Abstract
Androgens and androgen receptors (AR) play a pivotal role in expression of the male phenotype. Several diseases, such as androgen insensitivity syndrome (AIS) and prostate cancer, are associated with alterations in AR functions. Indeed, androgen blockade by drugs that prevent the production of androgens and/or block the action of the AR inhibits prostate cancer growth. However, resistance to these drugs often occurs after 2–3 years as the patients develop castration-resistant prostate cancer (CRPC). In CRPC, a functional AR remains a key regulator. Early studies focused on the functional domains of the AR and its crucial role in the pathology. The elucidation of the structures of the AR DNA binding domain (DBD) and ligand binding domain (LBD) provides a new framework for understanding the functions of this receptor and leads to the development of rational drug design for the treatment of prostate cancer. An overview of androgen receptor structure and activity, its actions in prostate cancer, and how structural information and high-throughput screening have been or can be used for drug discovery are provided herein.
Similar content being viewed by others
Introduction
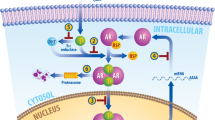
The androgen receptor (AR) (NR3C4, nuclear receptor subfamily 3, group C, gene 4) belongs to the steroid hormone group of nuclear receptors with the estrogen receptor (ER), glucocorticoid receptor (GR), progesterone receptor (PR) and mineralocorticoid receptor (MR)1,2,3. The AR is a ligand-dependent transcription factor that controls the expression of specific genes. The binding of the AR to its native ligands 5α-dihydrotestosterone (DHT) and testosterone initiates male sexual development and differentiation. The current model of action of androgens and the AR is depicted in Figure 1A. Androgens such as testosterone are synthesized primarily by the Leydig cells in the testes, under the regulation of luteinizing hormone (LH) produced by the anterior pituitary gland. LH secretion is in turn regulated by gonadotropin-releasing hormone (GnRH). Once produced, testosterone mostly circulates bound to serum sex hormone-binding globulin (SHBG) and albumin4,5. Only the free form enters prostate cells. Intracellularly, testosterone is converted into a more potent 5α-reduced metabolite of testosterone, 5α-dihydrotestosterone (DHT), which promotes the growth and survival of prostate cells. DHT binds to the AR with high affinity, displaces heat-shock proteins from the AR, drives the interaction between the N and C termini of the AR, and binds importin-α to translocate the AR into the nucleus6. In the nucleus, receptor dimers bind to androgen response elements (AREs) in the promoter regions of target genes, such as prostate-specific antigen (PSA) and transmembrane protease serine 2 (TMPRSS2), etc, to which they recruit various coregulatory proteins to facilitate transcription, leading to responses such as growth and survival7,8,9,10,11,12. Male sexual differentiation fails to occur in the absence of androgens or without a functioning AR. A complete loss of AR function in males results in complete androgen insensitivity syndrome13,14. The role of the AR in the development and progression of prostate cancer has led to increasing interest in this nuclear receptor. Prostate cancer is predicted to be the leading cause of cancer-related death in men over the next decade in the United States15. The development and progression of prostate cancer depends on androgenic stimulation11. As such, prostate cancer is treated by depriving tumors of androgens such as DHT and testosterone or blocking their actions. However, the effect of this type of treatment is transient, as patients relapse after developing a castration-resistant form of the disease that is usually due to increased levels of AR expression or mutations that cause the AR to be resistant to antiandrogens. Many studies have focused on providing new insights into the mechanisms of AR action in prostate cancer. The determination of the three-dimensional crystal structures of the AR DNA binding domain (DBD) and ligand binding domain (LBD) has helped expand our understanding of this receptor by revealing fine molecular details. The importance of the AR has also led to the development of many predictive models of compound binding. Structural information and biochemical experiments have been used to aid rational drug design to improve existing drugs and develop new treatments for the disease. This review summarizes AR structure-function relationships, describes the role of the AR in prostate cancer, and integrates a discussion on how structural information has been used to help guide rational drug design for treatment of prostate cancer.
Androgen and AR action. Genome organization of the human androgen receptor gene and the functional domain structure of the androgen receptor protein. (A) Androgen and AR signaling in prostate cells. After testicular synthesis, testosterone is transported to target tissues such as the prostate and becomes converted to dihydrotestosterone (DHT) by 5-α-reductase. DHT binds to the ligand-binding pocket and promotes the dissociation of heat-shock proteins (HSPs) from the AR. The AR then translocates into the nucleus, dimerizes and binds to the androgen response element (ARE) in the promoter region of target genes such as prostate-specific antigen (PSA) and TMPRSS2. At the promoter, the AR is able to recruit members of the basal transcription machinery [such as TATA-box-binding protein (TBP) and transcription factor IIF (TFIIF)] in addition to other coregulators such as members of the p160 family of coactivators and cAMP-response element-binding protein (CREB)-binding protein (CBP). SHBG: serum sex hormone-binding globulin. (B) The androgen receptor gene has been mapped to the long arm of the X-chromosome (locus: Xq11-q12). It contains eight exons interrupted by introns of varying lengths (0.7–2.6 kb) and codes for a protein of 919 amino acids consisting of several functional domains (N-terminal domain (NTD), DNA binding domain (DBD) and ligand binding domain (LBD); amino acid residue numbers are indicated above the AR protein domain map). Exon 1 codes for the NTD, exons 2 and 3 encode the DBD, and exons 4 to 8 encode both the hinge and LBD.
AR structure and activity
The AR gene is located on the X chromosome at the locus Xq11-Xq12 (Figure 1B)16,17,18. The protein coding region has 2757 nucleotides and spans eight exons, with introns that vary in size from 0.7 to 2.6 kb. The AR gene encodes a 110-kDa protein consisting of 919 amino acids (Figure 1B)19. Like other members of the nuclear receptor family, the AR consists of three major functional domains: (i) the N-terminal domain (NTD) (residues 1–555), followed by (ii) the DNA binding domain (DBD) (residues 555–623), and (iii) the C-terminal ligand binding domain (LBD) (residues 665–919), which is connected to the DBD by a flexible hinge region (residues 623–665) (Figure 1B)1. All three domains are important for receptor function. The highly conserved DBD tethers the AR to promoter and enhancer regions of AR-regulated genes by direct DNA binding to allow the activation functions of the NTD and LBD to stimulate transcription of these genes. The activation function 1 (AF1, residues 142–485) in the NTD is constitutively active20, whereas the activation function 2 (AF2), a hydrophobic surface composed of helices 3, 4, and 12 located in the LBD, is ligand dependent21. Currently, there is no structural information for the full length AR receptor. However, the structures of both the DBD and LBD have been solved separately, revealing critical details of the mechanism of action of this receptor.
NTD
The NTD accounts for more than half of the size of the AR (residues 1–555), and its entirety is encoded by exon 1. The sequence and lengths of the polyglutamine (CAG) and polyglycine (GGC) repeats of the AR NTD are highly variable in the human population22,23,24. The length of the poly-Q repeat region has been shown to affect the folding and structure of the AR-NTD25. Through biochemical and biophysical approaches, it was demonstrated that the removal of poly-Q repeats from the AR leads to a reduction in α-helical structure, whereas increasing the length of poly-Q repeats led to a modest increase in α-helical structure. The conformational changes in the NTD were proposed to have a concomitant impact on protein-protein interactions that is likely to explain the dependency of AR transcriptional activity on the repeat length26. As has been observed for many other poly-Q repeat-containing proteins, shorter repeats generally impose a higher AR transactivation activity, whereas longer repeats cause reduced activity27. The deletion of the poly-Q tract causes a four-fold increase in AR activation function compared with the wild-type protein28. Thus far, the structure of the NTD has not been defined through X-ray crystallography: this is likely due to its highly disordered structure as described by circular dichroism spectroscopy studies and structure prediction algorithms25,29,30,31,32. It has been proposed that intrinsically disordered proteins may undergo folding induced by the formation of specific protein-protein interactions29,30. Taken together, the structural plasticity of a partially folded state is hypothesized to be an intrinsic property of the NTD that allows interactions with many structurally diverse binding partners32,35, such as coactivators of the P160 family36, the basal transcription factors TATA-box binding protein and transcription factor IIF37, and intramolecular interactions with the LBD. Cooperative interactions with multiple binding partners enable high specificity by low-affinity interactions, which appear to be crucial for AR activity32,35.
Deletion mutagenesis showed that, within the NTD region, a domain is required for full transcriptional activity38. Deletion studies have also helped to further demarcate the AF1 region as the primary effector region of the NTD. The AF1 contains two separable transcription activation units [Tau-1 (amino acids 100–370] and Tau-5 (amino acids 360–485)) that are indispensable for full activity of the AR (Figure 1B)39. Tau-1 contains a nuclear receptor box, FQNLF (amino acids 23–27), and Tau-5 contains the WHTLF motif (amino acids 433–437), both of which mediate direct ligand-dependent, interdomain interactions between the NTD and the LBD (termed an N/C interaction), which are important in regulating some, but not all, androgen-dependent genes40,41,42. The N/C interaction also helps to stabilize the AR dimer complex and to slow the rate of ligand dissociation43,44.
DBD and hinge
The DBD (residues 556–623) is a cysteine-rich region that is highly conserved among steroid hormone receptors (Figure 2A). According to the crystal structure of the AR DBD, each DBD monomer has a core composed of two zinc fingers (Figure 2B) (PDB: 1R4I), each of which consists of four cysteine residues that coordinate a zinc ion. The AR functions as a dimer that, like other steroid receptors, binds to promoter DNA response elements consisting of two equal, common hexameric half-sites (5′-AGAACA-3′) separated by a 3 base-pair spacer (IR3)45. The α-helix of the N-terminal zinc finger (the “recognition helix”) interacts directly with nucleotides in the hormone response element in the DNA major groove (Figure 2B). Three amino acid residues at the N terminus of this α-helix, named the P(roximal) box [glycine-serine-valine] (amino acids 577–581; GSCKV), are identical in the PR, GR and MR (Figure 2A and 2B) and are responsible for the specific recognition of the DNA response element12. A question that persisted was how steroid receptors achieve target specificity if the AR, PR, GR, and MR bind a common DNA response element. Studies have identified selective androgen response elements (AREs) (eg, 5′-GGTTCT-3′) that allow specific AR activation46,47,48. AREs have hexameric half-sites in a direct repeat orientation. Structural studies have confirmed that selectivity is achieved by receptor dimerization in a “head-to-head” fashion through the D(istal) box region (amino acid 596–600; ASRND), which allows the AR to bind to direct repeat half-sites in its promoter (Figure 2A and 2B)45. Because the DBD domains are highly conserved among the different steroid receptors, the reason why other steroid receptors do not recognize selective AREs is still a matter of debate. Based on crystallographic data, it was speculated that the AR contains an additional interface that stabilizes the AR dimer/ARE complex. In contrast, the dimerization strength of other steroid receptors would not be sufficient to retain stable binding to selective AREs45,49.
Structures of AR functional domains. (A) Sequence alignment of the DNA binding domain of the androgen receptor (AR), progesterone receptor (PR), mineralocorticoid receptor (MR), and glucocorticoid receptor (GR) performed using ClustalW. *indicates the conserved cysteines involved in coordinating the zinc atom. (B) Top, crystal structure of the AR DBD (pink) (PDB: 1R4I) complexed with its hormone response element (red/purple). The DBD contains two zinc fingers (grey). Each zinc ion is coordinated by four cysteines (yellow). One zinc finger is involved in direct DNA binding mediated by the P-box (orange), which recognizes the specific hormone response element half-site 5′-AGAACA-3′. The other zinc finger is involved in a “head-to-head” receptor dimerization through the D-box (green). Bottom, cartoon representation of the AR DBD. (C) Crystal structure of an AR nuclear localization signal (NLS) peptide (amino acid 621-635) (orange) complexed with importin-α (yellow) (PDB: 3BTR). Residues from the major NLS site 629-RKLKKL-634 contribute to importin-α binding. (D) Crystal structure of the AR ligand binding domain (purple) (LBD) (PDB: 1E3G). The LBD consists of 11 α-helices and two small, two-stranded β-sheets arranged in a typical three-layer antiparallel helical sandwich fold. The long flexible linker between helices 1 and 3 is colored blue.
The nuclear localization signal (NLS) (residues 617–633) is localized at the junction between the DBD and the hinge region (residues 624–665) and is responsible for nuclear import of the receptor50,51. Passive transport across the nuclear pore complex has been suggested ranging from 20–40 kDa52. In contrast, the AR, which is 110 kDa in size, requires help to be actively transported upon ligand binding. A recent study has suggested that androgen binding induces a switch that exposes the NLS and thereby allows the NLS to promote nuclear import through binding of importin-α53. Details of this interaction have been identified in the crystal structure of importin-α bound to the AR NLS (Figure 2C) (PDB: 3BTR)54. The NLS is composed of two clusters of basic amino acids separated by ten residues (617-RKCYEAGMTLGARKLKK-634), a motif that is highly conserved with that of the GR, MR and PR steroid hormone receptors. In the complex, importin-α adopts a banana-shaped conformation in which its inner concave surface makes charge interactions with the second basic amino acid cluster of the NLS (Figure 2C). Apart from nuclear localization, the hinge region, and in particular its 629-RKLKKL-634 motif, was also found to play a complex role in DNA binding, coactivator recruitment, and N/C interaction55,56 and is a target site for acetylation, ubiquitylation and methylation56.
LBD
In contrast to the NTD, the AR LBD (residues 666–919) has been structurally well characterized by crystallography. The crystal structure of the AR LBD was first solved in the year 200057, and subsequently, many other complex structures were deposited into the protein databank (PDB). The three-dimensional structure is arranged in a three-layer, antiparallel α-helical sandwich (PDB: 1E3G) fold that is characteristic of NR LBDs. The AR LBD consists of eleven α-helices and four short β strands forming two anti-parallel β-sheets (Figure 2D). The H1 and H3 helices form the first layer of the α-helical sandwich. Unlike other nuclear receptors, the AR does not have H2, which is instead replaced by a long flexible linker (Figure 2D). The middle layer is formed by H4, H5, the first β sheet, H8, and H9. The third layer is completed by H10 and H11. There is a ligand binding pocket (LBP) surrounded by the N termini of H3, H5, and H11. H12, which forms the core of the activation function 2 domain (AF2), acts as a lid to close the LBP upon agonist binding (Figure 2D).
AR Ligands
A large variety of small molecules has been discovered or engineered to interact with the AR58,59,60. The chemistry of these ligands has been extensively reviewed60,61. In general, AR ligands were identified through in vitro binding or reporter gene assays. They are largely classified as agonists or antagonists based on their ability to activate or inhibit transcription of AR target genes. All of these ligands modulate AR function by binding to the LBP in the LBD. The receptor is able to accommodate many different ligands by modifying the volume of its LBP by changing the position or orientation of amino acid side chains62.
AR agonists
The two most important endogenous androgens are testosterone and DHT (Figure 3A). The functions of androgens were first described in 1889, when French physiologist and Professor of Medicine Charles Edward Brown-Sequard first identified androgen action through self-injections of testicular extracts. Subsequently, in 1935, Prof Ernst LAQUEUR and collaborators from the Netherlands characterized and named the active ingredient in the extract testosterone63. Another hallmark in the history of androgen biochemistry was the discovery that a fraction of testosterone is metabolized to the more potent androgen DHT in a reaction catalyzed by 5α-reductase64,65. DHT differs from testosterone by the absence of a single double bond on ring A (Figure 3A)13, which increases its affinity for the AR two-fold and decreases the rate of dissociation five-fold relative to testosterone66, differences that account for essential, DHT-specific functions.
Structural basis of AR agonism. (A) Chemical structures of testosterone, dihydrotestosterone and R1881. (B) Structural overlay of the AR LBD complexed with testosterone (PDB: 2AM9, orange), dihydrotestosterone (PDB: 1I37, green), and R1881 (PDB: 1E3G, purple). (C) Comparison of the binding of R1881, testosterone, and dihydrotestosterone in the AR ligand-binding pocket. AR LBD residues within a distance of 4 Å are shown. Key residues (N705, Q711, R752, and T877) that form hydrogen bonds with ligands are labeled and shown in stick presentation. Hydrogen bonds are indicated by dotted lines. (D) Structure of the AR LBD (purple) in complex with an FxxLF motif-containing peptide (yellow) (Left panel). The middle panel shows the interface between the AR LBD and the FxxLF motif. Hydrogen bonds are indicated by dotted lines with key residues labeled. A surface view of the motif-binding hydrophobic pocket is shown. A sequence alignment of helices H3 and H12 of the androgen receptor (AR), estrogen receptor (ER), progesterone receptor (PR), mineralocorticoid receptor (MR) and glucocorticoid receptor (GR) was performed using ClustalW. (E) Structure of the AR LBD complexed with 3,3′,5-triiodothyroacetic acid (TRIAC) (PDB: 2PIT). Left: cartoon representation; right: surface view of the binding function (BF3) surface pocket (green).
The first structure of the AR LBD was solved with the AR in complex with the potent synthetic androgen R1881 (metribolone) (Figure 3A) and shows the AR in an agonist conformation with the ligand inside of the LBP formed by H3, H5, and H11 (PDB: 1E3G) (Figure 2D and 3B)57. Later, structures of the AR LBD complexed with its physiological agonists DHT (PDB: 1I37 or 2AMA) and testosterone (PDB: 2AM9) were solved (Figure 3B)67,68. These three AR LBD agonist structures have very similar overall conformations, revealed key ligand and receptor interaction sites, and helped to define the general structural requirements for the binding of ligands in the LBP. R1881, DHT, and testosterone have 18, 16, and 15 contact points, respectively, with the LBD at a van der Waals distance cutoff of 4 Å (Figure 3C). These residues are hydrophobic and interact mainly with the steroid scaffold. The remaining amino acids are polar and form hydrogen bonds with the polar atoms of the ligand. Notably, there are four hydrogen bonds formed between the LBD and DHT/Testosterone/R1881. As shown in Figure 3C, the keto group of ring A interacts with the side chains of amino acids Q711 and R752, whereas the hydroxyl group at the 17β-position hydrogen-bonds with the side chains of N705 and T877. The position of the side chains is perfectly conserved in all three ligands studied, suggesting that this interaction is particularly important for the binding of androgens. This may explain why the AR binds androgens with a strong affinity in the low nanomolar range. A gross comparison of the AR complexed with DHT and testosterone showed no major differences in the protein structure able to account for the differences in DHT and testosterone physiological activity. A close analysis of the structure of DHT and testosterone showed that the presence of an unsaturated bond between C4–C5 in testosterone results in a change in the geometry of the A-ring, which changes the orientation of the ketone group at C3. This altered geometry changes the distance and angle of the hydrogen bond with residue R752 to favor the AR LBD interaction with DHT over that with testosterone68.
In an unbound state, the AR complexes with heat-shock, chaperone and co-chaperone proteins (such as HSP90, HSP70, and p23), which helps to maintain the apo state of the AR in a state competent to bind ligand (Figure 1A)73. However, in an agonist-bound state such as was seen in the crystal structure of DHT-bound AR (PDB: 1I37 or 2AMA), H12 was held near H3 and H4 (Figure 3D). This conformational change in the LBD has also been seen in other nuclear receptors, such as the retinoic acid receptor (RARγ) bound to all-trans retinoic acid (PDB: 2LBD)74, thyroid hormone receptor (TRα) bound to 3,5,3′-triiodothyronine (PDB: 3GWS)75,76 and peroxisome proliferator-activated receptor-α (PPARα) bound to the agonist ligand GW409544 (PDB: 1K7L)77. Hormone binding creates an 'active' conformation of the receptor that primes the receptor for coactivator binding to an AF2 region comprised of H3, H4, and H12. Many nuclear receptors retain full ligand-dependent transcriptional activity by binding to coactivators containing short leucine-rich LxxLL motifs (where “x” can be any amino acid) known as nuclear receptor (NR) boxes78. Binding to coactivators and association with histone acetyl transferase coactivator complexes such as p300/CBP results in potent histone acetyl transferase activity7,79. We and others have shown that the AR can bind with high affinity to some LxxLL motifs in p160 coactivators80,81. However, phage display demonstrates that the AR has a strong preference for the phenylalanine-rich motif FxxLF, or motifs with phenylalanine or tryptophan at positions +1, +5, or both82. Mutational changes of the +4 and +5 residues abolished the interaction of the motifs with the AR, which further confirms the importance of intact +1, +4, and +5 hydrophobic amino acids in the motif83. Structural studies have revealed that this strong preference is due to geometrical and conformational constraints that occur between a cofactor and a cofactor binding site on the LBD (PDB: 1T7R)82,84. The side chains of coactivator aromatic residues F+1, L+4, and F+5 insert into the hydrophobic cofactor binding groove on the LBD surface formed by helices 3, 4, and 12 (Figure 3D, left and middle panel). Amino acid side chains in the coregulator recognition site of the LBD can rearrange upon motif binding, a phenomenon known as the induced fit mechanism84. The charged amino acids lysine in H3 (K720 in AR) and glutamate in H12 (E897 in AR) are highly conserved among different nuclear receptors (Figure 3D, right panel). These residues are located at the end of the groove and electrostatically interact with the coactivator backbone, thus forming a “charge clamp” between the amide nitrogen of F+1 and E897 and the carbonyl group of F+5 and K720 (Figure 3D, middle panel). This type of electrostatic interaction adds specificity to the recognition of different coregulators80. The AF2 domain is important not only for forming the coregulator binding site; it also mediates the preferred N/C interaction mentioned earlier42,85. In recent years, an additional surface cleft called BF3 (binding function 3) was found to allosterically regulate AF2 coactivator binding (Figure 3E)86,87,88. This was a surprising finding to the group, as they initially set out to identify compounds that bound the AF2 pocket through a high-throughput screen. Interestingly, this site, localized by x-ray crystallography, reveals that compounds such as 3,3′,5-triiodothyroacetic acid (TRIAC) (PDB: 2PIT) and flufenamic acid (PDB: 2PIX) bind to a hydrophobic cleft at the junction of H1, the H3–H4 loop, and H9 (Figure 3E). The LBD has another important feature: the regulation of AR nuclear export. The AR is exported to the cytoplasm upon ligand withdrawal89. The presence of a nuclear export signal (NES) (residues 742–817) (Figure 1B) in the surrounding vicinity of the bound ligand can sense ligand withdrawal and thus helps to complete AR nuclear/cytoplasmic shuttling90.
Antiandrogens
Antiandrogens are AR ligands that antagonize the actions of androgens by competing for AR binding sites. Antiandrogens can be both steroidal and non-steroidal. Steroidal antiandrogens include cyproterone acetate, oxendolone, and spironolactone (Figure 4A). Steroidal antiandrogens have limited clinical applications due to their undesired side effects; this then led to the development of nonsteroidal antiandrogens. Toluidide derivatives such as flutamide, bicalutamide and nilutamide (Figure 4B) are pure antiandrogens without androgenic properties; this lack of androgenic properties makes them suitable for use in the treatment of prostate cancer60.
Structural model of AR antagonism. (A) Chemical structure of the steroidal antiandrogens cyproterone acetate, oxendolone and spironolactone. (B) Chemical structure of the non-steroidal antiandrogens flutamide, bicalutamide and nilutamide. (C) Nuclear receptor H12 helices can adopt different conformations. In an agonist state, the H12 of DHT-bound AR (PDB: 1I37 or 2AMA) is held near H3, H4, and H11, which form a groove for coactivator binding. In an antagonist state, H12 rotates clockwise toward H3 and blocks the coactivator binding site. (D) Structure of the PPARα LBD complexed with SRC-1 coactivator peptide (H12 in agonist conformation) and with the SMRT corepressor peptide (H12 in antagonist conformation). (E) Computer model of antagonist-bound AR shows the predicted displacement of H12.
Thus far, only the binding mechanism of agonists to the AR has been described (see section AR agonists), and the structural mechanism for antagonism of the AR remains unclear. In contrast to the AR, the structural basis of antagonism has been elucidated for other nuclear receptors, eg, from the structures of 4-hydroxyltamoxifen (4-OHT) bound to the ER91, RU-486 bound to the GR91,92 and the antagonist GW6471 bound to PPAR-α93. From these structures, we have learned that H12 becomes displaced over the coactivator binding pocket, which in turn prevents coactivator binding (Figure 4C). In the case of the ER, the size and structure of 4-OHT prevent the molecule from being completely confined in the cavity, leaving its bulky side chain protruding against H12 and preventing H12 from adopting a position essential for coactivator interaction. The schematic diagram in Figure 4C shows the different positions of NR AF2 in both the agonist-bound and antagonist-based states71. In the structure of PPAR-α bound to the antagonist GW6471, H12 rotates clockwise toward H3 and blocks the coactivator binding site (Figure 4C and 4D). In addition to blocking coactivator binding, H12 repositioning allows the recruitment of corepressors such as nuclear receptor co-repressor 1 (NCoR) and silencing mediator of retinoic acid and thyroid hormone receptor (SMRT) (PDB: 1KKQ) (Figure 4D)93.
It is unclear whether the mode of antagonism observed in other nuclear receptors is relevant to the AR as the structure of the antagonist form of the AR LBD has not been solved. However, functional studies do suggest that the H12 displacement model is likely applicable to the AR94,95. It is believed that bicalutamide binding blocks AF2 function. Computer modeling suggests that the displacement of H12 by bicalutamide would not be as dramatic as those seen for the ER. The model depicts a shift of the N terminus of helix 12 upwards and away from H3 and H4, which is believed to be sufficient for the distortion of AF2-coactivator binding (Figure 4E)95. Antagonist-induced conformational changes were analyzed using limited trypsinization, demonstrating that the binding of agonists and antagonists result in different conformational changes96. This modulation of protein surface topology was proposed to enable the recruitment of a different repertoire of regulators. It was shown that antagonists induce recruitment of the corepressors NCoR and SMRT7,94,97,98,99. In the absence of conflicting evidence, the AF2 displacement model holds as the mechanism for AR antagonism. However, the precise mechanism remains unclear, and a crystal structure of the AR LBD in the antagonist-bound conformation would be required to provide a better basis for the structure-based design of AR antagonists for the treatment of AR disorders.
AR and physiological disorders
Although the AR is involved in diverse activities, its primary functions are related to male physiology, such as sex differentiation and sex-specific pathology100. Defects in the AR gene can prevent the normal development of both internal and external male structures in 46, XY individuals and result in androgen insensitivity syndrome, which is the partial or complete inability of cells to respond to androgens13,101,102. Defects in the AR gene can be caused by four types of mutations: (i) Single point mutations resulting in substitutions or premature stop codons, (ii) nucleotide insertions and deletions resulting in frameshifts, (iii) complete or partial deletion of the gene, or (iv) intronic mutations that affect AR RNA splicing103. Currently, 1029 distinct mutations have been identified in the human AR gene and are distributed predominantly over the AR DBD- and LBD-coding regions. These mutations are well documented in the Androgen Receptor Gene Mutations Database World Wide Web Server at the Lady Davis Institute for Medical Research, available at http://androgendb.mcgill.ca104. Several investigations have associated the polymorphic polyglutamine repeats in the NTD with Kennedy's Disease, also known as spinobulbar muscular atrophy, a progressive neurodegenerative condition105,106,107. There are an increasing number of studies relating the action of the AR to breast108, larynx109, liver110, and testicular cancers111.
AR and prostate cancer
AR activity is intimately linked to prostate cancer, which is by far the most commonly diagnosed cancer among American men and the second leading cause of cancer deaths112,113. In 2010, direct medical costs for prostate cancer were projected to reach $12 billion and are expected to further increase by 2020114. Of the 1029 mutations found in gene that encodes the AR, 159 mutations predispose males to prostate cancer104. In addition, previous work has suggested that the length of the repeats in the NTD influences prostate cancer risk in men23,115,116. A meta-analysis of 19 studies including Caucasian, African-American and Asian subjects predicted an increased risk of prostate cancer in men with shorter (≤21) CAG repeats. However, a Swedish study suggests that men with shorter AR CAG lengths (eg, ≤22 repeats) are at a greater risk of developing prostate cancer. Other studies found no association between the AR CAG repeat length and prostate cancer risk117. Although evidence that mutations in the AR predispose men to prostate cancer is undisputed, AR NTD CAG repeat length association with prostate cancer risk thus remains controversial.
Prostate cancer cells, similar to normal prostate cells, require androgens to grow and survive. Growth of prostate cancer depends on the ratio of the rate of cell proliferation to the rate of cell death118. In prostate cancer, the rate of proliferation is higher than that of death, resulting in continuous net growth. Androgens and the AR are the main regulators of this ratio. More than 70 years ago, Charles HUGGINS demonstrated that androgen deprivation by orchiectomy (removal of the testes) caused regression of prostate cancer119,120. Increased serum levels of the important biomarker PSA suggest that AR activity is elevated in prostate cancer patients. According to the American Cancer Society, a PSA level above 4 ng/mL has been recognized to be abnormal, and these patients are advised to undergo a biopsy113,121.
The initiation of prostate cancer can in many cases be attributed to the activation of distinct growth-promoting pathways. One prominent example is the androgen-dependent upregulation of members of the E-twenty-six (ETS) family of transcription factors by gene fusions between the AR-regulated TMPRSS2 gene promoter and the coding region of the ETS family members erythroblast transformation-specific (ERG) and ETS variant 1 (ETV1), which have been estimated to occur in ∼50% of prostate tumors122,123,124. These fusions confer androgen responsiveness to ETS transcription factors, which leads to cell-cycle progression. Interestingly, the induction of this fusion is itself dependent on the DHT/AR-stimulated recruitment of three DNA-directed enzymes, activation-induced cytidine deaminase (AID), LINE-1 repeat-encoded ORF2 endonuclease, and topoisomerase II beta (TOP2B), that trigger chromosomal translocation125,126. Other signaling pathways shown to be involved in prostate cancer initiation and progression include the PI3K and RAS/RAF pathways; dysregulation of these pathways in both early and late stage prostate cancer was implicated through genomic profiling127. In this study, analysis of the AR signaling pathway revealed a greater alteration compared with the other pathways, indicating that the AR is still the “master regulator” of prostate cancer.
AR pathway perturbation is the mechanistic rationale for the use of androgen suppression methods to treat prostate cancer. Initial treatment includes androgen suppression via castration through surgical (orchiectomy) or chemical (gonadotropin-releasing hormone (GnRH) analogues such as leuprolide and goserelin) means128. GnRH agonists desensitize the GnRH receptor by interrupting its physiological intermittent stimulation, whereas the GnRH antagonist degarelix blocks GnRH stimulation directly129. Patients are then placed on androgen deprivation therapy, which is usually combined with leuprolide for total androgen blockade130,131. Deprivation is typically achieved by oral treatment with nonsteroidal antiandrogens, such as flutamide (Figure 3A), which was approved for the treatment of prostate cancer in 1989, and the newer, structurally related compounds bicalutamide and nilutamide (Figure 3A).
AR and castration-resistant prostate cancer (CRPC)
Patients on androgen deprivation therapy remain in long-term remission of the disease. However, the development of a castration-resistant disease is inevitable132,133. This form of prostate cancer is lethal, and patients no longer respond to first-line androgen deprivation therapy. CRPC patients are usually treated with chemotherapy including the anti-mitotic compound docetaxel, which has been demonstrated to confer a survival advantage134,135. The mechanisms of castration resistance remain unclear but are thought to be diverse. For a comprehensive review of the mechanisms of CRPC development, the reader is referred to other excellent reviews133,136,137,138,139. Briefly, there are four possible mechanisms of CRPC development: 1) Increased sensitivity of the AR to its agonists, 2) AR mutations that render the receptor responsive to alternate, non-androgen ligands, 3) ligand-independent AR activation, and 4) AR-independent mechanisms (Figure 5).
Androgen and AR action in castration-resistant prostate cancer. Mechanism of castration-resistant prostate cancer. Several mechanisms promote the progression of castration-resistant prostate cancer: (1) AR overexpression coupled with continued tumor steroidogenesis. (2) Promiscuous binding and activation of mutant AR by alternative ligands, such as estrogen (E2), progesterone (P), glucocorticoids (C) and flutamide (F). (3) Ligand-independent mechanisms of AR activation via crosstalk with Akt, HER2, and Ack1 kinases that phosphorylate the AR and via long non-coding RNAs (eg, PCGEM1) that bind to the AR to stimulate transcription of AR target genes. (4) AR-independent pathways, in which cancer cell survival and growth are directed by Stat3 signaling or by upregulation of anti-apoptotic Bcl-2. Glucocorticoid receptor (GR) was found to activate a similar set of AR target genes necessary for survival of cancer cells.
Patients on androgen deprivation therapy have lower levels of circulating androgens, which initially curb prostate cancer cell proliferation; however, the opposite happens in CRPC patients, who have increased tumor cell proliferation. One of the underlying mechanisms of CRPC is an increase in the expression of AR in the cell. Koivisto et al showed that 28% of androgen-independent tumors that developed after androgen deprivation therapy had increased AR expression due to AR gene amplification140. These results indicate that CRPC cells may not be strictly androgen independent, but rather, they become more sensitive due to a lowered threshold for androgens. Even under androgen deprivation therapy, androgen levels are sufficiently high to activate overexpressed AR, which is due to intratumoral in-situ synthesis141 and residual synthesis in the adrenal gland142, along with decreased levels of the androgen inactivating enzymes CYP3A4, CYP3A5, and CYP3A7 in patient tissue samples143.
Another mechanism for the development of CRPC is ligand promiscuity, which results from AR gene mutations that cause amino acid substitutions in the LBD that decrease specificity and selectivity for ligands (eg, T877A, L701H, W741L, and F876L). These mutant AR proteins bind to other steroid hormones, such as estrogen, progesterone and glucocorticoids, which induce the activation of AR transcriptional activity resulting in prostate cancer growth144. In certain situations, AR mutations cause antagonists to induce an agonist conformation, resulting in AR activation rather than inhibition. Early examples have been found in patients on flutamide treatment in combination with androgen blockade. Five of these 16 patients had the AR mutation T877A. Through luciferase assays, it was shown that the antagonist flutamide behaves like an agonist for these AR mutant proteins145, and it was suggested that flutamide exerts a strong selective pressure for AR mutations.
The third mechanism of CRPC development is AR activation through ligand-independent mechanisms146. Studies have shown that tyrosine kinase receptor-activating ligands, such as insulin-like growth-factor-1 (IGF-1), keratinocyte growth factor (KGF), and epidermal growth factor (EGF), can activate the AR as a consequence of activating the downstream PI3K/AKT/mTOR pathway, thus creating an 'outlaw receptor'. IGF-1 was able to cause AR activation, inducing a five-fold increase in PSA levels in LNCaP cells cultured in serum-free medium147. Activation of the AR complex can also occur via crosstalk with other signaling pathways, such as those mediated by the non-receptor tyrosine kinases Src and Ack1139,148,149,150,151. Recently, various groups have described AR activation by binding of long non-coding RNAs (eg, PCGEM1 and PRNCR1) to the AR that can result in castration-resistant prostate cancer152. In addition, several AR variants that lack the LBD and act as negative regulators of the NTD have been described in CRPC. The AR NTD is constitutively active in the absence of the LBD and thus can promote androgen depletion-resistant growth138,153.
The last pathway leading to CRPC bypasses AR signaling completely. It has been shown that castration therapy in mice triggers an inflammatory response released by the dying cells. Proinflammatory factors produced by dying prostate cancer cells cause the infiltration of B and T cells. Infiltrating B cells produce lymphotoxin and factors that increase Stat3 signaling, which is vital for promoting hormone-free survival of prostate cancer cells154. Similarly, upregulation of the anti-apoptotic protein Bcl-2 protects cancer cells from castration-induced apoptosis155. Very recently, CRPC tumors were found to upregulate the expression of GR, another member of the nuclear receptor family. GR was shown to drive the expression of a subset of AR target genes necessary for cancer cell survival156. The various pathways mentioned may operate simultaneously to enhance AR activity157. Most evidence suggests that ADT failure may not result from a loss of androgen signaling but rather from the acquisition of genetic changes that lead to aberrant activation of the AR and its signaling axis104,137,158. Thus, the AR remains a potential therapeutic target for prostate cancer therapy39.
In 2012, the second-generation AR antagonist enzalutamide (Figure 6) (MDV3100) was approved by the FDA for use in prostate cancer159,160,161. Enzalutamide is a more potent antagonist than bicalutamide because it binds to the AR ligand-binding pocket with higher affinity than first generation antagonists, such as bicalutamide, and also prevents the translocation of the receptor into the nucleus162. Recent studies have shown that abiraterone acetate (Zytiga), an inhibitor of the cytochrome P450 enzyme CYP17, impedes androgen synthesis and thus lowers the level of circulating ligand, improving overall survival in well-powered, randomized phase III studies163,164. In addition, a series of new agents are in clinical development, including the AR antagonists ARN-50944 and EPI-001 that directly target the AR, as well as orteronel (TAK-700)45,46 and galeterone (TOK-001)47 that indirectly target the AR. Collectively, the development of enzalutamide, abiraterone acetate, and other new agents validate the importance of the AR as an important therapeutic target.
Chemical structure of enzalutamide.
Structural understanding of disease/drug resistance-related androgen receptor mutations through X-ray crystallography and structural modeling
Because the AR remains the driving force in the progression of more aggressive castration-resistant prostate cancers139, it is important to understand how each genetic aberration affects the pharmacology of the AR and its ligands. Through the use of X-ray crystallography and computer modeling, biochemical observations can be understood at the atomic level, which is expected to greatly aid the design of improved drugs to overcome current clinical problems.
Structural basis of mutation-induced AR promiscuity
DHT-bound AR LBD T877A
The single point mutation T877A has been found in LNCaP cells and prostate cancer patients to confer abnormal binding characteristics to the AR. This mutation has been shown to cause a significant increase in the affinity of the AR for binding to estrogens and progesterone. In addition, this mutation also allows the AR to be activated by antiandrogens such as flutamide145,165. To understand the structural basis of this abnormal binding characteristic, Sack et al solved the crystal structure of the wild-type (WT) (PDB: 1I37) and T877A (PDB: 1I38) AR LBDs complexed with DHT67. A comparison of both structures revealed similar overall conformations. Interactions between DHT and both forms of the AR occur at almost identical points of contact, except at the mutated residue (Figure 7A). Interestingly, the introduction of the T877A mutation created additional space around the D-ring of DHT. This increase in volume allows bulkier ligands to enter the pocket, and this may induce AR promiscuity for other hormones and analogs, such as estrogens and progesterone. Conversely, the presence of T877 limits the pocket size and is thought to be important for ligand specificity. The concept that an increase in the LBP may increase ligand promiscuity was further supported by biochemical studies in which threonine was replaced by the larger amino acid aspartic acid, which prevented androgen binding, presumably by steric hindrance, making the mutant receptor unresponsive to androgen activation166.
Structural understanding of disease/drug resistance-related androgen receptor mutations. (A) Structural comparison of wild-type (PDB: 1I37) and mutant T877A (PDB: 1I38) AR LBDs in complex with dihydrotestosterone. Key residues involved in hydrogen bonding have been highlighted, with hydrogen bonds indicated by black dotted lines. (B) Structure of the AR LBD double mutant L701H/T877A complexed with 9α-fluorocortisol (PDB: 1GS4). (C) Structural overlay of androgen receptor complexed with FxxLF (PDB: 1XOW) and LxxLL (PDB: 1T7F) motif-containing peptides. Residues V730, M734, and I737 are involved in forming hydrophobic contacts with coactivator peptides. Residue V730 was mutated to M730 to demonstrate an enhanced binding of LxxLL peptides. (D) Structure of AR LBD W741L complexed with bicalutamide (PDB: 1Z95). Residues L704, N705, Q711, and R752 form hydrogen bonds with bicalutamide (indicated by dotted lines). Also shown is the wild-type W741 residue (white) to illustrate a possible steric clash between tryptophan and the B-ring of bicalutamide.
9α-fluorocortisol-bound AR LBD L701H/T877A
Two mutations, L701H and T877A, were identified in the androgen-independent prostate tumor cell lines MDA 2a and 2b. The double mutation allows the AR to be activated in the absence of androgens. Both mutations are located in the LBD and strongly increase AR sensitivity to cortisol and cortisone, which in turn leads to the promotion of prostate cancer cell growth167. Not surprisingly, the double mutant also renders the receptor responsive to progesterone and estrogen due to the presence of the T877A mutation, most likely for the reason mentioned in the above section. However, the T877A mutation alone is insufficient to promote receptor binding and activation by cortisol. Matias et al provided a structural basis for the glucocorticoid responsiveness of the AR LBD double-mutant L701H/T877A (Figure 7B)168. The overall fold of the L701H/T877A double-mutant AR LBD complexed with 9α-fluorocortisol (PDB: 1GS4) is similar to previous agonist structures. The main structural difference is the formation of a favorable hydrogen bonding network between the D- and C-rings of 9α-fluorocortisol and A877, H701, and S778 of the mutant receptor. Specifically, hydrogen bonding between S778 and H701 correctly positioned the imidazole ring of H701 to form a hydrogen bond with the hydroxyl group on the D-ring to form a hydrogen bond with H701, whereas A877 was able to form a close Van der Waals contact with the D-ring of 9α-fluorocortisol. In contrast, a threonine residue at position 877 would be extremely unfavorable for 9α-fluorocortisol binding, and the hydrophobic leucine at position 701 in the wild-type receptor would be unable to engage in the formation of this stabilizing hydrogen bond network.
The V730M mutation alters coactivator selectivity
V730M is a somatic mutation that was detected in an advanced-stage prostate carcinoma that resulted in an increase in AR activation by androgens169. The crystal structure of the wild-type AR illustrated the reason why the LxxLL motifs of typical NR coactivators fail to hydrogen bond with Glu897 of H12 and make fewer and less optimized hydrophobic contacts with the AF2 pocket compared with FxxLF-containing peptides, explaining the lower affinity of AF2 for the LXXLL-containing p160-type coactivators82. In contrast, cell-based reporter gene assays and in vitro binding assays showed that V730M increases LxxLL binding without affecting FxxLF binding, resulting in increased overall AR transcriptional activity84. In three-dimensional space, V730 is located near the coactivator binding site84 and, along with M734 and I737 in the hydrophobic groove, allows the formation of a smoother and flatter surface that permits greater complementarity to FxxLF compared with the LxxLL motif (Figure 7C). A mutation of V730 to methionine would alter the interaction surface to become more favorable to binding to LxxLL motifs and thereby to the recruitment of LxxLL motif-containing coactivators, such as the SRC coactivators, which are commonly associated with the AR in recurrent prostate cancer7,170,171.
Mutations alter drug antagonist properties
Bicalutamide-bound AR LBD W741L
The administration of antiandrogens is the standard approach to treat prostate cancer. Examples of first generation antiandrogens include bicalutamide, flutamide, and nilutamide, which inhibit androgen action by binding to the androgen receptor in a competitive fashion, ie, as an antagonist. Unfortunately, these first-generation drugs demonstrated agonist properties in LNCaP cells, which overexpress the AR, a state that mimics castration-resistant prostate cancer. Hara et al reported two LBD mutations, W741L and W741C, in LNCaP cells that appeared during bicalutamide treatment and caused the receptor to be activated by bicalutamide172. The structure of the AR LBD W741L bound to bicalutamide shed light on how the mutation imparts agonistic properties leading to bicalutamide resistance173. In the structure, bicalutamide adopts a bent conformation within the ligand-binding pocket (Figure 7D). Although hydrophobic interactions account for the majority of the contact points between bicalutamide and the AR LBD, hydrogen bonds form in two different regions. Similar to the previous structures of the AR bound to R1881 and DHT, the cyano group of the bicalutamide A ring forms hydrogen bonds with Q711 and R752. Additional hydrogen bonding was observed between the amide nitrogen and the chiral hydroxyl group of bicalutamide and amino acids L704 and N705. Unlike in the structures of R1881- and DHT-bound AR, T877 is not involved in hydrogen bonding (Figure 7D), which may explain the lower binding affinity of bicalutamide compared with R1881 or DHT. In the presence of the W741L mutation, the bulkier side chain of tryptophan is replaced by the smaller side chain of leucine. This provides more space to accommodate the B-ring of bicalutamide, allowing the AR to maintain a similar fold as that seen in other agonist-bound structures and thus favoring the formation of an active state of AF2. In other words, this increase in space in the LBP would allow bicalutamide to bind without displacing H12, which is the basis for its antagonist activity.
MDV3100 and AR LBD F876L
The recent FDA approval of enzalutamide (Figure 6) and the development of ARN-509 reiterate the importance of targeting AR signaling for CRPC treatment160,162. Despite the success of enzalutamide at causing a significant drop in the serum PSA levels of patients, responses to enzalutamide are often short-lived. Some causes of resistance to enzalutamide have been identified174,175. Through a reporter-based mutagenesis screen, Balbas et al identified the AR mutation F876L. In contrast to wild-type AR, enzalutamide functions as an agonist for AR F876L and causes the mutant receptor to bind enzalutamide six times more effectively than wild-type receptor176. To understand the effect of the F876L mutation, the group also mutated F876 to a bulkier residue, tyrosine, or to another aliphatic amino acid, isoleucine. Only the F876I mutation caused enzalutamide to behave as an agonist, similar to F876L, indicating that resistance is due to a clear structural change in the drug-receptor complex. As the structure of enzalutamide-bound AR has not been reported, the Sawyer group modeled the AR-enzalutamide complex through ligand docking and molecular dynamics simulations. Docking suggested that enzalutamide interacts with the wild-type AR differently than with bicalutamide. As shown in Figure 5D, the B-ring of bicalutamide makes contact with H12. However, enzalutamide does not interact with helix 12; instead, its C-ring interacts with the C terminus of helix 11 and the loop region between helix 11 and helix 12. It was proposed that this accommodation allows enzalutamide to directly contact F876, resulting in a conformational rearrangement in helix 11 that prevents helix 12 from adopting the agonist conformation required for coactivator binding. In contrast, in the presence of the F876L mutation, leucine lacks the favorable contact with enzalutamide that is predicted to be necessary for helix 12 displacement. Hence, helix 12 is thought to assume an agonist-like conformation that allows coactivator recruitment.
Rational drug discovery in progress
A growing amount of evidence suggests that mutations in the AR-encoding gene occur spontaneously in prostate cancer and eventually result in the relapse of patients. As a consequence, the effects of drugs change, and patients are no longer responsive to treatment. These gain-of-function mutations present a scientific challenge to pharmacologically overcome the mechanisms of drug resistance and further highlight the importance of identifying advanced compounds to inhibit AR activity. Rational drug design, which is the application of a structure-function relationship, is now widely used in modern medicinal chemistry for developing exquisitely selective ligands.
Can the available structural information on AR-ligand interactions based on x-ray crystallography and modeling be exploited for the rational design of next-generation antiandrogens to overcome problems with antiandrogen resistance? Available structures of the wild-type and mutant AR in complex with ligands reveal a clear explanation for how changes in a single residue can result in dramatic changes in ligand-binding properties and pinpoint the key determinants of receptor-ligand specificity and affinity. This information will greatly facilitate the development of new antagonists for the treatment of prostate cancer. Hence, this section will provide an overview of the development of new antiandrogens.
Targeting the NTD
Hypothetically, there are numerous regulatory sites that can be therapeutically exploited. As discussed earlier in this review, various functional domains, such as the NTD, DBD and LBD, can be targeted by small molecules. Targeting the intrinsically unstructured NTD remains a challenge. Nevertheless, it is an attractive target for developing novel therapeutics, especially in light of increasing reports indicating that constitutively active AR splice variants that lack the LBD are found in prostate cancer patients. A number of NTD inhibitors have been reported in the literature, with decoy proteins providing the first proof-of-concept evidence that targeting the NTD is a viable method to pre-clinically control the growth of CRPC177. NTD peptide (amino acids 1–558) was overexpressed to competitively block binding of interacting proteins or the N/C interaction. In another study, Anderson et al reported the identification and characterization of the small molecule EPI-001, which was isolated from the marine sponge Geodia lindgreni and targets the NTD-AF1 domain. This compound also reduces the interaction between the NTD and the known AF1-interacting coactivator CBP. EPI-001 was effective in mice bearing LNCaP xenografts, with no apparent toxicity detected. Thus far, this is the best characterized compound shown to effectively target the AR NTD for treatment of CRPC178. Recently, this group has continued to design new EPI-001 analogs that target the NTD and are currently under clinical development for human study179. More high-throughput screens have been used to identify new classes of compounds, such as the glycerol ethers niphatenones isolated from the sponge Niphates digitalis, which are proposed to covalently bind the AF1 and may serve as lead compounds for further drug development180.
Targeting the DBD
Fewer strategies targeting the DBD have been explored to date. In early work, a hairpin pyrrole-imidazole polyamide was designed to target the ARE to disrupt AR DBD binding and was effective at inhibiting androgen-induced PSA expression in LNCaP cells181. Building on this work, affinity and specificity were further improved by creating a cyclic polyamide with a greater effect on decreasing PSA mRNA levels182. Although this was an ingenious approach, allosterically affecting the conformation of double-stranded DNA to prevent the AR DBD from binding has limited clinical applicability. This is because each polyamide can only target a subset of AR-dependent genes, instead of simultaneously targeting all AR-dependent genes important for disease progression. To overcome this shortcoming, a recent report described a high-throughput screen to target the AR protein rather than the DNA to reduce AR binding to the promoter and enhancer regions of PSA and TMPRSS2. Of ∼160 000 molecules, the authors identified 1-[3-(2-chlorophenoxy)propyl]-1H-indole-3carbonitrile (CPIC) as a compound that was able to reduce AR-specific DNA binding, although it remains unclear whether this compound works directly by binding to the DBD183. Given that the structure of the AR DBD has been solved, one might have expected the development of further DBD-targeted drugs. However, this has not been the case. One factor that may be limiting progress in this area of research lies in limited specificity, given that the DBD has high sequence homology among all members of the nuclear receptor family. This issue needs to be addressed before DBD-targeted drugs will enter the market.
Targeting the LBD
Targeting AF2 and BF3
It is generally accepted that AR activation requires the formation of a functional AF2 region for interactions with both cofactors and N/C. This makes the AR AF-2 surface an attractive target to modulate AR activity. Compounds that bind the AF2 would hypothetically prevent intramolecular association between the AR LBD and NTD and interactions of the LBD with coregulators. Selective peptide inhibitors have been developed for ligand-bound ER and TR184. A similar approach could be adopted for the AR. Hydrophobicity, size, and complementarity all substantially contribute to differential binding affinity and to the selective high-affinity binding of the AR AF2 to phenylalanines at the +1 and +5 positions in coactivator recognition helices. Hence, inhibitors that are designed to bind to the hydrophobic groove at the +1 or +5 sites to disrupt coactivator interaction may provide promising leads80. The feasibility of targeting AR in its coactivator binding pocket was demonstrated using peptide antagonists185. Phenylalanine-rich peptide antagonists were able to inhibit AR gene reporter activation without affecting PR-mediated activation, demonstrating some level of specificity for the androgen receptor. However, as different nuclear receptors have evolved to bind hydrophobic LxxLL consensus motifs through similar binding mechanisms, it remains important that mimetics are designed in a way that conveys greater selectivity to the AR. Improving mimetics by including specific flanking sequences is expected to improve selectivity, as studies have shown that the sequences immediately flanking the consensus motif confer specificity in vivo83,186,187,188,189. Further characterization of the unique requirements for androgen receptor-specific coactivator binding may be useful for the design of peptide antagonists. The effects of AR peptide antagonists have been evaluated in mammalian cell-based assays; however, none has been further evaluated in animal models, and thus, it remains unclear whether they will be effective in vivo. The use of peptide antagonists may be difficult as drug delivery of small peptides can be a rather challenging task to overcome. In place of peptide antagonists, Axerio-Cilies et al employed methods of computer-aided drug discovery to discover small molecule inhibitors of the AF2 using the ZINC database, which currently consists of ten million purchasable compounds in the market. Six lead compounds were found to inhibit AR transcriptional activity. These authors confirmed by X-ray crystallography that compound 5 binds specifically to the AF2 site (PDB: 2YHD)190.
The same group adopted this method to identify inhibitors that target the BF3191. Several X-ray structures (PDB: 2YLP, 2YLO, 4HLW) confirmed the presence of the BF3 site and its importance in regulating the AF2. Both AF2 and BF3 inhibitors provide new therapeutic avenues that could potentially help overcome the gain-of-function mutations that are selected for in the presence of current antiandrogens. By targeting a different site on the AR, AF2, and BF3 inhibitors can be concurrently taken with antiandrogens to prolong time to cancer remission.
Targeting the LBP
Finally, drugs can be designed to bind to the ligand-binding pocket. Over the past decade, most research efforts have been devoted to the design of small molecules that target the LBP, which is well ordered in crystal structures. Current research is focused on designing AR inhibitors with high affinity and specificity. Antiandrogens that have a high enough binding affinity to sufficiently displace DHT from the ligand-binding pocket need to be designed. Based on the agonist-AR LBD complex structure, it is believed that favorable H bonding between the ligand and AR T877, N705, Q711, and R752 and a hydrophobicity within a proper range are critical for ligands to bind to the AR with high affinity. Computer modeling of the binding of AR antagonists such as flutamide, nilutamide and bicalutamide also suggested that both polar and hydrophobic interactions are essential for proper binding to anchor those compounds into the pocket192. A structural comparison of the wild-type AR bound to DHT and the agonist-converted AR-bicalutamide complex demonstrated that the sites that bind the A and B rings of DHT are similar to the sites bound by bicalutamide. Only the C6, C7, and C8 atoms of the B ring and the C15 and C16 atoms in the D ring of DHT bind to locations not bound by bicalutamide173. Additional van der Waals interactions by these atoms are likely responsible for the much higher binding affinity of DHT. Hence, designing antiandrogens with increased bulk in this region may enhance binding affinity. Enzalutamide was developed by ligand-based drug design, which relied on the pharmacophores of known drugs, to bind and inhibit the AR with a much improved binding affinity (IC50=21 nmol/L, compared with 160 nmol/L for bicalutamide)162. Specifically, enzalutamide was developed by starting with the AR agonist RU59063 and systematically modifying its chemical groups while maintaining the steroid fold162.
Specificity is another key factor to consider in the design of antiandrogens. Although enzalutamide generally seems to be well tolerated in humans, there was a low incidence of adverse events where patients developed seizures193. This effect was postulated to be a result of inhibition of GABA-gated chloride channels, as shown in animal models194. Such off-target effects can be minimized or eliminated by ensuring tissue selectivity. Entry of selective steroid receptor antagonists into different target tissues would be a new avenue to explore to avoid neurological side effects. However, to date, structural features that are essential for achieving uptake selectivity have not been determined. Building on the success of enzalutamide, the Sawyer group developed the compound ARN-509 by maintaining the strong affinity of enzalutamide while reducing its off-target effects. ARN-509 has greater efficacy compared with enzalutamide in laboratory animals and is currently undergoing phase I clinical trials195,196. Notably, the levels of ARN-509 in the brain are lower than those of enzalutamide at therapeutic doses, and phase I clinical trials have demonstrated an excellent safety profile and efficacy at decreasing PSA levels for ARN-509. Although the mechanism of tissue specificity remains unclear, there is clearly a need for further research in this area.
The search for new AR antagonists is ongoing. In the absence of a structure showing the AR in antagonist conformation, the use of computer modeling has provided an entry point for structure-based drug design197. Recent drug discovery strategies are based on two criteria: first, compound structures that are distinct from bicalutamide and enzalutamide to avoid cross-resistance and, second, compounds that retain antagonist properties even for known AR mutant proteins that adopt agonist conformations when bound to current AR antagonists (also known as pan-antagonists). Small-molecule in silico screens have identified molecules that fulfill both criteria. For example, the compound DIMN [(6-13,4-dihydro-1H-isoquinolin-2-yl)-N-(6-methylpyrindin-2yl)nicotinamide] has shown promising in vitro antagonistic activity198. Similarly, Shen et al identified another series of structurally defined AR antagonists, chemotype A–F compounds, that function in the micromolar range and remain antagonistic for AR T877A and AR W741C. Interestingly, these compounds also impair nuclear localization and enhance AR degradation, thus providing a multi-pronged approach that might overcome all new somatic mutations that arise during the course of the disease199. Other candidates, such as compound 3 from the NCI-3D database200, DIMN analogs termed 7AU and 7BB201, and MEL-3202, all showed encouraging in vitro activities and need to be tested in animal models.
In silico models of drug-AR LBD complexes have provided a path for rational drug design to overcome the effects of agonist-converting mutations and restore the clinical efficacy of currently used drugs. In the structure of AR LBD W741L complexed with bicalutamide, the 4-fluorophenyl sulfone group of bicalutamide is located between residues of H12 and the side chain of L741. A tryptophan at position 741 would result in a steric clash that would block H12 from adopting its agonist conformation (Figure 5C). McGinkey and Koh predicted that creating derivatives of bicalutamide with an expanded aryl sulfone core would extend farther toward H12 and would thus interfere with H12 adopting an agonist conformation even in the presence of the L741 mutation203. They validated their model with the compound PLM1, which exhibited no agonist properties in both wild-type and mutant conditions. Likewise, the flutamide derivative SC333 was shown to retain antagonistic properties toward AR T877A204. Recently, the Sawyer group has shown that it is possible to use structural and modeling data to chemically design and guide modifications of the enzalutamide structure to restore antagonism in the presence of the F876L mutation176. In their model, position 4 of the enzalutamide B-ring comes into closer contact with H12 in AR F876L. They predicted that contact with H12 may help reposition the AF2 into an antagonist conformation. To test this hypothesis, they synthesized a series of analogs with bulkier and more complex B rings. Of those compounds, DR103, DR105 and DR106 were able to overcome the effect of the F876L mutation and retain antagonistic properties. Molecular docking simulations suggested that DR103 would be capable of displacing the N-terminal residues of H12, thus imposing an antagonist-like conformation. Efforts from all groups not only led to the design of a new pan-antagonist but also helped to confirm the proposed model for AR antagonism as a result of the displacement of H12.
Conclusion and perspective
The success of the Sawyer group and others in predicting drug actions through molecular dynamics simulations is promising and raises hopes for the development of improved antiandrogens. However, it is important to note that flaws in the docking method do persist, as it employs rigid protein structures, which limit accurate predictions. It would therefore be reassuring to verify these computational models by crystallographic studies. Clearly, crystallography of the AR LBD antagonist conformation has been challenging86,173, as the AR LBD remains unstable in the absence of an agonist during protein expression and purification. In addition, the antagonist-bound AR LBD remains complexed with the heat-shock proteins groEL, likely due to improper receptor folding. Methods to stabilize the AR in an antagonist conformation should be explored for crystallizing the protein.
Although the AR LBP remains the primary target of small molecules, the focus of screens is beginning to shift to other functionally significant domains of the protein, such as the NTD, DBD, AF2, and BF3. Recently, using an in silico method, Helsen et al provided insights into the existence and importance of physical connectivity between the AR DBD and LBD205. This information will give researchers another plausible interface for drug intervention. More detailed information on the structure of the full length AR would provide a complete picture necessary to understand interdomain interactions. Crystallization of full length nuclear receptors remains a formidable challenge, and currently, only two structures of nuclear receptors that include all functional domains have been solved by the Rastinejad group (HNF-4α homodimers (PDB: 4IQR)34 and PPAR/RXR heterodimers33 complexed with their DNA elements and coactivator peptides). These structures have greatly expanded our understanding of the physical connectivity between the LBD and DBD, which can be utilized for the design of small molecule inhibitors.
With rapid advances in protein engineering and data collection/diffraction methods, including X-ray free-electron lasers206, we expect that the full length structure of the AR in antagonist conformation will eventually be solved. We believe that by capitalizing on various critical contributions to the understanding of the AR structure and function and advancements in crystallography techniques, the future of rational drug design for better antiandrogens for the treatment of prostate cancer remains bright.
References
Mangelsdorf DJ, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K, et al. The nuclear receptor superfamily: the second decade. Cell 1995; 83: 835–9.
Tsai MJ, O'Malley BW . Molecular mechanisms of action of steroid/thyroid receptor superfamily members. Annu Rev Biochem 1994; 63: 451–86.
Nuclear Receptors Nomenclature Committee. A unified nomenclature system for the nuclear receptor superfamily. Cell 1999; 97: 161–3.
Rosner W, Hryb DJ, Khan MS, Nakhla AM, Romas NA . Sex hormone-binding globulin: anatomy and physiology of a new regulatory system. J Steroid Biochem Mol Biol 1991; 40: 813–20.
Baker ME . Albumin, steroid hormones and the origin of vertebrates. J Endocrinol 2002; 175: 121–7.
Srinivas-Shankar U, Wu FC . Drug insight: testosterone preparations. Nat Clin Pract Urol 2006; 3: 653–65.
Shang Y, Myers M, Brown M . Formation of the androgen receptor transcription complex. Mol Cell 2002; 9: 601–10.
Dehm SM, Tindall DJ . Molecular regulation of androgen action in prostate cancer. J Cell Biochem 2006; 99: 333–44.
Wang Q, Carroll JS, Brown M . Spatial and temporal recruitment of androgen receptor and its coactivators involves chromosomal looping and polymerase tracking. Mol Cell 2005; 19: 631–42.
van Royen ME, van Cappellen WA, de Vos C, Houtsmuller AB, Trapman J . Stepwise androgen receptor dimerization. J Cell Sci 2012; 125: 1970–9.
Heinlein CA, Chang C . Androgen receptor in prostate cancer. Endocr Rev 2004; 25: 276–308.
Beato M, Herrlich P, Schutz G . Steroid hormone receptors: many actors in search of a plot. Cell 1995; 83: 851–7.
Brinkmann AO . Molecular basis of androgen insensitivity. Mol Cell Endocrinol 2001; 179: 105–9.
McPhaul MJ, Marcelli M, Tilley WD, Griffin JE, Wilson JD . Androgen resistance caused by mutations in the androgen receptor gene. FASEB J 1991; 5: 2910–5.
Siegel R, Naishadham D, Jemal A . Cancer statistics, 2013. CA Cancer J Clin 2013; 63: 11–30.
Lubahn DB, Joseph DR, Sullivan PM, Willard HF, French FS, Wilson EM . Cloning of human androgen receptor complementary DNA and localization to the X chromosome. Science 1988; 240: 327–30.
Brown CJ, Goss SJ, Lubahn DB, Joseph DR, Wilson EM, French FS, et al. Androgen receptor locus on the human X chromosome: regional localization to Xq11–12 and description of a DNA polymorphism. Am J Hum Genet 1989; 44: 264–9.
Migeon BR, Brown TR, Axelman J, Migeon CJ . Studies of the locus for androgen receptor: localization on the human X chromosome and evidence for homology with the Tfm locus in the mouse. Proc Natl Acad Sci U S A 1981; 78: 6339–43.
Gelmann EP . Molecular biology of the androgen receptor. J Clin Oncol 2002; 20: 3001–15.
McEwan IJ . Molecular mechanisms of androgen receptor-mediated gene regulation: structure-function analysis of the AF-1 domain. Endocr Relat Cancer 2004; 11: 281–93.
He B, Kemppainen JA, Voegel JJ, Gronemeyer H, Wilson EM . Activation function 2 in the human androgen receptor ligand binding domain mediates interdomain communication with the NH2-terminal domain. J Biol Chem 1999; 274: 37219–25.
Sasaki M, Kaneuchi M, Sakuragi N, Fujimoto S, Carroll PR, Dahiya R . The polyglycine and polyglutamine repeats in the androgen receptor gene in Japanese and Caucasian populations. Biochem Biophys Res Commun 2003; 312: 1244–7.
Hsing AW, Gao YT, Wu G, Wang X, Deng J, Chen YL, et al. Polymorphic CAG and GGN repeat lengths in the androgen receptor gene and prostate cancer risk: a population-based case-control study in China. Cancer Res 2000; 60: 5111–6.
Chang CS, Kokontis J, Liao ST . Structural analysis of complementary DNA and amino acid sequences of human and rat androgen receptors. Proc Natl Acad Sci U S A 1988; 85: 7211–5.
Davies P, Watt K, Kelly SM, Clark C, Price NC, McEwan IJ . Consequences of poly-glutamine repeat length for the conformation and folding of the androgen receptor amino-terminal domain. J Mol Endocrinol 2008; 41: 301–14.
Werner R, Holterhus PM, Binder G, Schwarz HP, Morlot M, Struve D, et al. The A645D mutation in the hinge region of the human androgen receptor (AR) gene modulates AR activity, depending on the context of the polymorphic glutamine and glycine repeats. J Clin Endocrinol Metab 2006; 91: 3515–20.
Choong CS, Kemppainen JA, Zhou ZX, Wilson EM . Reduced androgen receptor gene expression with first exon CAG repeat expansion. Mol Endocrinol 1996; 10: 1527–35.
Callewaert L, Christiaens V, Haelens A, Verrijdt G, Verhoeven G, Claessens F . Implications of a polyglutamine tract in the function of the human androgen receptor. Biochem Biophys Res Commun 2003; 306: 46–52.
Lavery DN, McEwan IJ . Structural characterization of the native NH2-terminal transactivation domain of the human androgen receptor: a collapsed disordered conformation underlies structural plasticity and protein-induced folding. Biochemistry 2008; 47: 3360–9.
Reid J, Kelly SM, Watt K, Price NC, McEwan IJ . Conformational analysis of the androgen receptor amino-terminal domain involved in transactivation. Influence of structure-stabilizing solutes and protein-protein interactions. J Biol Chem 2002; 277: 20079–86.
Lavery DN, McEwan IJ . The human androgen receptor AF1 transactivation domain: interactions with transcription factor IIF and molten-globule-like structural characteristics. Biochem Soc Trans 2006; 34: 1054–7.
Lavery DN, McEwan IJ . Structure and function of steroid receptor AF1 transactivation domains: induction of active conformations. Biochem J 2005; 391: 449–64.
Chandra V, Huang P, Hamuro Y, Raghuram S, Wang Y, Burris TP, et al. Structure of the intact PPAR-gamma-RXR- nuclear receptor complex on DNA. Nature 2008; 456: 350–6.
Chandra V, Huang P, Potluri N, Wu D, Kim Y, Rastinejad F . Multidomain integration in the structure of the HNF-4alpha nuclear receptor complex. Nature 2013; 495: 394–8.
Uversky VN . Multitude of binding modes attainable by intrinsically disordered proteins: a portrait gallery of disorder-based complexes. Chem Soc Rev 2011; 40: 1623–34.
Bevan CL, Hoare S, Claessens F, Heery DM, Parker MG . The AF1 and AF2 domains of the androgen receptor interact with distinct regions of SRC1. Mol Cell Biol 1999; 19: 8383–92.
McEwan IJ, Gustafsson J . Interaction of the human androgen receptor transactivation function with the general transcription factor TFIIF. Proc Natl Acad Sci U S A 1997; 94: 8485–90.
Simental JA, Sar M, Lane MV, French FS, Wilson EM . Transcriptional activation and nuclear targeting signals of the human androgen receptor. J Biol Chem 1991; 266: 510–8.
Callewaert L, Van Tilborgh N, Claessens F . Interplay between two hormone-independent activation domains in the androgen receptor. Cancer Res 2006; 66: 543–53.
Doesburg P, Kuil CW, Berrevoets CA, Steketee K, Faber PW, Mulder E, et al. Functional in vivo interaction between the amino-terminal, transactivation domain and the ligand binding domain of the androgen receptor. Biochemistry 1997; 36: 1052–64.
He B, Kemppainen JA, Wilson EM . FXXLF and WXXLF sequences mediate the NH2-terminal interaction with the ligand binding domain of the androgen receptor. J Biol Chem 2000; 275: 22986–94.
Wilson EM . Analysis of interdomain interactions of the androgen receptor. Methods Mol Biol 2011; 776: 113–29.
Zhou ZX, Lane MV, Kemppainen JA, French FS, Wilson EM . Specificity of ligand-dependent androgen receptor stabilization: receptor domain interactions influence ligand dissociation and receptor stability. Mol Endocrinol 1995; 9: 208–18.
Langley E, Kemppainen JA, Wilson EM . Intermolecular NH2-/carboxyl-terminal interactions in androgen receptor dimerization revealed by mutations that cause androgen insensitivity. J Biol Chem 1998; 273: 92–101.
Shaffer PL, Jivan A, Dollins DE, Claessens F, Gewirth DT . Structural basis of androgen receptor binding to selective androgen response elements. Proc Natl Acad Sci U S A 2004; 101: 4758–63.
Claessens F, Alen P, Devos A, Peeters B, Verhoeven G, Rombauts W . The androgen-specific probasin response element 2 interacts differentially with androgen and glucocorticoid receptors. J Biol Chem 1996; 271: 19013–6.
Verrijdt G, Schoenmakers E, Haelens A, Peeters B, Verhoeven G, Rombauts W, et al. Change of specificity mutations in androgen-selective enhancers. Evidence for a role of differential DNA binding by the androgen receptor. J Biol Chem 2000; 275: 12298–305.
Schoenmakers E, Verrijdt G, Peeters B, Verhoeven G, Rombauts W, Claessens F . Differences in DNA binding characteristics of the androgen and glucocorticoid receptors can determine hormone-specific responses. J Biol Chem 2000; 275: 12290–7.
Claessens F, Denayer S, Van Tilborgh N, Kerkhofs S, Helsen C, Haelens A . Diverse roles of androgen receptor (AR) domains in AR-mediated signaling. Nucl Recept Signal 2008; 6: e008.
Jenster G, Trapman J, Brinkmann AO . Nuclear import of the human androgen receptor. Biochem J 1993; 293: 761–8.
Zhou ZX, Sar M, Simental JA, Lane MV, Wilson EM . A ligand-dependent bipartite nuclear targeting signal in the human androgen receptor. Requirement for the DNA-binding domain and modulation by NH2-terminal and carboxyl-terminal sequences. J Biol Chem 1994; 269: 13115–23.
Marte B . Passage through the nuclear pore. Nat Cell Biol 2001; 3: E135.
Ni L, Llewellyn R, Kesler CT, Kelley JB, Spencer A, Snow CJ, et al. Androgen induces a switch in the androgen receptor from cytoplasmic retention to nuclear import. Mol Cell Biol 2013; 33: 4766–78.
Cutress ML, Whitaker HC, Mills IG, Stewart M, Neal DE . Structural basis for the nuclear import of the human androgen receptor. J Cell Sci 2008; 121: 957–68.
Haelens A, Tanner T, Denayer S, Callewaert L, Claessens F . The hinge region regulates DNA binding, nuclear translocation, and transactivation of the androgen receptor. Cancer Res 2007; 67: 4514–23.
Clinckemalie L, Vanderschueren D, Boonen S, Claessens F . The hinge region in androgen receptor control. Mol Cell Endocrinol 2012; 358: 1–8.
Matias PM, Donner P, Coelho R, Thomaz M, Peixoto C, Macedo S, et al. Structural evidence for ligand specificity in the binding domain of the human androgen receptor. Implications for pathogenic gene mutations. J Biol Chem 2000; 275: 26164–71.
Fang H, Tong W, Branham WS, Moland CL, Dial SL, Hong H, et al. Study of 202 natural, synthetic, and environmental chemicals for binding to the androgen receptor. Chem Res Toxicol 2003; 16: 1338–58.
Sathya G, Chang CY, Kazmin D, Cook CE, McDonnell DP . Pharmacological uncoupling of androgen receptor-mediated prostate cancer cell proliferation and prostate-specific antigen secretion. Cancer Res 2003; 63: 8029–36.
Singh SM, Gauthier S, Labrie F . Androgen receptor antagonists (antiandrogens): structure-activity relationships. Curr Med Chem 2000; 7: 211–47.
Gao W, Bohl CE, Dalton JT . Chemistry and structural biology of androgen receptor. Chem Rev 2005; 105: 3352–70.
Cantin L, Faucher F, Couture JF, de Jesus-Tran KP, Legrand P, Ciobanu LC, et al. Structural characterization of the human androgen receptor ligand-binding domain complexed with EM5744, a rationally designed steroidal ligand bearing a bulky chain directed toward helix 12. J Biol Chem 2007; 282: 30910–9.
VC M . The history of clinical endocrinology: a comprehensive account of endocrinology from earliest times to the present day. Carnforth: Parthenon Press; 1993.
Baulieu EE, Lasnizki I, Robel P . Metabolism of testosterone and action of metabolites on prostate glands grown in organ culture. Nature 1968; 219: 1155–6.
Bruchovsky N, Wilson JD . The conversion of testosterone to 5-alpha-androstan-17-beta-ol-3-one by rat prostate in vivo and in vitro. J Biol Chem 1968; 243: 2012–21.
Grino PB, Griffin JE, Wilson JD . Testosterone at high concentrations interacts with the human androgen receptor similarly to dihydrotestosterone. Endocrinology 1990; 126: 1165–72.
Sack JS, Kish KF, Wang C, Attar RM, Kiefer SE, An Y, et al. Crystallographic structures of the ligand-binding domains of the androgen receptor and its T877A mutant complexed with the natural agonist dihydrotestosterone. Proc Natl Acad Sci U S A 2001; 98: 4904–9.
Pereira de Jesus-Tran K, Cote PL, Cantin L, Blanchet J, Labrie F, Breton R . Comparison of crystal structures of human androgen receptor ligand-binding domain complexed with various agonists reveals molecular determinants responsible for binding affinity. Protein Sci 2006; 15: 987–99.
Schwabe JW . Transcriptional control: how nuclear receptors get turned on. Curr Biol 1996; 6: 372–4.
Li Y, Lambert MH, Xu HE . Activation of nuclear receptors: a perspective from structural genomics. Structure 2003; 11: 741–6.
Bourguet W, Germain P, Gronemeyer H . Nuclear receptor ligand-binding domains: three-dimensional structures, molecular interactions and pharmacological implications. Trends Pharmacol Sci 2000; 21: 381–8.
Wang Z, Benoit G, Liu J, Prasad S, Aarnisalo P, Liu X, et al. Structure and function of Nurr1 identifies a class of ligand-independent nuclear receptors. Nature 2003; 423: 555–60.
Cano LQ, Lavery DN, Bevan CL . Mini-review: Foldosome regulation of androgen receptor action in prostate cancer. Mol Cell Endocrinol 2013; 369: 52–62.
Renaud JP, Rochel N, Ruff M, Vivat V, Chambon P, Gronemeyer H, et al. Crystal structure of the RAR-gamma ligand-binding domain bound to all-trans retinoic acid. Nature 1995; 378: 681–9.
Wagner RL, Apriletti JW, McGrath ME, West BL, Baxter JD, Fletterick RJ . A structural role for hormone in the thyroid hormone receptor. Nature 1995; 378: 690–7.
Nascimento AS, Dias SM, Nunes FM, Aparicio R, Ambrosio AL, Bleicher L, et al. Structural rearrangements in the thyroid hormone receptor hinge domain and their putative role in the receptor function. J Mol Biol 2006; 360: 586–98.
Xu HE, Lambert MH, Montana VG, Plunket KD, Moore LB, Collins JL, et al. Structural determinants of ligand binding selectivity between the peroxisome proliferator-activated receptors. Proc Natl Acad Sci U S A 2001; 98: 13919–24.
Heery DM, Kalkhoven E, Hoare S, Parker MG . A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature 1997; 387: 733–6.
Ogryzko VV, Schiltz RL, Russanova V, Howard BH, Nakatani Y . The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell 1996; 87: 953–9.
Estebanez-Perpina E, Moore JM, Mar E, Delgado-Rodrigues E, Nguyen P, Baxter JD, et al. The molecular mechanisms of coactivator utilization in ligand-dependent transactivation by the androgen receptor. J Biol Chem 2005; 280: 8060–8.
Zhou XE, Suino-Powell KM, Li J, He Y, Mackeigan JP, Melcher K, et al. Identification of SRC3/AIB1 as a preferred coactivator for hormone-activated androgen receptor. J Biol Chem 2010; 285: 9161–71.
Hur E, Pfaff SJ, Payne ES, Gron H, Buehrer BM, Fletterick RJ . Recognition and accommodation at the androgen receptor coactivator binding interface. PLoS Biology 2004; 2: E274.
He B, Minges JT, Lee LW, Wilson EM . The FXXLF motif mediates androgen receptor-specific interactions with coregulators. J Biol Chem 2002; 277: 10226–35.
He B, Gampe RT Jr, Kole AJ, Hnat AT, Stanley TB, An G, et al. Structural basis for androgen receptor interdomain and coactivator interactions suggests a transition in nuclear receptor activation function dominance. Mol Cell 2004; 16: 425–38.
Slagsvold T, Kraus I, Bentzen T, Palvimo J, Saatcioglu F . Mutational analysis of the androgen receptor AF-2 (activation function 2) core domain reveals functional and mechanistic differences of conserved residues compared with other nuclear receptors. Mol Endocrinol 2000; 14: 1603–17.
Estebanez-Perpina E, Arnold LA, Nguyen P, Rodrigues ED, Mar E, Bateman R, et al. A surface on the androgen receptor that allosterically regulates coactivator binding. Proc Natl Acad Sci U S A 2007; 104: 16074–9.
Grosdidier S, Carbo LR, Buzon V, Brooke G, Nguyen P, Baxter JD, et al. Allosteric conversation in the androgen receptor ligand-binding domain surfaces. Mol Endocrinol 2012; 26: 1078–90.
Buzon V, Carbo LR, Estruch SB, Fletterick RJ, Estebanez-Perpina E . A conserved surface on the ligand binding domain of nuclear receptors for allosteric control. Mol Cell Endocrinol 2012; 348: 394–402.
Tyagi RK, Lavrovsky Y, Ahn SC, Song CS, Chatterjee B, Roy AK . Dynamics of intracellular movement and nucleocytoplasmic recycling of the ligand-activated androgen receptor in living cells. Mol Endocrinol 2000; 14: 1162–74.
Saporita AJ, Zhang Q, Navai N, Dincer Z, Hahn J, Cai X, et al. Identification and characterization of a ligand-regulated nuclear export signal in androgen receptor. J Biol Chem 2003; 278: 41998–2005.
Shiau AK, Barstad D, Loria PM, Cheng L, Kushner PJ, Agard DA, et al. The structural basis of estrogen receptor/coactivator recognition and the antagonism of this interaction by tamoxifen. Cell 1998; 95: 927–37.
Kauppi B, Jakob C, Farnegardh M, Yang J, Ahola H, Alarcon M, et al. The three-dimensional structures of antagonistic and agonistic forms of the glucocorticoid receptor ligand-binding domain: RU-486 induces a transconformation that leads to active antagonism. J Biol Chem 2003; 278: 22748–54.
Xu HE, Stanley TB, Montana VG, Lambert MH, Shearer BG, Cobb JE, et al. Structural basis for antagonist-mediated recruitment of nuclear co-repressors by PPARalpha. Nature 2002; 415: 813–7.
Hodgson MC, Shen HC, Hollenberg AN, Balk SP . Structural basis for nuclear receptor corepressor recruitment by antagonist-liganded androgen receptor. Mol Cancer Ther 2008; 7: 3187–94.
Osguthorpe DJ, Hagler AT . Mechanism of androgen receptor antagonism by bicalutamide in the treatment of prostate cancer. Biochemistry 2011; 50: 4105–13.
Kuil CW, Mulder E . Mechanism of antiandrogen action: conformational changes of the receptor. Mol Cell Endocrinol 1994; 102: R1–5.
Zhu P, Baek SH, Bourk EM, Ohgi KA, Garcia-Bassets I, Sanjo H, et al. Macrophage/cancer cell interactions mediate hormone resistance by a nuclear receptor derepression pathway. Cell 2006; 124: 615–29.
Chen CD, Welsbie DS, Tran C, Baek SH, Chen R, Vessella R, et al. Molecular determinants of resistance to antiandrogen therapy. Nat Med 2004; 10: 33–9.
Dotzlaw H, Moehren U, Mink S, Cato AC, Iniguez Lluhi JA, Baniahmad A . The amino terminus of the human AR is target for corepressor action and antihormone agonism. Mol Endocrinol 2002; 16: 661–73.
Quigley CA, De Bellis A, Marschke KB, el-Awady MK, Wilson EM, French FS . Androgen receptor defects: historical, clinical, and molecular perspectives. Endocr Rev 1995; 16: 271–321.
Hughes IA, Davies JD, Bunch TI, Pasterski V, Mastroyannopoulou K, MacDougall J . Androgen insensitivity syndrome. Lancet 2012; 380: 1419–28.
Hiort O . Clinical and molecular aspects of androgen insensitivity. Endocr Dev 2013; 24: 33–40.
McPhaul MJ . Androgen receptor mutations and androgen insensitivity. Mol Cell Endocrinol 2002; 198: 61–7.
Gottlieb B, Beitel LK, Nadarajah A, Paliouras M, Trifiro M . The androgen receptor gene mutations database: 2012 update. Hum Mutat 2012; 33: 887–94.
La Spada AR, Wilson EM, Lubahn DB, Harding AE, Fischbeck KH . Androgen receptor gene mutations in X-linked spinal and bulbar muscular atrophy. Nature 1991; 352: 77–9.
Zajac JD, Fui MN . Kennedy's disease: clinical significance of tandem repeats in the androgen receptor. Adv Exp Med Biol 2012; 769: 153–68.
Finsterer J . Bulbar and spinal muscular atrophy (Kennedy's disease): a review. Eur J Neurol 2009; 16: 556–61.
Lobaccaro JM, Lumbroso S, Belon C, Galtier-Dereure F, Bringer J, Lesimple T, et al. Androgen receptor gene mutation in male breast cancer. Hum Mol Genet 1993; 2: 1799–802.
Goulioumis AK, Varakis J, Goumas P, Papadaki H . Androgen receptor in laryngeal carcinoma: could there be an androgen-refractory tumor? ISRN Oncol 2011; 2011: 180518.
Chen PJ, Yeh SH, Liu WH, Lin CC, Huang HC, Chen CL, et al. Androgen pathway stimulates microRNA-216a transcription to suppress the tumor suppressor in lung cancer-1 gene in early hepatocarcinogenesis. Hepatology 2012; 56: 632–43.
Garolla A, Ferlin A, Vinanzi C, Roverato A, Sotti G, Artibani W, et al. Molecular analysis of the androgen receptor gene in testicular cancer. Endocr Relat Cancer 2005; 12: 645–55.
Siegel R, DeSantis C, Virgo K, Stein K, Mariotto A, Smith T, et al. Cancer treatment and survivorship statistics, 2012. CA Cancer J Clin 2012; 62: 220–41.
Hoffman RM . Clinical practice. Screening for prostate cancer. N Engl J Med 2011; 365: 2013–9.
Mariotto AB, Yabroff KR, Shao Y, Feuer EJ, Brown ML . Projections of the cost of cancer care in the United States: 2010–2020. J Natl Cancer Inst 2011; 103: 117–28.
Silva Neto B, Koff WJ, Biolchi V, Brenner C, Biolo KD, Spritzer PM, et al. Polymorphic CAG and GGC repeat lengths in the androgen receptor gene and prostate cancer risk: analysis of a Brazilian population. Cancer Invest 2008; 26: 74–80.
Schleutker J . Polymorphisms in androgen signaling pathway predisposing to prostate cancer. Mol Cell Endocrinol 2012; 360: 25–37.
Price DK, Chau CH, Till C, Goodman PJ, Baum CE, Ockers SB, et al. Androgen receptor CAG repeat length and association with prostate cancer risk: results from the prostate cancer prevention trial. J Urol 2010; 184: 2297–302.
Denmeade SR, Lin XS, Isaacs JT . Role of programmed (apoptotic) cell death during the progression and therapy for prostate cancer. Prostate 1996; 28: 251–65.
Huggins C . Endocrine-induced regression of cancers. Cancer Res 1967; 27: 1925–30.
Huggins C, Hodges CV . Studies on prostatic cancer: I. The effect of castration, of estrogen and of androgen injection on serum phosphatases in metastatic carcinoma of the prostate. 1941. J Urol 2002; 168: 9–12.
Zeliadt SB, Buist DS, Reid RJ, Grossman DC, Ma J, Etzioni R . Biopsy follow-up of prostate-specific antigen tests. Am J Prev Med 2012; 42: 37–43.
Rubin MA, Maher CA, Chinnaiyan AM . Common gene rearrangements in prostate cancer. J Clin Oncol 2011; 29: 3659–68.
Marx J . Medicine. Fused genes may help explain the origins of prostate cancer. Science 2005; 310: 603.
Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science 2005; 310: 644–8.
Lin C, Yang L, Tanasa B, Hutt K, Ju BG, Ohgi K, et al. Nuclear receptor-induced chromosomal proximity and DNA breaks underlie specific translocations in cancer. Cell 2009; 139: 1069–83.
Haffner MC, Aryee MJ, Toubaji A, Esopi DM, Albadine R, Gurel B, et al. Androgen-induced TOP2B-mediated double-strand breaks and prostate cancer gene rearrangements. Nat Genet 2010; 42: 668–75.
Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell 2010; 18: 11–22.
Palmbos PL, Hussain M . Non-castrate metastatic prostate cancer: have the treatment options changed? Semin Oncol 2013; 40: 337–46.
Van Poppel H, Klotz L . Gonadotropin-releasing hormone: an update review of the antagonists versus agonists. Int J Urol 2012; 19: 594–601.
Crawford ED, Eisenberger MA, Mcleod DG, Spaulding JT, Benson R, Dorr FA, et al. A controlled trial of leuprolide with and without flutamide in prostatic-carcinoma. New Engl J Med 1989; 321: 419–24.
Hellerstedt BA, Pienta KJ . The current state of hormonal therapy for prostate cancer. CA Cancer J Clin 2002; 52: 154–79.
Macfarlane RJ, Chi KN . Research in castration-resistant prostate cancer: what does the future hold? Curr Oncol 2010; 17: S80–6.
Harris WP, Mostaghel EA, Nelson PS, Montgomery B . Androgen deprivation therapy: progress in understanding mechanisms of resistance and optimizing androgen depletion. Nat Clin Pract Urol 2009; 6: 76–85.
Petrylak DP, Tangen CM, Hussain MH, Lara PN Jr, Jones JA, Taplin ME, et al. Docetaxel and estramustine compared with mitoxantrone and prednisone for advanced refractory prostate cancer. N Engl J Med 2004; 351: 1513–20.
Tannock IF, de Wit R, Berry WR, Horti J, Pluzanska A, Chi KN, et al. Docetaxel plus prednisone or mitoxantrone plus prednisone for advanced prostate cancer. N Engl J Med 2004; 351: 1502–12.
Feldman BJ, Feldman D . The development of androgen-independent prostate cancer. Nat Rev Cancer 2001; 1: 34–45.
Nyquist MD, Dehm SM . Interplay between genomic alterations and androgen receptor signaling during prostate cancer development and progression. Horm Cancer 2013; 4: 61–9.
Sharifi N . Mechanisms of androgen receptor activation in castration-resistant prostate cancer. Endocrinology 2013; 154: 4010–7.
Green SM, Mostaghel EA, Nelson PS . Androgen action and metabolism in prostate cancer. Mol Cell Endocrinol 2012; 360: 3–13.
Koivisto P, Kononen J, Palmberg C, Tammela T, Hyytinen E, Isola J, et al. Androgen receptor gene amplification: a possible molecular mechanism for androgen deprivation therapy failure in prostate cancer. Cancer Res 1997; 57: 314–9.
Montgomery RB, Mostaghel EA, Vessella R, Hess DL, Kalhorn TF, Higano CS, et al. Maintenance of intratumoral androgens in metastatic prostate cancer: a mechanism for castration-resistant tumor growth. Cancer Res 2008; 68: 4447–54.
Zhu H, Garcia JA . Targeting the adrenal gland in castration-resistant prostate cancer: a case for orteronel, a selective CYP-17 17,20-lyase inhibitor. Curr Oncol Rep 2013; 15: 105–12.
Mitsiades N, Sung CC, Schultz N, Danila DC, He B, Eedunuri VK, et al. Distinct patterns of dysregulated expression of enzymes involved in androgen synthesis and metabolism in metastatic prostate cancer tumors. Cancer Res 2012; 72: 6142–52.
Buchanan G, Greenberg NM, Scher HI, Harris JM, Marshall VR, Tilley WD . Collocation of androgen receptor gene mutations in prostate cancer. Clin Cancer Res 2001; 7: 1273–81.
Taplin ME, Bubley GJ, Ko YJ, Small EJ, Upton M, Rajeshkumar B, et al. Selection for androgen receptor mutations in prostate cancers treated with androgen antagonist. Cancer Res 1999; 59: 2511–5.
Jenster G . Ligand-independent activation of the androgen receptor in prostate cancer by growth factors and cytokines. J Pathol 2000; 191: 227–8.
Culig Z, Hobisch A, Cronauer MV, Radmayr C, Trapman J, Hittmair A, et al. Androgen receptor activation in prostatic tumor cell lines by insulin-like growth factor-I, keratinocyte growth factor, and epidermal growth factor. Cancer Res 1994; 54: 5474–8.
Mellinghoff IK, Vivanco I, Kwon A, Tran C, Wongvipat J, Sawyers CL . HER2/neu kinase-dependent modulation of androgen receptor function through effects on DNA binding and stability. Cancer Cell 2004; 6: 517–27.
Mahajan NP, Liu Y, Majumder S, Warren MR, Parker CE, Mohler JL, et al. Activated Cdc42-associated kinase Ack1 promotes prostate cancer progression via androgen receptor tyrosine phosphorylation. Proc Natl Acad Sci U S A 2007; 104: 8438–43.
Liu Y, Karaca M, Zhang Z, Gioeli D, Earp HS, Whang YE . Dasatinib inhibits site-specific tyrosine phosphorylation of androgen receptor by Ack1 and Src kinases. Oncogene 2010; 29: 3208–16.
Kraus S, Gioeli D, Vomastek T, Gordon V, Weber MJ . Receptor for activated C kinase 1 (RACK1) and Src regulate the tyrosine phosphorylation and function of the androgen receptor. Cancer Res 2006; 66: 11047–54.
Yang L, Lin C, Jin C, Yang JC, Tanasa B, Li W, et al. LncRNA-dependent mechanisms of androgen-receptor-regulated gene activation programs. Nature 2013; 500: 598–602.
Guo Z, Yang X, Sun F, Jiang R, Linn DE, Chen H, et al. A novel androgen receptor splice variant is up-regulated during prostate cancer progression and promotes androgen depletion-resistant growth. Cancer Res 2009; 69: 2305–13.
Ammirante M, Luo JL, Grivennikov S, Nedospasov S, Karin M . B-cell-derived lymphotoxin promotes castration-resistant prostate cancer. Nature 2010; 464: 302–5.
Setlur SR, Rubin MA . Current thoughts on the role of the androgen receptor and prostate cancer progression. Adv Anat Pathol 2005; 12: 265–70.
Arora VK, Schenkein E, Murali R, Subudhi SK, Wongvipat J, Balbas MD, et al. Glucocorticoid receptor confers resistance to antiandrogens by bypassing androgen receptor blockade. Cell 2013; 155: 1309–22.
Mitsiades N . A road map to comprehensive androgen receptor axis targeting for castration-resistant prostate cancer. Cancer Res 2013; 73: 4599–605.
Steinkamp MP, O'Mahony OA, Brogley M, Rehman H, Lapensee EW, Dhanasekaran S, et al. Treatment-dependent androgen receptor mutations in prostate cancer exploit multiple mechanisms to evade therapy. Cancer Res 2009; 69: 4434–42.
Bishr M, Saad F . Overview of the latest treatments for castration-resistant prostate cancer. Nat Rev Urol 2013; 10: 522–8.
Scher HI, Fizazi K, Saad F, Taplin ME, Sternberg CN, Miller K, et al. Increased survival with enzalutamide in prostate cancer after chemotherapy. N Engl J Med 2012; 367: 1187–97.
Antonarakis ES, Eisenberger MA . Expanding treatment options for metastatic prostate cancer. N Engl J Med 2011; 364: 2055–8.
Tran C, Ouk S, Clegg NJ, Chen Y, Watson PA, Arora V, et al. Development of a second-generation antiandrogen for treatment of advanced prostate cancer. Science 2009; 324: 787–90.
Ryan CJ, Smith MR, de Bono JS, Molina A, Logothetis CJ, de Souza P, et al. Abiraterone in metastatic prostate cancer without previous chemotherapy. N Engl J Med 2013; 368: 138–48.
de Bono JS, Logothetis CJ, Molina A, Fizazi K, North S, Chu L, et al. Abiraterone and increased survival in metastatic prostate cancer. N Engl J Med 2011; 364: 1995–2005.
Veldscholte J, Ris-Stalpers C, Kuiper GG, Jenster G, Berrevoets C, Claassen E, et al. A mutation in the ligand binding domain of the androgen receptor of human LNCaP cells affects steroid binding characteristics and response to anti-androgens. Biochem Biophys Res Commun 1990; 173: 534–40.
Risstalpers C, Verleunmooijman MCT, Trapman J, Brinkmann AO . Threonine on amino-acid position-868 in the human androgen receptor is essential for androgen-binding specificity and functional-activity. Biochem Biophys Res Commun 1993; 196: 173–80.
Zhao XY, Malloy PJ, Krishnan AV, Swami S, Navone NM, Peehl DM, et al. Glucocorticoids can promote androgen-independent growth of prostate cancer cells through a mutated androgen receptor. Nat Med 2000; 6: 703.
Matias PM, Carrondo MA, Coelho R, Thomaz M, Zhao XY, Wegg A, et al. Structural basis for the glucocorticoid response in a mutant human androgen receptor (AR(ccr)) derived from an androgen-independent prostate cancer. J Med Chem 2002; 45: 1439–46.
Culig Z, Hobisch A, Cronauer MV, Cato ACB, Hittmair A, Radmayr C, et al. Mutant androgen receptor detected in an advanced-stage prostatic-carcinoma Is activated by adrenal androgens and progesterone. Mol Endocrinol 1993; 7: 1541–50.
Culig Z, Klocker H, Bartsch G, Hobisch A . Androgen receptors in prostate cancer. Endocr Relat Cancer 2002; 9: 155–70.
Xu J, Wu RC, O'Malley BW . Normal and cancer-related functions of the p160 steroid receptor co-activator (SRC) family. Nat Rev Cancer 2009; 9: 615–30.
Hara T, Miyazaki J, Araki H, Yamaoka M, Kanzaki N, Kusaka M, et al. Novel mutations of androgen receptor: a possible mechanism of bicalutamide withdrawal syndrome. Cancer Res 2003; 63: 149–53.
Bohl CE, Gao W, Miller DD, Bell CE, Dalton JT . Structural basis for antagonism and resistance of bicalutamide in prostate cancer. Proc Natl Acad Sci U S A 2005; 102: 6201–6.
Golshayan AR, Antonarakis ES . Enzalutamide: an evidence-based review of its use in the treatment of prostate cancer. Core Evid 2013; 8: 27–35.
Joseph JD, Lu N, Qian J, Sensintaffar J, Shao G, Brigham D, et al. A clinically relevant androgen receptor mutation confers resistance to second-generation antiandrogens enzalutamide and ARN-509. Cancer Discov 2013; 3: 1020–9.
Balbas MD, Evans MJ, Hosfield DJ, Wongvipat J, Arora VK, Watson PA, et al. Overcoming mutation-based resistance to antiandrogens with rational drug design. Elife 2013; 2: e00499.
Quayle SN, Mawji NR, Wang J, Sadar MD . Androgen receptor decoy molecules block the growth of prostate cancer. Proc Natl Acad Sci U S A 2007; 104: 1331–6.
Andersen RJ, Mawji NR, Wang J, Wang G, Haile S, Myung JK, et al. Regression of castrate-recurrent prostate cancer by a small-molecule inhibitor of the amino-terminus domain of the androgen receptor. Cancer Cell 2010; 17: 535–46.
Myung JK, Banuelos CA, Fernandez JG, Mawji NR, Wang J, Tien AH, et al. An androgen receptor N-terminal domain antagonist for treating prostate cancer. J Clin Invest 2013; 123: 2948–60.
Meimetis LG, Williams DE, Mawji NR, Banuelos CA, Lal AA, Park JJ, et al. Niphatenones, glycerol ethers from the sponge niphates digitalis block androgen receptor transcriptional activity in prostate cancer cells: structure elucidation, synthesis, and biological activity. J Med Chem 2012; 55: 503–14.
Nickols NG, Dervan PB . Suppression of androgen receptor-mediated gene expression by a sequence-specific DNA-binding polyamide. Proc Natl Acad Sci U S A 2007; 104: 10418–23.
Chenoweth DM, Harki DA, Phillips JW, Dose C, Dervan PB . Cyclic pyrrole-imidazole polyamides targeted to the androgen response element. J Am Chem Soc 2009; 131: 7182–8.
Cherian MT, Wilson EM, Shapiro DJ . A competitive inhibitor that reduces recruitment of androgen receptor to androgen-responsive genes. J Biol Chem 2012; 287: 23368–80.
Geistlinger TR, McReynolds AC, Guy RK . Ligand-selective inhibition of the interaction of steroid receptor coactivators and estrogen receptor isoforms. Chem Biol 2004; 11: 273–81.
Chang CY, Abdo J, Hartney T, McDonnell DP . Development of peptide antagonists for the androgen receptor using combinatorial peptide phage display. Mol Endocrinol 2005; 19: 2478–90.
Needham M, Raines S, McPheat J, Stacey C, Ellston J, Hoare S, et al. Differential interaction of steroid hormone receptors with LXXLL motifs in SRC-1a depends on residues flanking the motif. J Steroid Biochem Mol Biol 2000; 72: 35–46.
Ko L, Cardona GR, Iwasaki T, Bramlett KS, Burris TP, Chin WW . Ser-884 adjacent to the LXXLL motif of coactivator TRBP defines selectivity for ERs and TRs. Mol Endocrinol 2002; 16: 128–40.
Heery DM, Hoare S, Hussain S, Parker MG, Sheppard H . Core LXXLL motif sequences in CREB-binding protein, SRC1, and RIP140 define affinity and selectivity for steroid and retinoid receptors. J Biol Chem 2001; 276: 6695–702.
Ren Y, Behre E, Ren Z, Zhang J, Wang Q, Fondell JD . Specific structural motifs determine TRAP220 interactions with nuclear hormone receptors. Mol Cell Biol 2000; 20: 5433–46.
Axerio-cilies P, Lack NA, Nayana MRS, Chan KH, Yeung A, Leblanc E, et al. Inhibitors of androgen receptor activation function-2 (AF2) site identified through virtual screening. J Med Chem 2011; 54: 6197–205.
Lack NA, Axerio-Cilies P, Tavassoli P, Han FQ, Chan KH, Feau C, et al. Targeting the binding function 3 (BF3) site of the human androgen receptor through virtual screening. J Med Chem 2011; 54: 8563–73.
Bisson WH, Cheltsov AV, Bruey-Sedano N, Lin B, Chen J, Goldberger N, et al. Discovery of antiandrogen activity of nonsteroidal scaffolds of marketed drugs. Proc Natl Acad Sci U S A 2007; 104: 11927–32.
Berruti A, Generali D, Tampellini M . Enzalutamide in prostate cancer after chemotherapy. N Engl J Med 2012; 367: 2448.
Foster WR, Car BD, Shi H, Levesque PC, Obermeier MT, Gan J, et al. Drug safety is a barrier to the discovery and development of new androgen receptor antagonists. Prostate 2011; 71: 480–8.
Clegg NJ, Wongvipat J, Joseph JD, Tran C, Ouk S, Dilhas A, et al. ARN-509: a novel antiandrogen for prostate cancer treatment. Cancer Res 2012; 72: 1494–503.
Rathkopf DE, Morris MJ, Fox JJ, Danila DC, Slovin SF, Hager JH, et al. Phase I study of ARN-509, a novel antiandrogen, in the treatment of castration-resistant prostate cancer. J Clin Oncol 2013; 31: 3525–30.
Villoutreix BO, Eudes R, Miteva MA . Structure-based virtual ligand screening: recent success stories. Comb Chem High Throughput Screen 2009; 12: 1000–16.
Song CH, Yang SH, Park E, Cho SH, Gong EY, Khadka DB, et al. Structure-based virtual screening and identification of a novel androgen receptor antagonist. J Biol Chem 2012; 287: 30769–80.
Shen HC, Shanmugasundaram K, Simon NI, Cai C, Wang H, Chen S, et al. In silico discovery of androgen receptor antagonists with activity in castration resistant prostate cancer. Mol Endocrinol 2012; 26: 1836–46.
Liu B, Geng G, Lin R, Ren C, Wu JH . Learning from estrogen receptor antagonism: structure-based identification of novel antiandrogens effective against multiple clinically relevant androgen receptor mutants. Chem Biol Drug Des 2012; 79: 300–12.
Yang SH, Song CH, Van HT, Park E, Khadka DB, Gong EY, et al. SAR based design of nicotinamides as a novel class of androgen receptor antagonists for prostate cancer. J Med Chem 2013; 56: 3414–8.
Voet A, Helsen C, Zhang KY, Claessens F . The discovery of novel human androgen receptor antagonist chemotypes using a combined pharmacophore screening procedure. ChemMedChem 2013; 8: 644–51.
McGinley PL, Koh JT . Circumventing anti-androgen resistance by molecular design. J Am Chem Soc 2007; 129: 3822–3.
Zhou J, Liu B, Geng G, Wu JH . Study of the impact of the T877A mutation on ligand-induced helix-12 positioning of the androgen receptor resulted in design and synthesis of novel antiandrogens. Proteins 2010; 78: 623–37.
Helsen C, Dubois V, Verfaillie A, Young J, Trekels M, Vancraenenbroeck R, et al. Evidence for DNA-binding domain — ligand-binding domain communications in the androgen receptor. Mol Cell Biol 2012; 32: 3033–43.
Boutet S, Lomb L, Williams GJ, Barends TRM, Aquila A, Doak RB, et al. High-resolution protein structure determination by serial femtosecond crystallography. Science 2012; 337: 362–64.
Acknowledgements
This work was supported by the Singapore National Medical Research Council (R-174-000-137-275), the Jay and Betty Van Andel Foundation (H Eric XU and Karsten MELCHER), Amway (China) Limited (H Eric XU), and the National Institutes of Health 5R01DK071662-08 (H Eric XU) and GM102545 and GM104212 (Karsten MELCHER). MH Eileen TAN is supported by a PhD scholarship from the NUS Graduate School for Integrative Sciences & Engineering.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
This work is licensed under the Creative Commons Attribution-NonCommercial-No Derivative Works 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0
About this article
Cite this article
Tan, M., Li, J., Xu, H. et al. Androgen receptor: structure, role in prostate cancer and drug discovery. Acta Pharmacol Sin 36, 3–23 (2015). https://doi.org/10.1038/aps.2014.18
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/aps.2014.18
Keywords
This article is cited by
-
New advances of the androgen receptor in prostate cancer: report from the 1st International Androgen Receptor Symposium
Journal of Translational Medicine (2024)
-
Androgen drives melanoma invasiveness and metastatic spread by inducing tumorigenic fucosylation
Nature Communications (2024)
-
Identification of novel metabolites of abiraterone in human serum and their metabolic pathways
Analytical Sciences (2024)
-
DeepAR: a novel deep learning-based hybrid framework for the interpretable prediction of androgen receptor antagonists
Journal of Cheminformatics (2023)
-
Caprylic Acid (FFA C8:0) promotes the progression of prostate cancer by up-regulating G protein-coupled receptor 84/ Krüppel-like factor 7
BMC Cancer (2023)