Abstract
Aim:
To identify novel serum biomarkers for lung cancer diagnosis using magnetic bead-based surface-enhanced laser desorption/ionization time-of-flight mass spectrum (SELDI-TOF-MS).
Methods:
The protein fractions of 121 serum specimens from 30 lung cancer patients, 30 pulmonary tuberculosis patients and 33 healthy controls were enriched using WCX magnetic beads and subjected to SELDI-TOF-MS. The spectra were analyzed using Bio-marker Wizard version 3.1.0 and Biomarker Patterns Software version 5.0. A diagnostic model was constructed with the marker proteins using a linear discrimination analysis method. The validity of this model was tested in a blind test set consisted of 8 randomly selected lung cancer patients, 10 pulmonary tuberculosis patients and 10 healthy volunteers.
Results:
Seventeen m/z peaks were identified, which were significantly different between the lung cancer group and the control (tuberculosis and healthy control) groups. Among these peaks, the 6445, 9725, 11705, and 15126 m/z peaks were selected by the Biomarker Pattern Software to construct a diagnostic model for lung cancer. This four-peak model established in the training set could discriminate lung cancer patients from non-cancer patients with a sensitivity of 93.3% (28/30) and a specificity of 90.5% (57/63). The diagnostic model showed a high sensitivity (75.0%) and a high specificity (95%) in the blind test validation. Database searching and literature mining indicated that the featured 4 peaks represented chaperonin (M9725), hemoglobin subunit beta (M15335), serum amyloid A (M11548), and an unknown protein.
Conclusion:
A lung cancer diagnostic model based on bead-based SELDI-TOF-MS has been established for the early diagnosis or differential diagnosis of lung cancers.
Similar content being viewed by others
Introduction
Lung cancer is among the leading causes of cancer death in the world, especially in developing countries. In the past several years, the morbidity and mortality related to lung cancer have been on the rise, mostly due to late diagnosis and metastasis1, 2, 3. Therefore, early diagnosis is an important factor for improving the prognosis of lung cancer patients. The vast majority of lung cancer patients in an early stage exhibit no symptoms, and the cancer is commonly detected as an abnormal shadow on a chest roentgenogram or a chest computed tomography (CT) scan4. It is therefore urgent to identify better methods that can provide more information for lung cancer diagnosis, especially during the early stages. The currently used biomarkers for the early diagnosis of lung cancer in the clinic are carcinoembryonic antigen (CEA), cytokeratin-19 fragments (CYFRA-211) and neuron-specific enolase (NSE)5, 6, 7. However, all of these biomarkers have a poor positive predictive value for lung cancer patients, especially for those in an early stage, and some of these biomarkers are not specific to lung cancer.
Applications of proteomic technologies in recent years have significantly broadened our understanding of the molecular mechanisms of numerous diseases and have aided in biomarker screening and drug target discovery. SELDI-TOF-MS is considered a high-efficiency comparative proteomic approach that possesses diverse advantages over traditional protein separation and identification techniques8. Moreover, SELDI-TOF-MS and pattern recognition software have been successfully used in the identification of specific markers for certain diseases, such as prostate cancer, ovarian cancer and rheumatism8, 9, 10.
In this article, serum specimens from lung cancer patients and non-cancer controls were subjected to magnetic bead-based SELDI-TOF-MS. Characteristic peaks for lung cancer patients were identified, and a diagnostic model was constructed. The diagnostic model was validated using a blind test set, which demonstrated clinical significance of this model in lung cancer diagnosis.
Materials and methods
Patient recruitment and sample collection
Serum specimens from 30 lung cancer patients (CA), 30 pulmonary tuberculosis (benign lung disease) patients (TB), and 33 healthy individuals (HC) were collected. Another 8 randomly selected patients with lung cancer, 10 cases of pulmonary tuberculosis and 10 healthy volunteers were enrolled to test the validity of the diagnostic model constructed in this study. The demographic characteristics of the lung cancer patients and the control subjects are shown in Table 1. Informed consent was obtained from every participant, and the study received ethical committee approval. The blood samples were collected in 4 mL BD vacutainer tubes without anticoagulant, allowed to clot at room temperature for up to 1 h, and centrifuged at 4 °C for 5 min at 1000×g. The pooled sera were frozen and stored at -80 °C for future analysis.
Proteomic fraction preparation and SELDI-TOF-MS assay
Serum samples were denatured in the presence of U9 lysis buffer (9 mol/L urea, 2% CHAPS, 50 mmol/L Tris-HCl, pH 9.0; Bio-Rad) and then mixed with WCX-2 (NaAC, pH 4.0) buffer. The sample was then incubated with WCX-2-pretreated magnetic beads (Changchun Bokun Science and Technology, China) for 60 min. The beads were then washed twice with NaAC and eluted with 1% TFA (trifluoroacetic acid)11. The protein fraction was then lyophilized and prepared for SELDI-TOF-MS (BioRad, USA) according to the manufacturer's instructions. Masses were acquired from m/z 1000-50 000.
Protein identification and bioinformatics analysis
Protein database searching was performed with the MASCOT search engine (http://www.matrixscience.com/; Matrix Science, London, UK), which compared the monoisotopic peaks against the NCBI nonredundant protein database (http://www.ncbi.nlm.nih.gov/). The allowed mass tolerance was less than 0.05%12, 13, 14. Proteins with MASCOT scores greater than 63 were considered significant (P<0.05).
Data processing and pattern recognition
Principal component analysis (PCA) was performed to cluster the samples (MATLAB, MathWorks, USA). The score plots aided in visualizing the data, and a diagnostic model was constructed with the marker proteins using a linear discrimination analysis method. The classification performance (specificity and sensitivity) was assessed using the AUC values of the ROC curves15, 16. The mass spectrometry analysis system was applied to identify the characteristic molecules corresponding to the featured peaks in the Metlin (http://metlin.scripps.edu/) and HMDB (http://www.hmdb.ca/) databases. The protein hits were verified using text mining techniques.
Results
Identification of the serum protein profile
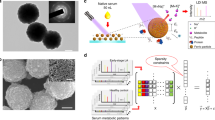
The protein fractions of the serum samples from 30 lung cancer patients, 30 pulmonary tuberculosis patients and 33 healthy controls were enriched using WCX magnetic beads, which is a particularly effective method for the detection of low-molecular-weight proteins and peptides, and were analyzed by SELDI-TOF-MS. Following baseline correction and peak alignment, protein signals were obtained for all samples. Representative mass chromatogram data for lung cancer patients, pulmonary tuberculosis patients and healthy controls are shown in Figure 1.
Protein fingerprint analysis of serum protein from lung cancer patients
The mass spectrum data of the 121 samples were analyzed using Biomarker Wizard (version 3.1.0) to identify the peaks that were different between the lung cancer patients and the control individuals. The Shapiro-Wilk test was used to evaluate the normality of the distribution of the peaks, and the homogeneity of the variance was calculated by the Levene's test. All of the peaks were sorted using the P value from the ANOVA and the Student-Newman-Keuls post-hoc test using SPSS 16.0 (SPSS Inc, USA). Seventeen significant discriminating m/z peaks (4188.21, 4548.39, 4763.26, 4983.18, 5069.19, 5351.19, 5486.84, 6212.41, 6445.26, 6573.47, 9725.37, 11705.4, 11769.7, 15126.9, 15335.2, 15938.7, and 19790.5) were found between the lung cancer group and the control groups (P<0.05), and are shown in Table 2. The importance of these peaks determined by the Biomarker Pattern Software is listed as well. The most important peak was assigned an importance index of 100. The importance of other peaks was calculated relative to that of the top peak, and a value below 100 was conferred to each peak. The peaks at 4763.26, 5069.19, 5351.19, 5486.84, 6212.41, 11705.4, 11769.7, 15335.2, and 15938.7 Da were higher in the lung cancer patients than that in the control groups, and the peaks at 4188.21, 4548.39, 4983.18, 6445.26, 6573.47, 9725.37, 15126.9, and 19790.5 Da were lower in the lung cancer patients than that in the control groups.
Diagnostic model construction and validation
The 17 m/z peaks that could discriminate between lung cancer group and the control groups were identified by Biomarker Patterns Software Version 5.0 and analyzed to select peaks for the establishment of a diagnostic biomarker pattern. The m/z peaks at 6445, 9725, 11705, and 15126 were selected by the pattern recognition software as the best markers to construct a diagnostic model for lung cancer (Figure 2). This four-peak model established in the training set could discriminate lung cancer patients from healthy individuals as well as pulmonary tuberculosis patients with a sensitivity of 93.3% (28/30) and a specificity of 90.5% (57/63). The decision tree is presented in Figure 3, and the characteristics of the diagnostic model are shown in Figure 4. The prediction accuracy was validated using a blind test set consisting of 28 randomly selected individuals. The sensitivity and specificity of the prediction are shown in Table 3. We combined database searching with literature mining to determine the identities of the proteins corresponding to the featured peaks. Three of the featured proteins were identified as chaperonin (M9725), hemoglobin subunit beta (M15335) and serum amyloid A (M11548). There was no protein match for the 6445 Da peak in the searched databases, indicating it might be a novel protein.
Four characteristic peaks in lung cancer patients. Protein spectrum of serum samples from two different lung cancer patients (CA), two pulmonary tuberculosis patients (TB) and two healthy controls (HC). The x-axis represents the molecular mass calculation (m/z), and the y-axis represents the relative intensity.
Boosting decision tree classification of the participants. The root node (top) and descendant nodes were shown as ellipses and the terminal nodes (Nodes 1–7) were shown as rectangles. The mass value in the nodes was followed by lower or equal to intensity value. If the answer to the question in a node of the tree is yes, proceed down to the left node, otherwise (ie no), proceed down to the right node. When proceeding to the terminal nodes, the decision tree assigned samples to three groups. Samples in terminal nodes 2, 3, 4, and 6 were assigned to TB, terminal node 1 was to CA and terminal nodes 5 and 7 were to HC. The numbers in rectangles represent the actual clinical diagnosis of samples assigned to this terminal node by decision tree (ie in terminal node 1, decision tree assigned 31 samples to CA, but actually 28 of them were CA according to the clinical diagnosis).
To explore the clinical significance of the constructed model, the validity of the model was tested by a blind test set consisting of 8 randomly selected lung cancer patients, 10 pulmonary tuberculosis patients and 10 healthy volunteers. The sensitivity of the diagnostic model was 75.0% (6/8), and the specificity was 95% (19/20).
Discussion
During the last several decades, the identification of novel biomarkers for complex diseases has become increasingly successful because of the emergence of high-throughput proteomic techniques such as SELDI-TOF-MS17. Biomarkers, especially biomarker patterns, are considered to be reliable and powerful tools for the early diagnosis, differential diagnosis, and therapy of some diseases18, 19. Analysis of serum proteins by SELDI-TOF-MS provides new information about small proteins with high serum abundances20, 21. Magnetic beads with large surfaces are better able to enrich proteins from serum than other materials. Joint application of magnetic beads and SELDI-TOF-MS might be a more powerful strategy to discover novel serum biomarkers with low abundances.
In this study, the protein fingerprints in the sera from 30 lung cancer patients, 30 pulmonary tuberculosis patients and 33 healthy controls were analyzed using magnetic beads and SELDI-TOF-MS, and seventeen characteristic m/z peaks were identified using Biomarker Wizard. Theoretically, multiple markers are much more powerful and reliable in the diagnosis of a disease than a single marker alone. In our study, the 17 discriminating m/z peaks were analyzed by Biomarker Patterns Software, and only 4 protein peaks, those at 6445, 9725, 11705, and 15126 m/z, were capable of serving as markers for lung cancer diagnosis. This four-peak model established in the training set could discriminate lung cancer patients from healthy individuals and from pulmonary tuberculosis patients with a sensitivity of 93.3% (28/30) and a specificity of 90.5% (57/63). The validity of the model was tested using a blind test set, and the sensitivity of the diagnostic model was 75.0% (6/8), and the specificity was 95% (19/20). Despite the limitation of the test set size, these data indicate that our study provides a novel and potent tool to distinguish lung cancer patients from tuberculosis patients and healthy individuals using serum.
In conclusion, our findings suggest that the application of magnetic beads and SELDI-TOF-MS could be a potent strategy for the identification of serum biomarkers. More importantly, the diagnostic model we constructed using the protein peaks at 6445, 9725, 11705, and 15126 m/z could successfully distinguish lung cancer patients from tuberculosis patients and normal controls, which might be of clinical significance in the early diagnosis of lung cancer. Database searching with literature mining showed that the featured peaks were chaperonin (M9725), hemoglobin subunit beta (M15335), serum amyloid A (M11548) and an unknown protein. A previous report has illustrated that serum amyloid A could be a promising serum biomarker for lung cancer, consistent with the results of our study. However, the clinical significance of chaperonin and hemoglobin subunit beta in lung cancer diagnosis deserves further investigation.
Author contribution
Qi-bin SONG and Hua-zong ZENG designed research; Wei-guo HU and Yi YAO performed research; Peng WANG contributed new reagents and samples; Wei-guo HU and Peng WANG analyzed data; and Qi-bin song, Wei-guo HU, and Hua-zong ZENG wrote the paper.
References
Spira A, Ettinger DS . Multidisciplinary management of lung cancer. N Engl J Med 2004; 350: 379–92.
Herbst RS, Heymach JV, Lippman SM . Lung Cancer. N Engl J Med 2008; 359: 1367–80.
Singh N, Agarwal R, Aggarwal AN . Quality-of-life assessment in trials of lung cancer. Lancet 2007; 370: 933.
Petricoin EF, Zoon KC, Kohn EC, Barrett JC, Liotta LA . Clinical proteo-mics: translating benchside promise into bedside reality. Nat Rev Drug Discov 2002; 1: 683–95.
Schneider J, Velcovsky HG, Morr H, Katz N, Neu K, Eigenbrodt E . Comparison of the tumor markers tumor M2-PK, CEA, CYFRA21-1, NSE and SCC in the diagnosis of lung cancer. Anticancer Res 2000; 20: 5053–8.
Ardizzoni A, Cafferata MA, Tiseo M, Filiberti R, Marroni P, Grossi F, et al. Decline in serum carcinoembryonic antigen and cytokeratin 19 fragment during chemotherapy predicts objective response and survival in patients with advanced nonsmall cell lung cancer. Cancer 2006; 107: 2842–9.
Holdenrieder S, von Pawel J, Dankelmann E, Duell T, Faderl B, Markus A, et al. Nucleosomes and CYFRA 21-1 indicate tumor response after one cycle of chemotherapy in recurrent non-small cell lung cancer. Lung Cancer 2009; 63: 128–35.
Petricoin EF, Ardekani AM, Hitt BA, Levine PJ, Fusaro VA, Steinberg SM, et al. Use of proteomic patterns in serum to identify ovarian cancer. Lancet 2002; 359: 572–7.
Zhang Z, Bast RC Jr, Yu Y, Li J, Sokoll LJ, Rai AJ, et al. Three biomarkers identified from serum proteomic analysis for the detection of early stage ovarian cancer. Cancer Res 2004; 64: 5882–90.
Goncalves A, Bertucci F, Birnbaum D, Borg JP . Protein profiling SELDI-TOF and breast cancer: clinical potential applications. Med Sci (Paris) 2007; 23: 23–6.
Baumann S, Ceglarek U, Fiedler GM, Lembcke J, Leichtle A, Thiery J . Standardized approach to proteome profiling of human serum based on magnetic bead separation and matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Clin Chem 2005; 51: 973–80.
Trygg J, Holmes E, Lundstedt T . Chemometrics in metabonomics. J Proteome Res 2007; 6: 469–79.
Smith CA, Want EJ, O'Maille G, Abagyan R, Siuzdak G . XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal Chem 2006; 78: 779–87.
Tibshirani R, Hastie T, Narashiman B, Chu G . Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc Natl Acad Sci U S A 2002; 99: 6567–72.
Anthony ML, Rose VS, Nicholson JK, Lindon JC . Classification of toxin-induced changes in 1H NMR spectra of urine using an artificial neural network. J Pharm Biomed Anal 1995; 13: 205–11.
Beckonert O, Bollard ME, Ebbels T, Keun HC, Antti H, Holmes E, et al. NMR-based metabonomic toxicity classification: hierarchical cluster analysis and k-nearest. Anal Chim Acta 2003; 490: 3–15.
Fung ET, Wright GL Jr, Dalmasso EA . Proteomic strategies for biomarker identification: progress and challenges. Curr Opin Mol Ther 2000; 2: 643–50.
Skytt A, Thysell E, Stattin P, Stenman UH, Antti H, Wikström P . SELDI-TOF MS versus prostate specific antigen analysis of prospective plasma samples in a nested case control study of prostate cancer. Int J Cancer 2007; 121: 615–20.
Hundt S, Haug U, Brenner H . Blood markers for early detection of colorectal cancer: a systematic review. Cancer Epidemiol Biomarkers Prev 2007; 16: 1935–53.
Wu CC, Chien KY, Tsang NM, Chang KP, Hao SP, Tsao CH, et al. Cancer cell-secreted proteomes as a basis for searching potential tumor markers: nasopharyngeal carcinoma as a model. Proteomics 2005; 5: 3173–82.
Welsh JB, Sapinoso LM, Kern SG, Brown DA, Liu T, Bauskin AR, et al. Large-scale delineation of secreted protein biomarkers overexpressed in cancer tissue and serum. Proc Natl Acad Sci U S A 2003; 100: 3410–5.
Acknowledgements
The authors also thank Cluster Biotech Co Ltd for the assistance in bio-information analysis.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Song, Qb., Hu, Wg., Wang, P. et al. Identification of serum biomarkers for lung cancer using magnetic bead-based SELDI-TOF-MS. Acta Pharmacol Sin 32, 1537–1542 (2011). https://doi.org/10.1038/aps.2011.137
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/aps.2011.137
Keywords
This article is cited by
-
An Estimation Formula for Resonance Frequency Using Sex and Height for Healthy Individuals and Patients with Incurable Cancers
Applied Psychophysiology and Biofeedback (2024)
-
Circulating microvesicles and exosomes in small cell lung cancer by quantitative proteomics
Clinical Proteomics (2022)
-
Carbonic anhydrase 1 is a promising biomarker for early detection of non-small cell lung cancer
Tumor Biology (2016)
-
Serum peptidome patterns of hepatocellular carcinoma based on magnetic bead separation and mass spectrometry analysis
Diagnostic Pathology (2013)
-
The discovery and identification of a candidate proteomic biomarker of active tuberculosis
BMC Infectious Diseases (2013)