size (Figures 4c and d).
Kaplan–Meier survival curves showing EFS and OS according to the location and number of CEBPA mutations. Only patients with hotspot mutations predicted to cause p30 isoform translation and/or disruption or loss of the C-terminal bZIP domain were included in the analysis. (a) EFS and (b) OS in patients harboring a single N-terminal mutation (CEBPA-single N-term), patients harboring a single C-terminal mutation (CEBPA-single C-term), patients harboring both N and C-terminal mutations (CEBPA-double N+C-term), WT patients (excluding core-binding factor-acute myeloid leukemia (CBF-AML) cases (WT non-CBF)) and patients with CBF-AML. (c) EFS and (d) OS of AML patients with a normal karyotype. P-values were determined using the log-rank test.
Multivariate analysis
Multivariate Cox regression analysis, including age and white blood cell count at the time of diagnosis, was performed to examine whether CEBPA mutations were a favorable prognostic factor (Table 3). FLT3-ITD and NPM1 mutations were not included as variables owing to the small number of positive cases and because no statistically significant differences were detected by univariate analysis. For the total cohort (n=315), multivariate analysis identified both CEBPA-single and CEBPA-double as independent favorable prognostic factors for EFS (hazard ratio (HR): 0.47 and 0.33; P=0.02 and 0.01, respectively; upper column, Table 3). CEBPA-double was also identified as an independent favorable prognostic factor for OS (HR: 0.30; P=0.04). For the normal karyotype cohort (n=62), CEBPA-double was also identified as an independent prognostic factor for favorable EFS (HR: 0.28; P=0.04; lower column, Table 3). This may indicate that other factors, such as age and white blood cell count, had masked the benefit of CEBPA mutations in the univariate analysis.
Discussion
Here we examined CEBPA mutations in 315 pediatric AML patients enrolled in the AML-05 study. We detected CEBPA mutations in 47 patients (14.9%), which is comparable to the reported frequency in adult and pediatric AML patients (approximately 10%).8, 9, 10, 11, 12, 13, 14, 15, 16, 17 In all, 26 out of the 47 cases (55.3%) harbored a single CEBPA mutation; this percentage is higher than that reported in previous studies of pediatric AML patients.15, 16 We detected the HP196–197 insertion in 131/315 cases (41.6%). This well-known polymorphism was previously observed in approximately 10% of AML cases; thus the percentage identified in the present study was rather high.21, 22 Whether this polymorphism is also common in healthy Japanese populations remains to be seen. A recent study by a Korean group reported the incidence of this polymorphism as 30%; thus the frequency of this polymorphism may vary considerably according to geographical region.23 The majority of CEBPA-double patients comprised M1 or M2 French–American–British subtypes, which is in agreement with the findings of previous studies.15, 16 CEBPA mutations were predominant in the intermediate risk and normal karyotype group, which is also consistent with previous findings.15, 16, 17
With regard to prognosis, the results presented herein suggest that CEBPA mutations, especially CEBPA-double, are an independent favorable prognostic factor in pediatric AML patients. Multivariate analysis of the normal karyotype cohort identified CEBPA-double as an independent favorable prognostic factor for EFS, but not OS; this finding may be due to the small sample size. As the majority of pediatric AML patients lack markers that indicate a favorable or poor prognosis, it is important to identify prognostic markers in intermediate-risk patients. CEBPA mutations show promise as markers of a favorable prognosis in pediatric AML patients, because they are strongly associated with intermediate risk.
Consistent with our results, several studies (including one pediatric study) postulated that AML patients harboring double CEBPA mutations have a favorable prognosis.9, 10, 11, 12 Two different CEBPA mutations have a synergistic effect on AML development, and the mechanism underlying leukemogenesis is likely to be different from that in AML patients harboring a single CEBPA mutation.24, 25 We found that a combination of N-terminal and C-terminal mutations is essential for a better prognosis in CEBPA-double patients (data not shown), indicating that a favorable prognosis is restricted in patients who lack WT CEBPA p42 expression among CEBPA-double patients. Moreover, a recent study of a large cohort of adult AML patients suggests that patients harboring double CEBPA mutations belong to a genetically distinct subtype and should be clearly distinguished from patients harboring a single mutation.13 In this study, we could not examine the prognostic impact of concomitant molecular mutations because of their low incidence; therefore further analyses of pediatric AML patients is required.
In contrast to double CEBPA mutations, the prognostic value of single CEBPA mutation is currently under debate because of its small number. We detected a relatively large number of cases harboring a single CEBPA mutation in the total cohort, and multivariate analysis identified single mutation as an independent prognostic factor for favorable EFS (Table 3). Two adult AML studies (but no pediatric studies) showed that a single CEBPA mutation can be an independent favorable prognostic factor in patients harboring NPM1 mutations.26, 27 Indeed, the two patients in the present study that harbored both a single CEBPA mutation and an NPM1 mutation showed good long-term survival without any events. We also tried to examine the clinical significance of the location of the mutation in CEBPA-single patients but found no significant difference in outcomes for patients harboring N-terminal or C-terminal mutations. However, the CEBPA-single patients in the normal karyotype cohort who harbored a C-terminal mutation may have slightly poorer EFS and OS than those who harbored an N-terminal mutation (Figures 4c and d), which is not consistent with previous adult AML studies.12, 13 Gene expression profiling suggests that CEBPA-single patients harboring a C-terminal mutation are more similar to CEBPA-double patients than to CEBPA-single patients harboring N-terminal mutations.10 This latter study was performed in adult AML patients and needs to be validated in pediatric AML patients.
So far, the biological mechanisms underlying a favorable clinical outcome for AML patients harboring CEBPA mutations (including relative drug sensitivity) are not clear. Further studies of single and double CEBPA mutations and the underlying biology are required to enable better risk assessment and therapeutic approaches in pediatric AML.
Conclusion
This is the first nationwide study to examine the clinical significance of CEBPA mutations in Japanese pediatric AML patients. The results suggest that CEBPA mutations, especially double or triple CEBPA mutations, are an independent favorable prognostic factor for pediatric AML patients. CEBPA-double patients should be stratified into the favorable risk group, and the prognostic significance of these mutations should be validated prospectively.
References
Tenen DG, Hromas R, Licht JD, Zhang DE . Transcription factors, normal myeloid development, and leukemia. Blood 1997; 90: 489–519.
Porse BT, Pedersen TA, Xu X, Lindberg B, Wewer UM, Friis-Hansen L et al. E2F repression by C/EBPalpha is required for adipogenesis and granulopoiesis in vivo. Cell 2001; 107: 247–258.
Johansen LM, Iwama A, Lodie TA, Sasaki K, Felsher DW, Golub TR et al. c-Myc is a critical target for c/EBPalpha in granulopoiesis. Mol Cell Biol 2001; 21: 3789–3806.
Leroy H, Roumier C, Huyghe P, Biggio V, Fenaux P, Preudhomme C . CEBPA point mutations in hematological malignancies. Leukemia 2005; 19: 329–334.
Nerlov C . C/EBPalpha mutations in acute myeloid leukaemias. Nat Rev Cancer 2004; 4: 394–400.
Pabst T, Mueller BU, Zhang P, Radomska HS, Narravula S, Schnittger S et al. Dominant-negative mutations of CEBPA, encoding CCAAT/enhancer binding protein-alpha (C/EBPalpha), in acute myeloid leukemia. Nat Genet 2001; 27: 263–270.
Asou H, Gombart AF, Takeuchi S, Tanaka H, Tanioka M, Matsui H et al. Establishment of the acute myeloid leukemia cell line Kasumi-6 from a patient with a dominant-negative mutation in the DNA-binding region of the C/EBPalpha gene. Genes Chromosomes Cancer 2003; 36: 167–174.
Preudhomme C, Sagot C, Boissel N, Cayuela JM, Tigaud I, de Botton S et al. Favorable prognostic significance of CEBPA mutations in patients with de novo acute myeloid leukemia: a study from the Acute Leukemia French Association (ALFA). Blood 2002; 100: 2717–2723.
Pabst T, Eyholzer M, Fos J, Mueller BU . Heterogeneity within AML with CEBPA mutations; only CEBPA double mutations, but not single CEBPA mutations are associated with favourable prognosis. Br J Cancer 2009; 100: 1343–1346.
Wouters BJ, Löwenberg B, Erpelinck-Verschueren CA, van Putten WL, Valk PJ, Delwel R . Double CEBPA mutations, but not single CEBPA mutations, define a subgroup of acute myeloid leukemia with a distinctive gene expression profile that is uniquely associated with a favorable outcome. Blood 2009; 113: 3088–3091.
Dufour A, Schneider F, Metzeler KH, Hoster E, Schneider S, Zellmeier E et al. Acute myeloid leukemia with biallelic CEBPA gene mutations and normal karyotype represents a distinct genetic entity associated with a favorable clinical outcome. J Clin Oncol 2010; 28: 570–577.
Green CL, Koo KK, Hills RK, Burnett AK, Linch DC, Gale RE . Prognostic significance of CEBPA mutations in a large cohort of younger adult patients with acute myeloid leukemia: impact of double CEBPA mutations and the interaction with FLT3 and NPM1 mutations. J Clin Oncol 2010; 28: 2739–2747.
Fasan A, Haferlach C, Alpermann T, Jeromin S, Grossmann V, Eder C et al. The role of different genetic subtypes of CEBPA mutated AML. Leukemia 2013; 28: 794–803.
Liang DC, Shih LY, Huang CF, Hung IJ, Yang CP, Liu HC et al. CEBPalpha mutations in childhood acute myeloid leukemia. Leukemia 2005; 19: 410–414.
Ho PA, Alonzo TA, Gerbing RB, Pollard J, Stirewalt DL, Hurwitz C et al. Prevalence and prognostic implications of CEBPA mutations in pediatric acute myeloid leukemia (AML): a report from the Children's Oncology Group. Blood 2009; 113: 6558–6566.
Hollink IH, van den Heuvel-Eibrink MM, Arentsen-Peters ST, Zimmermann M, Peeters JK, Valk PJ et al. Characterization of CEBPA mutations and promoter hypermethylation in pediatric acute myeloid leukemia. Haematologica 2011; 96: 384–392.
Staffas A, Kanduri M, Hovland R, Rosenquist R, Ommen HB, Abrahamsson J et al. Presence of FLT3-ITD and high BAALC expression are independent prognostic markers in childhood acute myeloid leukemia. Blood 2011; 118: 5905–5913.
Mizushima Y, Taki T, Shimada A, Yui Y, Hiraumi Y, Matsubara H et al. Prognostic significance of the BAALC isoform pattern and CEBPA mutations in pediatric acute myeloid leukemia with normal karyotype: a study by the Japanese Childhood AML Cooperative Study Group. Int J Hematol 2010; 91: 831–837.
Tomizawa D, Tawa A, Watanabe T, Saito AM, Kudo K, Taga T et al. Excess treatment reduction including anthracyclines results in higher incidence of relapse in core binding factor acute myeloid leukemia in children. Leukemia 2013; 27: 2413–2416.
Tomizawa D, Tawa A, Watanabe T, Saito AM, Kudo K, Taga T et al. Appropriate dose reduction in induction therapy is essential for the treatment of infants with acute myeloid leukemia: a report from the Japanese Pediatric Leukemia/Lymphoma Study Group. Int J Hematol 2013; 98: 578–588.
Wouters BJ, Louwers I, Valk PJ, Löwenberg B, Delwel R . A recurrent in-frame insertion in a CEBPA transactivation domain is a polymorphism rather than a mutation that does not affect gene expression profiling-based clustering of AML. Blood 2007; 109: 389–390.
Biggio V, Renneville A, Nibourel O, Philippe N, Terriou L, Roumier C et al. Recurrent in-frame insertion in C/EBPalpha TAD2 region is a polymorphism without prognostic value in AML. Leukemia 2008; 22: 655–657.
Kim S, Kim DH, Jang JH, Jung CW, Jang MA, Ki CS et al. Novel mutations in CEBPA in Korean Patients with acute myeloid leukemia with a normal karyotype. Ann Lab Med 2012; 32: 153–157.
Bereshchenko O, Mancini E, Moore S, Bilbao D, Månsson R, Luc S et al. Hematopoietic stem cell expansion precedes the generation of committed myeloid leukemia-initiating cells in C/EBPalpha mutant AML. Cancer Cell 2009; 16: 390–400.
Somervaille TC, Cleary ML . Preview. Mutant CEBPA: priming stem cells for myeloid leukemogenesis. Cell Stem Cell 2009; 5: 453–454.
Dufour A, Schneider F, Hoster E, Benthaus T, Ksienzyk B, Schneider S et al. Monoallelic CEBPA mutations in normal karyotype acute myeloid leukemia: independent favorable prognostic factor within NPM1 mutated patients. Ann Hematol 2012; 91: 1051–1063.
Park SH, Chi HS, Cho YU, Jang S, Park CJ . CEBPA single mutation can be a possible favorable prognostic indicator in NPM1 and FLT3-ITD wild-type acute myeloid leukemia patients with intermediate cytogenetic risk. Leuk Res 2013; 37: 1488–1494.
Acknowledgements
This work was supported by a Grant for Clinical Cancer Research and a Grant-in-Aid for Cancer Research from the Ministry of Health, Labour and Welfare of Japan. We also thank to H Miyachi, N Kiyokawa, T Deguchi, Y Hayashi and all doctors participating in JPLSG AML-05 protocol.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Matsuo, H., Kajihara, M., Tomizawa, D. et al. Prognostic implications of CEBPA mutations in pediatric acute myeloid leukemia: a report from the Japanese Pediatric Leukemia/Lymphoma Study Group. Blood Cancer Journal 4, e226 (2014). https://doi.org/10.1038/bcj.2014.47
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/bcj.2014.47
This article is cited by
-
Sporadic and Familial Acute Myeloid Leukemia with CEBPA Mutations
Current Hematologic Malignancy Reports (2023)
-
Characterization of functional transposable element enhancers in acute myeloid leukemia
Science China Life Sciences (2020)
-
Prognostic value of genetic mutations in adolescent and young adults with acute myeloid leukemia
International Journal of Hematology (2018)
-
Purification of leukemic blast cells from blood smears using laser microdissection
International Journal of Hematology (2017)
-
NPM1, FLT3 and CEBPA mutations in pediatric patients with AML from Argentina: incidence and prognostic value
International Journal of Hematology (2016)