Abstract
Aim:
Dopamine receptors are present in the nervous system and also widely distributed in the periphery. The aim of this study was to investigate the role of D1 subtype dopamine receptors (DRD1) in the regulation of dehydroepiandrosterone sulfotransferase (SULT2A1) in HepG2 cells.
Methods:
HepG2 cells were treated with DRD1 agonists with or without DRD1 antagonist for 9 d. DRD1 and SULT2A1 mRNA expression, protein expression, and SULT2A1 activity were detected using RT-PCR, Western blotting and HPLC, respectively. The level of cAMP was measured using a commercial kit.
Results:
All the 5 DR subtypes (DRD1–DRD5) were found to be expressed in HepG2 cells. Treatment of HepG2 cells with the specific DRD1 agonists SKF82958 (2.5 μmol/L) or SKF38393 (5 and 50 μmol/L) significantly increased the mRNA and protein expression of both DRD1 and SULT2A1, and increased SULT2A1 activity and cAMP levels. These effects were partially blocked by co-treatment with the specific DRD1 antagonist SCH23390 (2.5 μmol/L). In addition, transfection of HepG2 cells with DRD1-specific siRNAs decreased DRD1 mRNA expression by 40%, which resulted in the reduction of SULT2A1 mRNA expression by 60%, protein expression by 40%, and enzyme activity by 20%.
Conclusion:
DRD1 activation upregulates DRD1 and SULT2A1 expression and SULT2A1 activity in HepG2 cells, suggesting that the DRD1 subtype may be involved in the metabolism of drugs and xenobiotics through regulating SULT2A1.
Similar content being viewed by others
Introduction
Dopamine is the predominant catecholamine neurotransmitter in the human central nervous system, where it controls a variety of functions including cognition, emotion, locomotor activity, hunger and satiety, and endocrine system regulation. Dopamine exerts its functions by binding to dopamine receptors1. Dopamine receptors (DRs) are widely expressed in the brain, belong to the seven transmembrane domain-containing G protein-coupled receptor family, and include five different receptor subtypes (named DRD1–DRD5). There are distinct similarities between the DRD1 and DRD5 subtypes and among the DRD2, DRD3, and DRD4 subtypes, which has led to the classification as the D1-like and D2-like receptor subfamilies, respectively2. In recent years, it has been found that DRs are not restricted solely to the nervous system but are instead widely distributed in the periphery, mainly at the levels of the cardiovascular system, the hepatobiliary system, the gastrointestinal system and the kidney. In particular, dopamine receptors are differentially expressed along the nephron where they regulate renal hemodynamics, electrolyte and water transport, and rennin secretion3. Dopamine receptors are also known to influence vasodilation and to change cardiac contractility within the cardiovascular system4,5. Shannon et al demonstrated that DRD2 is expressed in rat cholangiocytes but that DRD1 and DRD3 were absent. However, this group did not thoroughly investigate the expression of all these DRs in hepatocytes and rat liver. Their results also indicated that the DRD2 agonist quinelorane has an inhibitory effect on ductal secretion in secretin-stimulated choleresis6. Zhang et al recently demonstrated that DRD1 mediates the inhibition of dopamine on distal colonic motility. The increased dopamine content and DRD1 receptor expression in the smooth muscle layer may be a compensatory mechanism to balance enhanced colonic motility in response to acute cold-restraint stress7. In addition, the results of a recent study published in Cell suggest that DRs are expressed in cancer stem cells and breast cancer cells and may serve as a biomarker for diverse malignancies8. The growing information regarding the roles and functions of DRs in the peripheral nervous system prompted us to address the following questions: First, are DRD1–DRD5 receptors expressed in hepatocytes? Second, do these DRs have certain functions that are involved in liver metabolism? Third, do agonists and antagonists of specific DR subtypes regulate drug-metabolizing enzymes?
No other studies have so far found the existence of dopamine receptors in liver tissue or in hepatocytes or described a role for these receptors in the hepatic system except for two reports. One study showed that the expression of dopamine receptors was significantly different between diabetic and normal rats9. The other report used immunohistochemistry to show that DRD2 was expressed in liver but did not investigate the expression of the other DRs6. However, the brain dopaminergic system has been demonstrated to regulate the expression of liver cytochrome P450 (CYP450) enzymes by altering the levels of pituitary hormones and cytokines10,11,12. Furthermore, no in vitro studies have been performed to investigate the role of dopamine receptors in the regulation of CYP450s. As one of the major superfamilies of phase II drug-metabolizing enzymes, the induction of SULTs by hormones and other endogenous molecules has been widely described13,14,15,16. However, neither the induction of SULT by the important neurotransmitter dopamine nor the role of DRs in the regulation of sulfotransferases (SULTs) has been well studied. To the best of our knowledge, there are only two reports on the regulation of SULTs by the dopamine analogue methamphetamine (METH), which is a potent modulator of dopamine signaling in the brain and can increase the synaptic levels of dopamine, serotonin and norepinephrine17,18,19,20,21,22. One study used microarrays to screen a series of candidate genes after single-dose METH treatment in rats and found that METH treatment induced rat brain SULT1A1 by 4.3-fold in the amygdala21. The other study was performed by our group and found that a 7-d treatment of METH significantly induced the expression of SULT1A1, SULT2A1, and SULT1E1 at the mRNA and protein levels in rat liver and brain23. However, there are currently no data to indicate the regulatory mechanism of SULT by METH in the rat liver and brain. Considering the role of the brain dopaminergic system in the regulation of CYP450 isoforms in rat liver and the tight relationship between the action mechanisms of METH, dopamine, and the dopaminergic system, we hypothesized that dopamine receptors play an important role in the liver in the regulation of drug metabolizing enzymes, including sulfotransferase isoforms.
Dehydroepiandrosterone (DHEA)-sulfotransferase (SULT2A1) is a highly abundant, cytosolic sulfo-conjugating enzyme that is found in first-pass enterohepatic tissues and in steroidogenic adrenal tissue24,25. Similar to all other members of the SULT family, SULT2A1 utilizes 3′-phosphoadenosine 5′-phosphosulfate (PAPS) as the sulfate-donating cofactor. SULT2A1 specifically sulfates hydroxysteroid DHEA and also catalyzes the sulfation of endogenous steroids (testosterone, estrogen and pregnenolone), clinically active drugs and environmental chemicals24,26. SULT2A1 has been reported to be a target of many receptors that are activated by endogenous and xenobiotic agents. It remains unknown whether dopamine receptors affect SULT2A1 expression and activity in organs that are involved in biotransformation.
The purpose of this study was to investigate the influence of dopamine D1 receptor activation on SULT2A1 expression and activity. To accomplish this goal, we first examined the mRNA expression of DRD1–DRD5 in HepG2 cells. The human hepatoma cell line HepG2 is widely used to study the induction of drug-metabolizing enzymes in vitro27. We then evaluated the effects of different dopamine receptor agonists that were combined with a specific DRD1 antagonist on both DRD1 expression and SULT2A1 expression and activity in HepG2 cells. Finally, we also investigated the effects of DRD1 knockdown on the levels of human cytosolic SULT2A1 mRNA, protein, and activity and the effects of DRD1 agonists on cAMP (cyclic adenosine monophosphate) levels. Our results reveal that the activation of the DRD1 subtype plays an important role in SULT2A1 regulation in HepG2 cells.
Materials and methods
Materials
The following chemicals were used in our studies: dopamine, SKF38393, SKF82958 and SCH23390 (Sigma-Aldrich, St Louis, USA); fetal bovine serum (Hyclone, UT, USA); penicillin/streptomycin (Invitrogen, USA); Dulbecco's modified Eagle's medium (DMEM, M&C Gene Tech Co Ltd, Beijing, China); 0.25% trypsin-EDTA (Beijing Dingguo Biotechnology Co, Ltd, China), 3′-phosphoadenosine 5′-phosphosulfate (PAPS) and DHEA (Hubei Jianyuan Chemical Co Ltd, Wuhan, China); and theophylline (National Institutes for Food and Drug Control, Beijing, China). Rabbit anti-human SULT2A1 antibody was a gift from Dr David RINGER (American Cancer Society). Rabbit anti-actin and anti-DR (DRD1–DRD5) antibodies were purchased from Beijing Biosynthesis Biotechnology Co, Ltd (Beijing, China). Horseradish peroxidase-linked anti-rabbit antibody was purchased from Cell Signaling Technology Inc (Danvers, MA, USA).
HepG2 cell culture and treatment
HepG2 cells were obtained from the Cancer Institute and Hospital of the Chinese Academy of Medical Sciences (Beijing, China). Cells were cultured in DMEM that was supplemented with 10% fetal bovine serum and 100 U/mL of penicillin and streptomycin. The cultures were maintained in a humidified atmosphere with 5% CO2 at 37 °C. The medium was changed every 2 or 3 d with sub-culturing. All experiments using HepG2 cells were performed between passages 8 and 16 during which phase II enzyme expression did not significantly change27. To treat the HepG2 cells with the DRD1 agonists and the specific DRD1 antagonist SCH23390, the cells were seeded in 10-cm plates at a density of 2.5×105 cells per plate. HepG2 cells were treated on the first day with the dopamine receptor agonist with or without the specific DRD1 antagonist at the indicated concentrations by adding the compounds to the culture media. The control treatments included cells that were maintained for the same period of time in medium that was supplemented with the chemical solvent. The medium was changed every 2 d with the addition of fresh agonist. The cells were harvested on d 9. To carry out the DRD1-small interfering RNA (siRNA) transfection experiments in HepG2 cells, the cells were seeded in 6-well plates at a density of 5×105 cells per well. Lipofectamine 2000 (Invitrogen) was used for siRNA transfections by following the manufacturer's instructions.
Western blot analysis
Cells were homogenized in ice-cold RIPA buffer (50 mmol/L Tris pH 7.4, 150 mmol/L NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, protease inhibitor and 1 mmol/L phenylmethanesulfonylfluoride [PMSF]). Protein concentrations were determined by using a modified Bradford protein assay kit (Beyotime Institute of Biotechnology, Haimen, China). Protein lysates (20 μg) were separated by 12% SDS-PAGE and transferred to polyvinylidene difluoride (PVDF) membranes (Pierce, Rockford, IL, USA). Membranes were then probed with the following antibodies: anti-human actin (1:1000), anti-human SULT2A1 (1:1000), and anti-human DR (1:500). Horseradish peroxidase-linked anti-rabbit antibody was used as the secondary antibody. The membrane was developed with SuperSignal Ultra (Pierce, Rockford, IL, USA). Individual protein bands were quantified by densitometry with an imaging system (Bio-Rad, USA) and normalized to β-actin. Immunoblotting was performed as previously described28.
Real-time reverse-transcription polymerase chain reaction (RT-PCR) analysis
Total RNA was purified from HepG2 cells using an CellAmp Direct RNA Prep Kit for Real-Time PCR (Takara, Japan) according to the manufacturer's protocol. The concentration and purity of the extracted RNA was assessed by measuring the 260/280 absorption ratio using a spectrophotometer. cDNA was synthesized from 2 μg of RNA using a reverse transcriptase M-MLV synthesis kit (Takara, Japan). RT-PCR was performed on the MyiQ5 real-time PCR detection system (Bio-Rad, USA) with SYBR Premix Ex Taq (Takara, Japan). The PCR products for human β-actin, SULT2A1, DRD1, DRD2, DRD3, DRD4, and DRD5 were synthesized with the primer pairs FP (forward primer) 5′-CGCGAGAAGATGACCCAGAT-3′/RP (reverse primer) 5′-TCACCGGAGTCCATCACGAT-3′ (hACTIN, Gene ID: 60), FP 5′-ATCCAATCTGTGCCCATCTG-3′/RP 5′-AATAACTGGATGGGGAGGTG-3′ (hSULT2A1, Gene ID: 6822), FP 5′-CAGCGAAGTCCACATTCC-3′/RP 5′-TTTCTGGTGGTGACAGGAG-3′ (hDRD1, Gene ID: 1812), FP 5′-CGAGCATCCTGAACTTGTG-3′/RP 5′-TGAGTCCGAAGAGGAGTGG-3′ (hDRD2, Gene ID: 1813), FP 5′-GGAGCCGAAGTGGTAAAC-3′/RP 5′-CACCTGTGGAGTTCTCTGC-3′ (hDRD3, Gene ID: 1814), FP 5′-GACCTCCTCCTCGCTCTC-3′/RP 5′-GAACCTGTCCACGCTGAT-3′ (hDRD4, Gene ID: 1815), FP 5′-CTCAACTGGCACAGGGAC-3′/RP 5′-GATGAGCGAGGAAGAGATG-3′ (hDRD5, Gene ID: 1816). All the primers were purchased from AuGCT DNA-SYN Biotechnology Co, Ltd (Beijing, China). Samples were run in triplicate under the following conditions: initial denaturation for 30 s at 95 °C and 45 continuous cycles of 15 s at 95 °C, 30 s at 60 °C and 33 s at 72 °C. The gene expression levels in each sample were normalized to human β-actin mRNA.
SULT2A1 activity assay
The activity of SULT2A1 in the cell cytosol was determined using a high performance liquid chromatographic (HPLC) method as previously described29. Briefly, all the enzymatic assays were performed in a total reaction volume of 250 μL that included 50 μg of protein from HepG2 cell cytosols as the enzyme source, 0.2 μmol/L of DHEA as the substrate and 10 μmol/L of PAPS as the cofactor. After a 30 min incubation at 37 °C in a shaking water bath, the reaction was stopped by adding 250 μL of methanol as the stop buffer. Data are presented as the average from three individual experiments.
siRNA-mediated knockdown of DRD1
Three sets of synthetic DRD1-siRNAs (Shanghai GenePharma Co Ltd, China) were used in parallel in six-well plates. Lipofectamine 2000 (Invitrogen) and DMEM were used for transfecting 50 nmol/L of the siRNA duplexes according to the manufacturer's instructions. To assess the knockdown efficiency of DRD1, the mRNA and protein levels were monitored at 24 h and 48 h, respectively, in cells that were transfected with DRD1-siRNA. HepG2 cells that were untreated, transfected with non-targeting siRNA (Shanghai GenePharma Co Ltd, China) or mock transfected with Lipofectamine 2000 served as controls. The siRNA that was used for the experiments decreased DRD1 expression by 40% (siRNA sense: 5′-GCCUGUCGAAUGUUCUCAATT-3′, anti-sense: 5′-UUGAGAACAUUCGACAGGCTT-3′, targeting position of bp 1690–1710, Shanghai GenePharma Co Ltd, China). Following transfection with DRD1-siRNA, SULT2A1 mRNA and protein levels were measured after 24 h and SULT2A1 activity was determined after 48 h. Cy-3 fluorescently labeled FAM non-targeting siRNA (Shanghai GenePharma Co Ltd, China) was used in the optimization of the transfection procedure.
cAMP assay
HepG2 cells were treated with dopamine (at 0, 0.25, and 2.5 μmol/L) and SKF38393 (0, 0.05, 0.5, and 5 μmol/L) for 7 d. After the treatment, the medium was aspirated and 1 mL of 0.1 mol/L HCl was added for every 35 cm2 of surface area. The cells were scraped off with a cell scraper and transferred to centrifuge tubes. The cells were then lysed using an ultrasonic cell disruption system and centrifuged at a speed of 12 000 rounds per minute for 10 min. Protein lysates were collected, homogenized and quantified by the BCA protein method. The amount of cAMP was determined by using the cAMP Direct Immunoassay Kit (Catalog #K371-100, Biovision) according to the recommended protocol. The concentration range was 0.039–10 pmol/well, and the accuracy and precision of this method was validated by the manufacturer.
Statistical analysis
The results were expressed as the mean±SD and were analyzed using analysis of variance followed by a paired comparison (Bonferroni). P<0.05 was taken as the minimal level of significance.
Results
The expression of DRD1–DRD5 in HepG2 cells
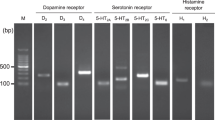
We first examined whether dopamine receptors are expressed in HepG2 cells. Indeed, we found that all five of the dopamine receptors subtypes (DRD1–DRD5) were expressed at different mRNA and protein levels in HepG2 cells. The mRNA expression levels of the DRD1-like family members were higher than the DRD2-like family members. The cycle threshold (Ct) values for DRD1, DRD2, DRD3, DRD4, and DRD5 were 29.18, 32.09, 35.18, 38.07, and 26.46, respectively. The Ct values for DRD1 and DRD5 were both lower than 30, and the values for the DRD2-like family members were higher than 30. Western blot results are shown in Figure 1, which indicate that all the DRD1–DRD5 subtypes are expressed in HepG2 cells. These results confirm that dopamine receptors are present in HepG2 cells and suggest that hepatic dopamine receptors may play a role in hepatic tissues.
Representative Western blots of dopamine receptors (DRD1/DRD2/DRD3/DRD4/DRD5) and β-actin in HepG2 cells. Each protein lysate (20 μg) was loaded twice in parallel.
Role of DRD1 activation in SULT2A1 induction
We then determined whether DRD1 activation induced SULT2A1 expression by treating HepG2 cells with the non-specific agonist dopamine and specific DRD1 agonists SKF82958 and SKF383931,30. As shown in Figure 2, the non-specific agonist dopamine moderately increased DRD1 mRNA expression by approximately 2-fold, whereas the specific agonists SKF82958 and SKF38393 induced a robust increase in DRD1 mRNA expression (∼30-fold and ∼8-fold, respectively) when HepG2 cells were treated with these compounds at the highest concentrations. The overall magnitude of SULT2A1 mRNA induction in HepG2 cells was consistent with the extent of DRD1 activation. The induction of SULT2A1 mRNA expression by dopamine was lower than with SKF38393 and SKF82958. The extent of SULT2A1 mRNA induction in SKF38393- and SKF82958-treated cells was approximately 10- and 40-fold, respectively. Western blot results corroborated the real-time PCR results by showing that dopamine and a high concentration of SKF82958 induced SULT2A1 protein expression. SKF38393 induced SULT2A1 protein expression in a concentration-dependent manner (Figure 2). The induction of SULT2A1 mRNA and protein levels correlated with the induction of its enzyme activity (Figure 2).
The effects of DRD1 subtype activation by the non-specific DRD1 agonist dopamine (A), and by the specific DRD1 agonists SKF82958 (B) and SKF38393 (C) on SULT2A1 and DRD1 subtype mRNA level (a), protein level (b) and enzyme activity (c) in HepG2 cells. HepG2 cells cultures were treated for 9 d with varying concentrations of DRD1 agonists as follows: (A) 0, 0.25, 2.5, and 25 μmol/L of dopamine; (B) 0, 0.025, 0.25, and 2.5 μmol/L of SKF82958; (C) 0, 0.05, 0.5, 5, and 50 μmol/L of SKF38393. SULT2A1 and DRD1 mRNA expression levels were determined by relative quantitative real-time PCR and were normalized to human β-actin mRNA. Protein lysate (20 μg) was used to confirm the increased expression of SULT2A1 and DRD1 by Western blot analysis. β-Actin was used as a loading control. SULT2A1 activity was measured as the rate of the specific reaction with the substrate DHEA (0.2 μmol/L). bP<0.05, cP<0.01 in comparison with the control treatment.
Role of repression of DRD1 activation in SULT2A1 induction
To assess the effect of repression of DRD1 activation on SULT2A1 induction, HepG2 cells were treated with dopamine receptor agonists in combination with the specific DRD1 antagonist SCH233901. As shown in Figure 3, the expression of SULT2A1 mRNA was induced 20-fold by treatment with a high concentration of SKF82958 in combination with the antagonist SCH23390 but only 2-fold by treatment with a high concentration of SKF38393 in combination with SCH23390. Notably, the mRNA levels of SULT2A1 that were induced by high concentrations of the DRD1 agonists in combination with the antagonist were much lower than upon induction with the specific DRD1 agonists alone. The induction of SULT2A1 by SKF82958 and SKF38393 decreased remarkably from 40- to 20-fold and from 10- to 2-fold, respectively. These observations were further confirmed by assessing the DRD1 and SULT2A1 protein levels by Western blotting and by assaying SULT2A1 enzyme activity under the conditions described above (Figure 3). The induction of SULT2A1 protein expression in HepG2 cells by the agonists in combination with the antagonist was also slightly lower than by the agonists alone. SULT2A1 enzyme activity was negligibly induced by SKF82958 in combination with SCH23390 and was even less induced by treatment with SKF38393 in combination with SCH23390. The decrease in SULT2A1 induction due to the specific DRD1 antagonist prompted us to investigate SULT2A1 induction upon siRNA-mediated knockdown of DRD1.
The effects of DRD1 subtype activation by the specific DRD1 agonists SKF82958 (A) and SKF38393 (B) in combination with a fixed concentration of the specific DRD1 antagonist SCH23390 on SULT2A1 and DRD1 subtype mRNA level (a), protein level (b) and enzyme activity (c) in HepG2 cells. HepG2 cell cultures were treated for 9 d with varying concentrations of DRD1 agonists in combination with the antagonist as follows: (A) 0, 0.025, 0.25, and 2.5 μmol/L of SKF82958 in combination with SCH23390 (2.5 μmol/L); (B) 0, 0.05, 0.5, 5, and 50 μmol/L of SKF38393 in combination with SCH23390 (2.5 μmol/L). Treatment with the antagonist SCH23390 (2.5 μmol/L) in the absence of the DRD1-specific agonist was designated as the control treatment. SULT2A1 and DRD1 mRNA expressions were determined by relative quantitative real-time PCR and were normalized to human β-actin mRNA. Protein lysate (20 μg) was used to confirm the increased expression of SULT2A1 and DRD1 by Western blot analysis. β-Actin was used as a loading control. SULT2A1 activity was measured as the rate of the specific reaction with the substrate DHEA (0.2 μmol/L). bP<0.05, cP<0.01 in comparison with the control treatment.
Role of DRD1 knockdown in SULT2A1 induction
We optimized the conditions for siRNA-mediated knockdown of DRD1 to result in a 40% down-regulation of DRD1 mRNA in HepG2 cells that were transfected with 50 nmol/L of the DRD1-specific siRNA. As shown in Figure 4, the results indicated that 50 nmol/L siRNA was non-cytotoxic and showed that DRD1 levels remained unchanged in mock-transfected and non-target siRNA-transfected hepatocytes. In addition, the cells that were transfected with DRD1 siRNA exhibited a 40% decrease in DRD1 mRNA and protein levels and a significantly lower induction of SULT2A1 than the cells receiving the control treatments. The magnitude of SULT2A1 induction at the mRNA, protein and activity levels was reduced by 60%, 40%, and 20%, respectively. It is important to note that SULT2A1 induction in the non-target siRNA-transfected cells and mock-transfected cells was not significantly different from the untreated control cells. These observations further corroborate that the extent of SULT2A1 induction is related to level of DRD1 expression and activity.
Effects of DRD1-siRNA on DRD1 mRNA and protein expression and SULT2A1 induction. (A) DRD1 and SULT2A1 mRNA expression (normalized to human β-actin mRNA expression) in HepG2 cells 24 h after transfection with the transfection reagent alone (Mock), the non-targeting siRNA duplexes (negative control, NC, 50 nmol/L) and the DRD1-siRNA specific duplexes (50 nmol/L). (B) DRD1 and SULT2A1 protein expression in HepG2 cells that were treated with the transfection reagent alone (Mock), the non-targeting siRNA duplexes (negative control, NC, 50 nmol/L) and the DRD1-siRNA specific duplexes (50 nmol/L) for 48 h. Protein lysate (20 μg) was used for further analysis with β-actin as a loading control. (C) The extent of SULT2A1 enzyme activity induction was determined after transfection with the transfection reagent alone (Mock), the non-targeting siRNA duplexes (negative control, NC, 50 nmol/L) and the DRD1-siRNA specific duplexes (50 nmol/L) for 48 h. Untreated cells were designated as the control sample (Un.). bP<0.05, cP<0.01 in comparison with the control treatment.
Effect of dopamine and SKF 38393 on cAMP levels
To further confirm the function of dopamine in this study, we investigated the effects of dopamine and SKF38393 on cAMP levels in HepG2 cells. As shown in Figure 5, the cAMP levels in HepG2 cells increased in a dose-dependent manner after treatment with dopamine and SKF38393, which demonstrated that the two compounds work through the activation of the DRD1 receptors.
Alterations in cAMP levels in HepG2 cells that were treated with varying doses of dopamine (A) or the DRD1-specific agonist SKF38393 (B) for 7 d (cP<0.01).
Discussion
Currently, five dopamine receptor subtypes (DRD1–DRD5) have been identified. Based on their function, the DR receptors can be classified as either D1-like receptors or D2-like receptors. The DRD1 and DRD5 subtypes are classified as D1-like subfamily, and DRD2, DRD3, and DRD4 are classified as members of the D2-like subfamily2. D1-like receptors generally stimulate cAMP accumulation, whereas D2-like receptors inhibit cAMP accumulation. DRD1 and DRD2 are the most-studied subtypes of the D1-like and D2-like receptor subfamilies, respectively. Similarly, agonists and antagonists of DRD1 and DRD2 are primarily used in studies that involve dopamine receptors regardless of whether the receptors are present in the central nervous system1 or in peripheral systems3,4,5,6,7,8. However, it should be noted that although DRD2, DRD3 and DRD4 subtypes have been identified, their specific agonists and antagonists are very limited. For example, no studies have been reported on the specific agonist or antagonist of DRD5. Therefore, we first screened the effects of both DRD1 and DRD2 agonists on the enzymatic activities of phenol sulfotransferase (SULT1A1), SULT2A1 and estrogen sulfotransferase (SULT1E1). We found that all the DRD1 agonists that were used in this study, including the non-specific agonist dopamine and the specific agonists SKF38393 and SKF82958, significantly increased the activities of these three SULT isoforms. However, we also found that when HepG2 cells were treated with varying doses of the specific DRD2 agonists quinpirole and bromocriptine1,31, the enzyme activities of these SULTs either increased slightly or did not change (Supplementary Figure S1). Thus, we focused our investigation on DRD1. Furthermore, the real-time PCR results showed that the mRNA expression of all three SULTs examined in this study was induced by the specific DRD1 agonists SKF38393 and SKF82958. The induction of SULT2A1 was found to be the most dramatic especially at high concentrations (Figure 2) and was induced to a much higher level than SULT1A1 and SULT1E1 (Supplementary Figure S2). Our preliminary results also showed that blockade of DRD1 had little effect on the expression and activities of both SULT1A1 and SULT1E1. Thus, compared with the other two isoforms, SULT2A1 exhibited a higher sensitivity to the activation of DRD1. For this reason, we mainly investigated the role of DRD1 in the regulation of SULT2A1 in the present study.
The results presented above provide the first direct evidence for the expression of dopamine receptors and the role of the DRD1-related signaling pathway in the regulation of SULT2A1 in HepG2 cells. In our study, we provided a detailed analysis of the induction of SULT2A1 by DRD1 activation. SULT2A1 expression is tissue-specific, with transcripts found in liver, adrenal glands, intestine and fetal adrenal glands26. SULT2A1 is of interest because of its ability to detoxify environmental xenobiotics and to sulfate endogenous steroids that have been linked to steroid-related cancers15. Reports suggest that SULT2A1 can be regulated by endogenous steroids through different nuclear receptors and that this regulation varies for different nuclear receptors, which is similar to how the CYP450 enzymes are regulated13,14,32,33. Estrogen-related receptor alpha has been shown to mediate the down-regulation of human SULT2A1 in HepG2 cells13. The treatment of primary cultured human hepatocytes with ciprofibrate produced an approximately 2-fold increase in SULT2A1 mRNA, protein, and enzyme activity, which suggests a role for the lipid-sensing PPAR-α (peroxisome proliferator activated receptor alpha) transcription factor in the transcriptional up-regulation of human hepatic SULT2A1 gene transcription33. In addition, human constitutive active receptor (hCAR) was shown to mediate the induction of hSULT2A1 in HepG2 cells by methotrexate. Similarly, human vitamin D receptor (hVDR) also up-regulated hSULT2A1 gene expression, while the human pregnane X receptor (hPXR) down-regulated it34. Moreover, hVDR was shown to compete with hCAR for the hSULT2A1 promoter, and hPXR suppressed hCAR-mediated induction of hSULT2A1 by methotrexate in Caco-2 cells. These results indicate that crosstalk occurs among the nuclear receptors in the hSULT2A1 signal transduction pathway and that interactions among nuclear receptors also depend on ligands (inducers)34. In this study, we did not investigate the regulation of SULT2A1 by the nuclear receptor pathway because DRs are typical membrane receptors. Whether the activation of the DRs further influences the nuclear receptor pathway needs further investigation. It has been suggested that the dopamine analogue methamphetamine (METH) induces SULT protein and mRNA expression in rat liver and brain. The induction of SULT in the brain was found to be much higher than in the liver23. However, there have been no studies that have described the mechanism of SULT2A1 regulation by the dopaminergic system.
In this study, we showed that SULT2A1 is regulated by a DRD1 non-specific agonist (dopamine) and by specific agonists (SKF38393 and SKF82958) in HepG2 cells. In general, SULT2A1 mRNA induction in HepG2 cells correlates with DRD1 activation by agonists. The order of magnitude of DRD1 activation (SKF82958>SKF38393>dopamine) matched the order of SULT2A1 induction because SKF82958 and SKF38393 induced SULT2A1 mRNA levels in HepG2 cells by 40-fold and 10-fold, respectively (Figure 2). SULT induction by this magnitude has rarely been described23,34,35. On the contrary, DRD1 activation was reported to induce the mRNA and protein expression of other SULT isoforms, including SULT1A1 and SULT1E1, to a lesser degree (by approximately three-fold) (Supplementary Figure S2). These results indicate that the effect of DRD1 activation on SULT is selective and specific. Interestingly, the activation of the brain dopaminergic pathway by SKF82958 did not significantly influence the expression of CYP450 isoforms11, whereas our results clearly showed that SKF82958-induced SULT2A1 expression in HepG2 cells. This finding implies that the induction of SULT2A1 by hepatic DRD1 activation may serve a function that has yet to be elucidated.
Based on the finding that there was no significant induction of SULT2A1 activity by the specific antagonist SCH23390 (Supplementary Figure S3), we hypothesized that DRD1 activation would be repressed when an antagonist competitively occupies the receptor binding sites. In the present study, we used agonists in combination with an antagonist to underscore the importance of DRD1 expression on SULT2A1 induction in HepG2 cells. The levels of SULT2A1 mRNA expression, protein expression and enzyme activity decreased dramatically when HepG2 cells were simultaneously treated with the specific DRD1 agonist and antagonist (Figure 3). This result confirms our hypothesis and suggests that a selective induction of SULT2A1 by DRD1 activation. Moreover, as a member of the G protein-coupled receptor family, the activation of the dopamine receptor may lead to the stimulation of downstream signaling transmission and to the up-regulation of SULT2A1 expression. These results also confirmed that the mechanism of SULT2A1 induction by agonists is primarily through DRD1 activation rather than through the direct effects of transcription factors in the nucleus after passive diffusion through the cell and nuclear membranes.
The critical role of DRD1 activation in SULT2A1 induction was further shown by using a siRNA that targeted DRD1 mRNA. Attenuation of DRD1 expression in HepG2 cells with siRNA caused a marked reduction in the overall extent of SULT2A1 down-regulation. For example, with a 40% reduction in DRD1 mRNA level in cells, we observed an approximate 60% reduction in SULT2A1 mRNA expression (Figure 4). Blockade of DRD1 reduced the modulation of adenylyl cyclase activity, which resulted in an inhibition of cyclic AMP accumulation. These findings indicate that G protein-coupled signaling was altered36. The DRD1 receptors can couple to and activate the G protein, which in turn activates adenylate cyclase (AC). This enzyme then catalyzes the formation of cAMP, which acts on various downstream effector molecules. Therefore, cAMP is a key molecule in the DRD1 signaling pathway37. As shown in Figure 5, both dopamine and SKF38393 significantly up-regulated cAMP levels in HepG2 cells, which demonstrates that the two drugs work through the activation of DRD1 receptors. This result provides direct evidence regarding the mechanism of DRD1 activation.
In conclusion, our results indicate that DRD1 activation altered SULT2A1 expression and activity. Without the influence of the central nervous system, dopamine receptor-specific agonists significantly induced SULT2A1 expression, and blockade of DRD1 mRNA expression reduced SULT2A1 expression in HepG2 cells. Our results also revealed that SULT2A1 regulation by DRD1 is dopamine receptor subtype-dependent. Understanding the mechanism of SULT2A1 induction by DRD1 is important to better characterize the role of DRD1 in the peripheral systems, such as the liver and the role of SULT2A1 in hormone regulation and chemotherapy. Ongoing research is currently focused on the characterization of SULT2A1 induction by DRD1 in vivo and on the molecular mechanisms that underlie this regulation.
Abbreviations
PAPS, 3′-phosphoadenosine 5′-phosphosulfate; DR, dopamine receptor; DRD1, dopamine receptor D1 subtype; SULT, sulfotransferase; SULT2A1, dehydroepiandrosterone sulfotransferase; DHEA, dehydroepiandrosterone; METH, methamphetamine; cAMP, cyclic adenosine monophosphate.
Author contribution
Tian-yan ZHOU and Jiao-jiao XU designed the research; Jiao-jiao XU, Si-yuan WANG, Ye CHEN, Xue-yan SHAO, and Liang LI performed the research; Guang-ping CHEN and Zai-quan LI contributed analytic tools; Tian-yan ZHOU, Jiao-jiao XU and Wei LU analyzed the data; Jiao-jiao XU wrote the paper.
References
Missale C, Nash SR, Robinson SW, Jaber M, Caron MG . Dopamine receptors: From structure to function. Physiol Rev 1998; 78: 189–225.
Lachowicz JE, Sibley DR . Molecular characteristics of mammalian dopamine receptors. Pharmacol Toxicol 1997; 81: 105–13.
Jose PA, Eisner GM, Felder RA . Renal dopamine receptors in health and hypertension. Pharmacol Ther 1998; 80: 149–82.
Horn PT, Murphy MB . Dopamine receptor agonists in cardiovascular medicine. Trends Cardiovasc Med 1991; 1: 103–7.
Contreras F, Fouillioux C, Bolívar A, Simonovis N, Hernández-Hernández R, Armas-Hernandez MJ, et al. Dopamine, hypertension and obesity. J Hum Hypertens 2002; 16: S13–7.
Glaser S, Alvaro D, Roskams T, Phinizy JL, Stoica G, Francis H, et al. Dopaminergic inhibition of secretin-stimulated choleresis by increased PKC-expression and decrease of PKA activity. Am J Physiol Gastrointest Liver Physiol 2003; 284: G683–94.
Zhang X, Guo H, Xu J, Li Y, Li L, Zhang X, et al. Dopamine receptor D1 mediates the inhibition of dopamine on the distal colonic motility. Transl Res 2012; 159: 407–14.
Sachlos E, Risueno RM, Laronde S, Shapovalova Z, Lee J, Russell J, et al. Identification of drugs including a dopamine receptor antagonist that selectively target cancer stem cells. Cell 2012; 149: 1–14.
Nassar CF, Karkaji EG, Habbal ZM, Nasser MG . Dopamine receptors in normal and diabetic liver plasma membrane. Gen Pharmacol 1986; 17: 367–70.
Wojcikowski J, Golembiowska K, Daniel WA . The regulation of liver cytochrome p450 by the brain dopaminergic system. Curr Drug Metab 2007; 8: 631–8.
Wojcikowski J, Golembiowska K, Daniel WA . Regulation of liver cytochrome P450 by activation of brain dopaminergic system: physiological and pharmacological implications. Biochem Pharmacol 2008; 76: 258–67.
Wojcikowski J . Potential role of the brain dopaminergic system in the regulation of CYP450. Pol J Pharmacol 2004; 56: 701–8.
Huang CQ, Zhou TY, Chen Y, Sun T, Zhang SF, Chen GP . Estrogen-related receptor ERRα-mediated downregulation of human hydroxysteroid sulfotransferase (SULT2A1) in Hep G2 cells. Chem Biol Interact 2011; 192: 264–71.
Kohalmy K, Vrzal R . Regulation of phase II biotransformation enzymes by steroid hormones. Curr Drug Metab 2011; 12: 104–23.
Gamage N, Barnett A, Hempel N, Duggleby RG, Windmill KF, Martin JL, et al. Human sulfotransferases and their role in chemical metabolism. Toxicol Sci 2006; 90: 5–22.
Kocarek TA, Duanmu Z, Fang HL, Runge-Morris M . Age- and sex-dependent expression of multiple murine hepatic hydroxysteroid sulfotransferase (SULT2A) genes. Biochem Pharmacol 2008; 76: 1036–46.
Gold MS, Kobeissy FH, Wang KK, Merlo LJ, Bruijnzeel AW, Krasnova IN, et al. Methamphetamine- and trauma-induced brain injuries: comparative cellular and molecular neurobiological substrates. Biol Psychiatry 2009; 66: 118–27.
Volz TJ, Fleckenstein AE, Hanson GR . Methamphetamine-induced alterations in monoamine transport: implications for neurotoxicity, neuroprotection and treatment. Addiction 2007; 102: 44–8.
Kish SJ . Pharmacologic mechanisms of crystal meth. Can Med Assoc J 2008; 178: 1679–82.
Kita T, Wagner GC, Nakashima T . Current research on methamphetamine-induced neurotoxicity: animal models of monoamine disruption. J Pharmacol Sci 2003; 92: 178–95.
Niculescu AB 3rd, Segal DS, Kuczenski R, Barrett T, Hauger RL, Kelsoe JR . Identifying a series of candidate genes for mania and psychosis: a convergent functional genomics approach. Physiol Genomics 2000; 4: 83–91.
Guilarte TR, Nihei MK, McGlothan JL, Howard AS . Methamphetamine-induced deficits of brain monoaminergic neuronal markers: distal axotomy or neuronal plasticity. Neuroscience 2003; 122: 499–513.
Zhou TY, Huang CQ, Chen Y, Shanbhag P, Chen GP . Methamphetamine regulation of sulfotransferases in rat liver and brain. AM J Pharmacol Toxicol 2010; 3: 125–32.
Falany CN . Enzymology of human cytosolic sulfotransferases. FASEB J 1997; 11: 206–16.
Weinshilboum RM, Otterness DM, Aksoy IA, Wood TC, Her C, Raftogianis RB . Sulfation and sulfotransferases 1: Sulfotransferase molecular biology: cDNAs and genes. FASEB J 1997; 11: 3–14.
Jancova P, Anzenbacher P, Anzenbacherova E . Phase II drug metabolizing enzymes. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub 2010; 154: 103–16.
Westerink WM, Schoonen WG . Phase II enzyme levels in HepG2 cells and cryopreserved primary human hepatocytes and their induction in HepG2 cells. Toxicol In Vitro 2007; 21: 1592–602.
Maiti S, Chen XR, Chen GP . All-trans retinoic acid induction of sulfotransferases. Basic Clin Pharmacol Toxicol 2005; 96: 44–53.
Xu JJ, Chen Y, Li L, Li ZQ, Wang CA, Zhou TY, et al. An improved HPLC method for the quantitation of 3′-phosphoadenosine 5′-phosphate (PAP) to assay sulfotransferase enzyme activity in HepG2 cells. J Pharm Biomed Anal 2012; 62: 182–6.
Gilmore JH, Watts VJ, Lawler CP, Noll EP, Nichols DE, Mailman RB . “Full” dopamine D1 agonists in human caudate: biochemical properties and therapeutic implications. Neuropharmacology 1995; 34: 481–8.
Hoffman DC, Dickson PR, Ricahrd JB . The dopamine D2 receptor agonists, quinpirole and bromocriptine produce conditioned place preferences. Prog Neuropsychopharmacol Biol Psychiatry 1988; 12: 315–22.
Runge-Morris M, Kocarek TA, Falany CN . Regulation of the cytosolic sulfotransferases by nuclear receptors. Drug Metab Rev 2013; 45: 15–33.
Fang HL, Strom SC, Cai H, Falany CN, Kocarek TA, Runge-Morris M . Regulation of human hepatic hydroxysteroid sulfotransferase gene expression by the peroxisome proliferatoractivated receptor alpha transcription factor. Mol Pharmacol 2005; 67: 1257–67.
Chen XR, Maiti S, Zhang JM, Chen GP . Nuclear receptor interactions in methotrexate induction of human dehydroepiandrosterone sulfotransferase (hSULT2A1). J Biochem Mol Toxicol 2006; 20: 309–17.
Huang CQ, Chen Y, Zhou TY, Chen GP . Sulfation of dietary flavonoids by human sulfotransferases. Xenobiotica 2009; 39: 312–22.
Pivonello R, Ferone D, Lombardi G, Colao A, Lamberts S, Hofland LJ . Novel insights in dopamine receptor physiology. Eur J Endocrinol 2007; 1561: 13–21.
Weinshenker D, Warren ST . Fragile dopamine. Nature 2008; 455: 607–8.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant 81072699) and by the Scientific Research Foundation for the Returned Overseas Chinese Scholars by the State Education Ministry (Grant [2011]508). The authors deeply appreciate the generous gift of human SULT2A1 antibody from Dr David RINGER of the American Cancer Society.
Author information
Authors and Affiliations
Corresponding author
Additional information
( Supplementary information is available at the Acta Pharmacologica Sinica's website.
Supplementary information
Supplementary information, Figure S1
The effect of DRD2 subtype activation by specific DRD2 agonist quinpirole (A) and bromocriptine (B) on SULT1A1 (a), SULT2A1 (b) and SULT1E1 (c) enzyme activities in HepG2 cells. (PDF 131 kb)
Supplementary information, Figure S2
Comparison results of DRD1 (a), SULT1A1 (b), SULT2A1 (c), and SULT1E1 (d) mRNA expression between using DRD1 agonist SKF82958 or SKF38393 alone and SKF82958 or SKF38393 in combination with DRD1 antagonist SCH23390 in HepG2 cells. (PDF 375 kb)
Supplementary information, Figure S3
The effect of specific DRD1 antagonist SCH23390 on SULT2A1 enzyme activities in HepG2 cells. (PDF 174 kb)
Rights and permissions
About this article
Cite this article
Xu, Jj., Wang, Sy., Chen, Y. et al. Dopamine D1 receptor activation induces dehydroepiandrosterone sulfotransferase (SULT2A1) in HepG2 cells. Acta Pharmacol Sin 35, 889–898 (2014). https://doi.org/10.1038/aps.2014.19
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/aps.2014.19