Abstract
Aim:
To develop a novel 3D-QSAR approach for study of the epidermal growth factor receptor tyrosine kinase (EGFR TK) and its inhibitors.
Methods:
One hundred thirty nine EGFR TK inhibitors were classified into 3 clusters. Ensemble docking of these inhibitors with 19 EGFR TK crystal structures was performed. Three protein structures that showed the best recognition of each cluster were selected based on the docking results. Then, a novel QSAR (ensemble-QSAR) building method was developed based on the ligand conformations determined by the corresponding protein structures.
Results:
Compared with the 3D-QSAR model, in which the ligand conformations were determined by a single protein structure, ensemble-QSAR exhibited higher R2 (0.87) and Q2 (0.78) values and thus appeared to be a more reliable and better predictive model. Ensemble-QSAR was also able to more accurately describe the interactions between the target and the ligands.
Conclusion:
The novel ensemble-QSAR model built in this study outperforms the traditional 3D-QSAR model in rationality, and provides a good example of selecting suitable protein structures for docking prediction and for building structure-based QSAR using available protein structures.
Similar content being viewed by others
Introduction
The epidermal growth factor receptor (EGFR) is one of the most well-known and validated drug targets for cancer therapy1. It is composed of three structural domains: an extracellular ligand-binding domain, a transmembrane domain, and an intracellular domain. When an agonist binds to its ligand binding site, dimerization of the EGFR is triggered, which activates the intrinsic kinase domain, leading to autophosphorylation on specific tyrosine residues in the C-terminus2,3. Subsequently, signal transduction cascades are initiated that promote DNA synthesis and cell proliferation. Hence, the EGFR is a key regulator for cell migration, adhesion, proliferation, and immune responses in several cell phenotypes in the human skin2,4.
Overexpression or hyperactivity of the EGFR is evidently associated with different types of cancers5. In fact, at least three human epithelial cancers, including lung cancer, anal cancer and glioblastoma multiforme, are marked by functional overactivation of EGFR. Thus, the EGFR has turned out to be an effective therapeutic target for these epithelial cancers6. Due to the development of some specific antagonists targeting the EGFR, especially when the activated ErbB pathway is the underlying cause of a given malignancy5, cancer patients have greatly benefited from such targeted therapy. Clinically, two classes of EGFR antagonists, anti-EGFR monoclonal antibodies and small-molecule EGFR tyrosine kinase inhibitors (TKIs), are currently being used. In this study, we focused on small-molecule EGFR TKIs, such as Erlotinib7 and Gefitinib8, which reversibly compete with ATP through binding to the EGFR tyrosine kinase (TK) and thus inhibit the EGFR autophosphorylation and downstream signaling1,9.
Most EGFR TKIs are quinazoline-derivatives with diverse functional groups on the quinazoline ring. After carefully analyzing the differences of the side chains on the quinazoline ring among EGFR TKIs in the BindingDB10, we found that most of the reversibly binding ligands can be roughly classified into three clusters (Figure 1): inhibitors possessing the dimethoxyethane side chain, for example, Erlotinib and Lapatinib; inhibitors containing a similar long side chain on the quinazoline ring and ending with a sulfonyl group; and several quinazoline-derivatives with a relatively small side chain and lower molecular weight.
Representative molecules in each cluster. Erlotinib represents the molecules with two 2-methoxyethoxy on the side chain in cluster A; lapatinib represents the molecules in cluster B which consist of a long side chain on the quinazoline ring and with a sulfonyl group in the tail; N4-(3-bromophenyl)quinazoline-4,6-diamine represents molecules in cluster C whose side chain on the quinazoline ring is small.
Along with the development of EGFR TKIs, several crystallographic structures of the EGFR TK were also resolved. These known inhibitors and crystal structures have greatly facilitated our development of novel inhibitors for cancer treatment. The application of computer-aided drug design methods, such as docking11,12,13,14,15 and Quantitative Structure-Activity Relationship (QSAR) models16,17,18,19, further promotes the development of such novel inhibitors. In particular, by combining docking and QSAR predictions, 3D-QSAR models based on the alignment from the docking results have been found to be advantageous and offer wide coverage20.
Most virtual screening studies are hampered by the selection of the appropriate docking programs or scoring functions based on a single crystal structure21. However, proteins are dynamically flexible macromolecules, and different structures of a protein may show different recognition abilities towards even the same ligand. Although ensemble docking has shown some advantages in improving the enrichment factor or diversity22,23, to our knowledge, all docking-based 3D-QSAR studies still use a single crystal structure for ligand pose prediction17,24,25. However, the recognition ability of a protein structure towards inhibitors with different chemical types is rarely explored. Such studies raise two questions regarding (i) the recognition ability of a single EGFR TK crystal structure for inhibitors of different chemical types and (ii) the selection of an appropriate protein structure for the pose prediction of ligands of different chemical types. Thus, those 3D-QSAR models based on a single protein structure might be unreliable to some degree.
In fact, structure-based 3D-QSAR models are not limited to the ligand alignment from just a single protein structure. We can build a type of 3D-QSAR model based on the ligand alignment from multiple protein structures (Herein we refer to such 3D-QSAR models as ensemble-QSAR models). To investigate the rationality of ensemble-QSAR models, we performed an ensemble docking study on multiple EGFR crystal structures. Based on the docking results, an ensemble-QSAR model, which uses the ligand conformations predicted from multiple protein structures, was explored to facilitate the development of novel inhibitors.
Methods
Protein preparation
A total of 19 EGFR TK structures that were co-crystallized with an inhibitor, including 9 wild-type and 10 mutant strains, were retrieved from the RCSB Protein Data Bank (PDB)26. Of these structures, 7 were in the inactive form, and 12 were in the active form. After aligning all structures to 1XKK, the greatest difference was found in the activation loop. A small helix in the activation loop was broken down when the EGFR TK changed from the inactive to the active form. In the meantime, displacement of the C-helix also appeared during the conformational change from the inactive form to the active form. The alignment also revealed that the N-lobe is a flexible structural unit while the C-lope is rather stable (Figure S1). With these structures, the mutated residues in the ATP site changed back to their wild-type structures with Maestro GUI27. Multiple chains in a crystal structure were separated into individual chains. During the protein structure preparation, the presence or absence of the conserved water (CW) molecule (acting as the water bridge between the hydroxyl group of THR790 and 3-N of the quinazoline moiety) was considered. With all of the above situations taken into account, a total of 52 EGFR TK structures were generated (Table 1).
The Protein Preparation Wizard from Schrodinger was used to prepare all these structures. Hydrogen atoms were added to the protein structures corresponding to a pH value of 7. If necessary, the side chain structures of Gln and Asn were flipped to maximize the hydrogen bond interactions. All-atom constrained energy minimizations were carried out using the Impact Refinement module28 with the inter-atomic interactions modeled by the OPLS-2005 force field29, which alleviated the obvious steric clashes in the original PDB structures. An energy minimization was terminated when the Root Mean Square Deviation (RMSD) was larger than 0.30 Å.
Ligand preparation
Most of the EGFR TKIs used in clinical research belong to quinazoline-derivatives. In this work, they were obtained from BindingDB, a database containing hundreds of compounds with IC50 values towards EGFR TK10. We removed all covalently binding ligands because the reaction groups in these ligands can provide biased information to the QSAR model. The FCFP-4 fingerprint was utilized to classify the inhibitors into the three clusters as mentioned in the introduction section. According to the general principle of structural diversity and representation, we manually selected 139 EGFR TKIs, which are classified into cluster A, cluster B and cluster C, including 56, 35, and 48 molecules, respectively. Even though compounds in the same cluster share a similar quinazoline–based scaffold, their side chains reflect their diversity. The inhibitory activities of these compounds, which were tested in substrate phosphorylation assays, span approximately five orders of magnitude, with IC50 values ranging from 0.1–10 μmol/L. The structure, ID number, activity value, and cluster information for the 139 ligands were provided in Figure S2.
Selection of inactive compounds (decoys) is of critical importance for the evaluation of virtual screening protocols. Here, we selected the Directory of Universal Decoys (DUD)30 as the source of decoys. In the DUD, there are a total of 15996 decoys that are believed to be non-binders of EGFR TK. Due to the time-consuming nature of docking studies with all decoys, we condensed the database of molecules containing the FCFP-4 fingerprint to a set of 3800 structurally diverse decoys. The annotated EGFR inhibitors were then seeded into these 3800 decoys to form the final validation set.
All small molecules (139 ligands and 3800 decoys) were prepared using LigPrep31 by generating low energy ionization and tautomeric states within the pH range of 7.0±2.0. They were further minimized using the OPLS-2005 force field, leading to a set of initial docking conformations.
Docking simulation
Flexible docking was performed using the standard precision (SP) mode in Glide32 with the default settings. Glide uses a grid file to describe the profile of a binding pocket, with the center of the grids defined by the co-crystallized ligand. Binding sites of all the prepared PDB structures (within 5 Å of the bound ligand) were superimposed onto the 1XKK reference structure with the Protein Structure Alignment tool in Maestro to allow direct QSAR building with the docked poses of the ligands. Moreover, to assess the docking accuracy, self-docking studies were carried out with the rearranged coordinates of the co-crystallized ligands.
Analysis of the docking results
The top-ranked poses, based on GlideScore, were used to analyze the docking results. Self-docking calculations were evaluated by the RMSD between the docked pose and the co-crystallized pose. Receiver operating characteristic (ROC)33 curves were utilized to evaluate the docking performance for virtual screening. The ROC plot was constructed by stepping sequentially through the list of the test-set compounds ranked in order of increasing GlideScore. At each point of a ROC curve, the true positive rate (TPR) and false positive rate (FPR) were collected to characterize the virtual screening performance. Much attention should be paid to the early performance of the ROC curve, as it would facilitate the evaluation of a virtual screening protocol.
Ligand similarity
Discovery Studio 2.1 was used to generate the FCFP-4 fingerprints for all the molecules. Thereafter, the Tanimoto coefficient was calculated to quantify the similarity:

where a and b are the numbers of bits in their respective molecules and c is the number of bits common to both molecules.
3D-QSAR model building
3D-QSAR models were built using PHASE34,35. Reliable ligand conformation generation is essential for constructing a robust 3D-QSAR model. To incorporate the information from both ligands and receptors, we used the docking–guided method for ligand alignment. Nevertheless, the ensemble docking results indicated that different protein structure possessed different abilities in recognizing ligands in different clusters, which means that a specific protein structure usually exhibits good recognition ability toward ligands in one or two clusters. In this work, we combined the ligand conformations regenerated by constraint docking studies from their respective most favorable protein structures to improve the pose accuracy (Table S2). Because the residues within 5 Å of the binding pocket were aligned before grid generation, docking poses from different structures could be collected easily for the ensemble-QSAR model building. Of the 139 inhibitors mentioned above, 109 inhibitors were selected as the training set based on the usual guidelines, with the remaining 30 compounds used as a test set.
Results
Self docking
The first step of our study was focused on the evaluation of the Glide self-docking towards EGFR TK. The performances of some known docking programs with the kinase have been evaluated by La Motta et al17. However, the Glide program was not covered in their study. As Glide is a popular docking method that has performed well in numerous evaluations36,37,38,39, in this study self-docking was carried out on the 52 TK structures using the Glide SP mode. Figure 2 demonstrated the self-docking results for all complexes. To facilitate the evaluation, a ligand binding pose was considered to be successfully predicted when the RMSD between the best-scored and co-crystallized poses was less than 2.50 Å. As shown in Figure 2, 38 of the 52 structures were successfully reproduced. For the 52 self-docking structures, an average RMSD value of 2.20 Å was obtained, which was much smaller than 2.70 Å obtained by La Motta et al, reflecting the reliability of Glide towards the EGFR TK. Considering its remarkable performance, Glide SP docking was used for the subsequent docking studies.
Root-Mean-Square-Deviation (RMSD) of each co-crystallized ligand obtained from the Glide SP self-docking.
Simulated virtual screening
2J6M_W demonstrated the highest TPR1% value (the TPR value when the FPR value is 1%) of 0.764 for cluster A (Figure 3A). It also appeared to be an acceptable structure for recognizing molecules in clusters B and C, with TPR1% values of 0.371 and 0.306, respectively. The area under the curve (AUC) was used as another indicator to evaluate the docking performance. Accordingly, large AUC values of 0.957, 0.909, and 0.949 were obtained for clusters A, B, and C, respectively.
The ROC curves of ensemble docking. The curves for clusters A, B, C, and all the ligands are colored in red, dark cyan, blue, and black, respectively. (A) The ROC curves of 2J6M_W. (B) The ROC curves of 1XKK. (C) The ROC curves of 2JIU_AW.
Both 1XKK and 1XKK_W demonstrated even better ability to recognize inhibitors in cluster B, with TPR1% values larger than 0.900. We can see a sharp increase at the beginning of the 1XKK ROC curve for cluster B from Figure 3B, which leads to a high AUC value of 0.987. However, 1XKK performed relatively poorly towards the ligands in clusters A and C, which represented the specificity of 1XKK towards cluster B. 2JIU_AW exhibited the best TPR1% value of 0.531 toward the ligands in cluster C. In addition, a remarkable TPR1% value of 0.540 for all the 139 inhibitors was also obtained with this structure, which is also the highest TPR1% value among all structures tested. The good recognition ability of 2JIU_AW toward all ligands was represented by the ROC curves in Figure 3C. Although the TPR1% values for clusters A and B generated from 2JIU_AW are not the best among the 52 structures, they are still acceptable for virtual screening calculations. Therefore, if limited computational resources are available or the ligands' chemotype is unclear, we suggest using this structure for a virtual screening study.
The other structures used in our study did not show representative results for recognizing ligands in the three clusters. All ROC curves obtained for the structures used in this work and TPR1% were provided in Figure S3 and Table S1.
3D-QSAR study
Two 3D-QSAR models with different ligand alignment methods were built in this study. The first model was constructed with the ligand conformations determined by the structure 2JIU_AW, which had the highest TPR1% towards all 139 active molecules; the second model was an ensemble-QSAR, in which the ligand poses were determined by the optimally selected protein structures. 2J6M_W, 1XKK, and 2JIU_AW, which exhibited the highest TPR1% values for compounds in clusters A, B, and C, respectively, were selected and used to determine the ligand conformations in the corresponding clusters.
Figure 4 shows the essential differences of the docking poses of the 139 compounds through different docking alignment approaches. Figure 4A shows the docking alignment of all compounds from a single structure, 2JIU_AW. Although this structure is superior to all other structures on TPR1%, it cannot ensure the rationality of the binding poses for all 139 chemicals. As shown in Figure 4A, the quinazoline rings of the compounds show obvious fluctuations around the hinge part of the kinase, where a stable hydrogen-bond interaction should be formed with the main chain amide of MET79340. Moreover, the methylsulfonyl group of some inhibitors is not positioned correctly. Thus, 2JIU_AW cannot provide reliable conformations for all 139 ligands for further 3D QSAR building. By contrast, the poses generated by ensemble docking appear to be more reasonable, with the quinazoline rings and the methylsulfonyl groups being more highly ordered (Figure 4B). Therefore, by combining the ligand poses in each cluster generated by 2J6M_W, 1XKK, and 2JIU_AW, the alignment becomes more reasonable.
Docking-guided superimposition of the training and test set compounds determined by a single kinase structure (A) and the optimally selected multiple structures (B).
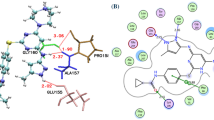
Two statistically significant 3D-QSAR models were constructed based on different ligand conformation alignments. Figure 5 shows the correlations between the experimental and predicted activities for the compounds in the training and test sets. For the single structure-based QSAR model, the correlation coefficients based on the training (R2) and test compounds (Q2) were 0.83 and 0.67 (factor 4), respectively. For the ensemble-QSAR model, values of 0.87 and 0.78 (factor 5) were obtained for R2 and Q2, respectively, indicating a clearly improved predictability. In addition to improving the statistical indicators, the ensemble-QSAR model has the potential to provide more reasonable biophysical knowledge because it was built based on enhanced docking poses. For example, the binding pose of compd-117 shows a more favorable interaction pattern in the ensemble-QSAR model than in the single structure-based QSAR model. As shown in Figure 6, the pose of the compound from the ensemble docking (colored in white) shows that it forms a hydrogen bond with the backbone of CYS797, and the iodine group experiences a favorable hydrophobic contact with the surrounding residues. These interactions were not found from the pose generated by the single structure 2JIU_AW (colored in yellow).
Experimental and predicted activities of the training (blue) and test (red) set compounds associated with the single structure-guided QSAR model (A) and the ensemble-QSAR model (B).
Comparison of the docking poses extracted from different kinase structures. The pose of compd-117 in white is from 2JIU_AW, and an alternative pose in yellow is from 2J6M_W. The figure was prepared with PyMol.
By analyzing the QSAR features, which include hydrogen bond donors, hydrophobic/nonpolar interactions, and electron-withdrawing property, significant advantages can be obtained from the ensemble 3D-QSAR. The two most important features, the hydrophobic (filed fraction: 0.55) and electron-withdrawing (filed fraction: 0.27) properties, of this QSAR model with two representative compounds are visualized in Figure 7. Figure 7A and 7B demonstrate the hydrophobic feature of the QSAR with the most potent inhibitor, compd-117, and the least active inhibitor, compd-94, respectively. Here, the green cubes indicate favorable hydrophobic regions that are helpful for improving the activity of an inhibitor, while unfavorable areas are shown in purple. Obviously, the green cubes match well with the hydrophobic regions of the protein (represented by yellow meshes). The iodine and chlorine groups of compd-117 occupy the two large green cubes. By contrast, the least active compound, compd-94, does not occupy the two large green cubes, and its methyl group on methylquinazoline is located in the unfavorable region. A similar case has been found for the electron-withdrawing features of the QSAR as well. As depicted in Figure 7C and Figure 7D, the carbonyl group of compd-117 is located in the favorable region (color in pale red), whereas compd-94 lacks the important electron-withdrawing feature.
Atom-based ensemble-QSAR model visualized with the most (compd-117 on the left) and least active (compd-94 on the right) compounds in the training set. (A) and (B) show the hydrophobic effect of the QSAR model associated with the most and the least active compounds, respectively. The green cubes indicate the favorable area and the purple cubes represent the unfavorable area. (C) and (D) demonstrate the electron-withdrawing effect of the QSAR model associated with the most and the least active compounds, respectively. The pale red cubes represent the favorable area and light green cubes indicate the unfavorable area.
Discussion
With many successful cases of virtual screening, molecular docking is becoming an increasingly popular tool for generating hit compounds in drug discovery41. However, several challenging issues remain, such as water-mediated protein-ligand interactions42 and protein flexibility43. Currently, docking-guided QSAR models are widely used for lead optimization, whereas protein flexibility has not been taken into account in most studies44. In this work, an ensemble docking-guided 3D-QSAR approach is proposed for the EGFR TK, in an effort to avoid the disadvantages of the single structure-guided QSAR models.
Ligand selectivity of the protein structures
According to the docking results, we can observe that a protein structure exhibits different capabilities in recognizing ligands of different chemical types (Figure S3), reflecting the fact that docking performances are sensitive to protein-ligand interactions and/or protein flexibility. Consequently, the selection of appropriate protein structures is essential when several protein structures are available during the virtual screening. For EGFR, the structures 2J6M_W, 1XKK, and 2JIU_AW performed best for the molecules in clusters A, B, and C, respectively. Therefore, if a virtual screening task is initiated, the full use of the three protein structures would be particularly helpful for enriching the active compounds.
When a docking-guided QSAR model is initiated, the ligand's alignment is crucial for generating a reliable QSAR model. From our docking results, we can see that a single protein crystal structure shows selectivity towards ligands in different clusters. Therefore, the predictability of the QSAR models based on a single crystal structure might be low. However, if multiple crystal structures are included and the most reasonable pose for each ligand is selected for building an ensemble-QSAR model, we can obtain a more reasonable QSAR model.
Conserved water molecule
One CW molecule appears in 13 of the 19 EGFR TK crystal structures. This CW molecule bridges the interactions between THR790 and the 3-N atom of quinazoline. Wissner et al tried to replace the water molecule with a 3-cyano group, but they found that the potency was not improved by this substitution45. In our docking calculations, the highest TPR1%All, TPRA1%, and TPRC1% values were obtained with the structures in the presence of the water molecule. For the inhibitors in cluster B, both 1XKK and 1XKK_W performed well during the docking study, with TPRB1% values of 0.971 and 0.943, respectively, indicating that the effect of the water molecule was not obvious in the docking of cluster B ligands. To further analyze the importance of this CW, we built a histogram and analyzed its function in the 13 crystal structures. As shown in Figure 8, when this CW was considered, the averaged TPR1% value increased in 11 of the 13 crystal structures. Therefore, we suggest that this water molecule should be retained during docking simulations if the ligands are not designed to replace it.
TPR1% values with and without the conserved water molecule in the 13 crystallography structures. The TPR1% values with this water taken into account are shown in red, while TPR1% values without the water are shown in black.
Ligand similarity
Based on the FCFP-4 fingerprint, we calculated the Tanimoto similarities between compounds in different clusters and co-crystallized ligands. The average similarity values and averaged TPR1% values for each crystal structure are shown in Table 2. This result shows that the ligands in 1XKK were similar to the molecules in cluster B with a similarity value of 0.73, and the highest average TPR1% value for cluster B was obtained with this protein crystal structure. This finding implies a high probability of obtaining an active ligand in a virtual screening when a binding pocket is shaped by a similar co-crystallized ligand. However, the docking performance is not merely determined by the ligand similarity, as exemplified by the results for compounds in cluster A. Though the co-crystallized ligand in 2ITZ exhibits a high similarity to cluster A ligands with a value of 0.65, a lower TPRA1% value is obtained, indicating the existence of some other factors affecting the docking performance. According to our study, the co-crystallized ligands in 2J6M (2J6M_W) and 2JIU (2JIU_AW) are not similar to the docked molecules in clusters A and C, respectively, but the highest TPR1% values were obtained for these clusters (Figure 3). A previously published paper showed that docking accuracy is related to ligand similarity, and higher similarity between the docked molecules and the co-crystallized ligand always leads to better docking accuracy46,47. We only obtain this type of correlation in our virtual screening study for the ligands in cluster B. As for the cluster A and C ligands, ligand similarity does not appear to work. We attribute this phenomenon to the smaller size of the ligands in clusters A and C than those in cluster B, and a similar ligand is not a prerequisite for shaping the binding pocket. Therefore, we can conclude that similarity can facilitate the virtual screening of the EGFR TK in limited cases, and careful evaluation of all available structures can be a prudent way of selecting target structures.
QSAR implications for designing new EGFR inhibitors
Currently, when the target protein structure is available, docking-guided 3D-QSARs are widely employed to detect the SARs of interested compounds and to obtain some clues for activity improvement. This type of alignment method not only reduces the computational resources used for ligand conformation sampling, but also fully utilizes the interaction information between ligands and the protein structures. It is also worth acknowledging that poor docking score performance when evaluating the activity of a series compounds could be addressed by the local QSAR model. However, for the proteins with plastic binding sites, the selection of a single structure seems to be questionable. Our study suggested a practical and rigorous approach to handle this problem. The enrichment factor and fingerprint-based clustering were used to optimally select a small number of representative protein structures, which are essentially important for the generation of reasonable conformations of diverse ligands.
As depicted in Figure 9 with compd-117 as a reference molecule, the combined effect of all five features from the QSAR model provides some clues for further modification of the inhibitors. The cubes from the ensemble-QSAR model directly relate the favorable (shown in blue) and unfavorable (shown in red) regions to the corresponding detailed protein-ligand interactions in the ATP binding site. The basic concept is that the designed compounds should occupy the blue cubes as much as possible while avoiding touching the red areas. At the top of the picture, a favorable hydrophobic region (horizontal arrow) indicates that the substitution with a small hydrophobic group, such as a methyl or halogen group, would improve the activity. Likewise, some modifications of the quinazoline (skew arrow) might contribute to the potency. In this favorable region, if a small hydrophobic group is tested, it might have some favorable interactions because of its hydrophobic contacts with the backbone of the P-loop and hinge-loop, while modification with a hydrogen bond donor group might form a hydrogen bond with the backbone carbonyl of MET793.
Combined effects of the ensemble 3D-QSAR visualized with the inhibitor Compd-22. The favorable and unfavorable regions are presented in blue and red cubes, respectively.
Conclusion
The identification capability of different EGFR TK crystal structures towards inhibitors of different chemical types has been comprehensively evaluated. These crystal structures exhibited different capabilities in binding pose prediction for inhibitors with different chemical types. According to the TPR1% values, we found that 2J6M_W, 1XKK, and 2JIU_AW performed best towards the active compounds of clusters A, B, and C, respectively. Based on the docking performances of the protein structures, we developed a practical method for building reliable QSAR models, the ensemble–QSAR model. This model can fully use the optimally selected structures relevant to different ligand clusters to improve the ligand alignment, and our study has shown that the model possess a remarkable predictability. We believe that the method developed in this work can also be applied to other targets when several crystal structures are available, though our study was only based on the EGFR TK system. Ensemble-QSAR thus provides a powerful tool for virtual screening and lead optimization.
Author contribution
Yun TANG, Yao-quan TU, and Xian-qiang SUN designed the research and revised the manuscript; Xian-qiang SUN, Lei CHEN, and Yao-zong LI conducted the research and wrote the paper; Wei-hua LI and Gui–xia LIU analyzed data.
References
Ciardiello F, Tortora G . EGFR antagonists in cancer treatment. N Engl J Med 2008; 358: 1160–74.
Citri A, Yarden Y . EGF-ERBB signalling: towards the systems level. Nat Rev Mol Cell Biol 2006; 7: 505–16.
Downward J, Parker P, Waterfield MD . Autophosphorylation sites on the epidermal growth factor receptor. Nature 1984; 311: 483–5.
Oda K, Matsuoka Y, Funahashi A, Kitano H . A comprehensive pathway map of epidermal growth factor receptor signaling. Mol Syst Biol 2005; 1: E11–E17.
Zhang H, Berezov A, Wang Q, Zhang G, Drebin J, Murali R, et al. ErbB receptors: from oncogenes to targeted cancer therapies. J Clin Invest 2007; 117: 2051–8.
Pao W, Chmielecki J . Rational, biologically based treatment of EGFR-mutant non-small-cell lung cancer. Nat Rev Cancer 2010; 10: 760–74.
Moyer JD, Barbacci EG, Iwata KK, Arnold L, Boman B, Cunningham A, et al. Induction of apoptosis and cell cycle arrest by CP-358,774, an inhibitor of epidermal growth factor receptor tyrosine kinase. Cancer Res 1997; 57: 4838–48.
Barker AJ, Gibson KH, Grundy W, Godfrey AA, Barlow JJ, Healy MP, et al. Studies leading to the identification of ZD1839 (IRESSA): an orally active, selective epidermal growth factor receptor tyrosine kinase inhibitor targeted to the treatment of cancer. Bioorg Med Chem Lett 2001; 11: 1911–4.
Sorscher SM . EGFR mutations and sensitivity to gefitinib. N Engl J Med 2004; 351: 1260–1; author reply 60–1.
Liu T, Lin Y, Wen X, Jorissen RN, Gilson MK . BindingDB: a web-accessible database of experimentally determined protein–ligand binding affinities. Nucleic Acids Res 2007; 35: D198–201.
Aparna V, Rambabu G, Panigrahi SK, Sarma JARP, Desiraju GR . Virtual screening of 4-anilinoquinazoline analogues as EGFR kinase inhibitors: Importance of hydrogen bonds in the evaluation of poses and scoring functions. J Chem Inf Model 2005; 45: 725–38.
Gundla R, Kazemi R, Sanam R, Muttineni R, Sarma JA, Dayam R, et al. Discovery of novel small-molecule inhibitors of human epidermal growth factor receptor-2: combined ligand and target-based approach. J Med Chem 2008; 51: 3367–77.
Li S, Sun X, Zhao H, Tang Y, Lan M . Discovery of novel EGFR tyrosine kinase inhibitors by structure-based virtual screening. Bioorg Med Chem Lett 2012; 22: 4004–9.
Chen Z, Li HL, Zhang QJ, Bao XG, Yu KQ, Luo XM, et al. Pharmacophore-based virtual screening versus docking-based virtual screening: a benchmark comparison against eight targets. Acta Pharmacol Sin 2009; 30: 1694–708.
Li J, Du Y, Liu X, Shen QC, Huang Al, Zheng MY, et al. Binding sensitivity of adefovir to the polymerase from different genotypes of HBV: molecular modeling, docking and dynamics simulation studies. Acta Pharmacol Sin 2013; 34: 319–28.
Pasha FA, Muddassar M, Srivastava AK, Cho SJ . In silico QSAR studies of anilinoquinolines as EGFR inhibitors. J Mol Model 2010; 16: 263–77.
La Motta C, Sartini S, Tuccinardi T, Nerini E, Da Settimo F, Martinelli A . Computational studies of epidermal growth factor receptor: docking reliability, three-dimensional quantitative structure-activity relationship analysis, and virtual screening studies. J Med Chem 2009; 52: 964–75.
Peng T, Pei J, Zhou J . 3D-QSAR and receptor modeling of tyrosine kinase inhibitors with flexible atom receptor model (FLARM). J Chem Inf Comput Sci 2003; 43: 298–303.
Cox R, Green DS, Luscombe C, Malcolm N, Pickett S . QSAR workbench: automating QSAR modeling to drive compound design. J Comput Aid Mol Des 2013; 27: 321–36.
Tuccinardi T, Ortore G, Santos MA, Marques SM, Nuti E, Rossello A, et al. Multitemplate alignment method for the development of a reliable 3D-QSAR model for the analysis of MMP3 inhibitors. J Chem Inf Model 2009; 49: 1715–24.
Huang SY, Grinter SZ, Zou XQ . Scoring functions and their evaluation methods for protein-ligand docking: recent advances and future directions. Phys Chem Chem Phys 2010; 12: 12899–908.
Osguthorpe DJ, Sherman W, Hagler AT . Exploring protein flexibility: incorporating structural ensembles from crystal structures and simulation into virtual screening protocols. J Phys Chem B 2012; 116: 6952–9.
Osguthorpe DJ, Sherman W, Hagler AT . Generation of receptor structural ensembles for virtual screening using binding site shape analysis and clustering. Chem Biol Drug Des 2012; 80: 182–93.
Lan P, Xie MQ, Yao YM, Chen WN, Chen WM . 3D-QSAR studies and molecular docking on [5-(4-amino-1H-benzoimidazol-2-yl)-furan-2-yl]-phosphonic acid derivatives as fructose-1,6-biphophatase inhibitors. J Comput Aid Mol Des 2010; 24: 993–1008.
Mouchlis VD, Mavromoustakos TM, Kokotos G . Molecular docking and 3D-QSAR CoMFA studies on indole inhibitors of GIIA secreted phospholipase A2. J Chem Inf Model 2010; 50: 1589–601.
Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, et al. The Protein Data Bank. Nucleic Acids Res 2000; 28: 235–42.
Maestro. Schordinger, LLC,New York, NY 2008.
Impact. Schrödinger, LLC, New York, NY, 2005.
Jorgensen WL, Maxwell DS, Tirado-Rives J . Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. J Am Chem Soc 1996; 118: 11225–36.
Huang N, Shoichet BK, Irwin JJ . Benchmarking sets for molecular docking. J Med Chem 2006; 49: 6789–801.
Ligprep. Schordinger, LLC,New York, NY 2008.
Friesner RA, Banks JL, Murphy RB, Halgren TA, Klicic JJ, Mainz DT, et al. Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J Med Chem 2004; 47: 1739–49.
Fawcett T . An introduction to ROC analysis. Pattern Recogn Lett 2006; 27: 861–74.
Dixon SL, Smondyrev AM, Knoll EH, Rao SN, Shaw DE, Friesner RA . PHASE: a new engine for pharmacophore perception, 3D QSAR model development, and 3D database screening: 1. Methodology and preliminary results. J Comput Aided Mol Des 2006; 20: 647–71.
Dixon SL, Smondyrev AM, Rao SN . PHASE: a novel approach to pharmacophore modeling and 3D database searching. Chem Biol Drug Des 2006; 67: 370–2.
Li X, Li Y, Cheng T, Liu Z, Wang R . Evaluation of the performance of four molecular docking programs on a diverse set of protein-ligand complexes. J Comput Chem 2010; 31: 2109–25.
Li Y, Shen J, Sun X, Li W, Liu G, Tang Y . Accuracy assessment of protein-based docking programs against RNA targets. J Chem Inf Model 2010; 50: 1134–46.
Sun X, Li Y, Li W, Xu Z, Tang Y . Computational investigation of interactions between human H2 receptor and its agonists. J Mol Graph Model 2011; 29: 693–701.
Cheng T, Li X, Li Y, Liu Z, Wang R . Comparative assessment of scoring functions on a diverse test set. J Chem Inf Model 2009; 49: 1079–93.
Yun CH, Boggon TJ, Li Y, Woo MS, Greulich H, Meyerson M, et al. Structures of lung cancer-derived EGFR mutants and inhibitor complexes: mechanism of activation and insights into differential inhibitor sensitivity. Cancer Cell 2007; 11: 217–27.
McInnes C . Virtual screening strategies in drug discovery. Curr Opin Chem Biol 2007; 11: 494–502.
Huang N, Shoichet BK . Exploiting ordered waters in molecular docking. J Med Chem 2008; 51: 4862–5.
B-Rao C, Subramanian J, Sharma SD . Managing protein flexibility in docking and its applications. Drug Discov Today 2009; 14: 394–400.
Tropsha A . Best practices for QSAR model development, Validation, and exploitation. Mol Inf 2010; 29: 476–88.
Wissner A, Overbeek E, Reich MF, Floyd MB, Johnson BD, Mamuya N, et al. Synthesis and structure-activity relationships of 6,7-disubstituted 4-anilinoquinoline-3-carbonitriles. The design of an orally active, irreversible inhibitor of the tyrosine kinase activity of the epidermal growth factor receptor (EGFR) and the human epidermal growth factor receptor-2 (HER-2). J Med Chem 2003; 46: 49–63.
Verdonk ML, Mortenson PN, Hall RJ, Hartshorn MJ, Murray CW . Protein-ligand docking against non-native protein conformers. J Chem Inf Model 2008; 48: 2214–25.
Tuccinardi T, Botta M, Giordano A, Martinelli A . Protein kinases: docking and homology modeling reliability. J Chem Inf Model 2010; 50: 1432–41.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant 21072059), the 863 Project (Grant 2012AA020308), the Fundamental Research Funds for the Central Universities (WY1113007) and the Shanghai Committee of Science and Technology (11DZ2260600). Xian-qiang Sun would like to thank China Scholarship Council for supporting his study in Sweden.
Author information
Authors and Affiliations
Corresponding authors
Additional information
The supplementary information was available on the APS website. Table S1 shows the TPR1% values of all protein structures in the virtual screen. Table S2 shows the constraints used for the docking to regenerate the docking poses. Figure S1 shows the alignment of the 19 EGFR structures to 1XKK. The data set for the 139 EGFR TK inhibitors is also provided in the supplementary information (Figure S2). Figure S3 shows the ROCs for all the structures used in our research.
Supplementary information
Figure S1
Alignment of all the 19 EGFR TK crystallography structures used in this work to 1XKK (DOCX 662 kb)
Figure S3
The ROC curves of ensemble docking. (JPG 1105 kb)
Table S1
TPR1% values of all protein structures in virtual screening. New Codes were represented with the same method as Table 1. (DOCX 20 kb)
Table S2
Constraint used for the docking to regenerated the docking poses. (DOCX 11 kb)
Rights and permissions
About this article
Cite this article
Sun, Xq., Chen, L., Li, Yz. et al. Structure-based ensemble-QSAR model: a novel approach to the study of the EGFR tyrosine kinase and its inhibitors. Acta Pharmacol Sin 35, 301–310 (2014). https://doi.org/10.1038/aps.2013.148
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/aps.2013.148
Keywords
This article is cited by
-
Drug design by machine-trained elastic networks: predicting Ser/Thr-protein kinase inhibitors’ activities
Molecular Diversity (2021)
-
QSAR study of human epidermal growth factor receptor (EGFR) inhibitors: conformation-independent models
Medicinal Chemistry Research (2019)
-
Screening of multi-targeted natural compounds for receptor tyrosine kinases inhibitors and biological evaluation on cancer cell lines, in silico and in vitro
Medical Oncology (2015)