Abstract
The emerging CRISPR/Cas9 system represents a promising platform for genome editing. However, its low transfection efficiency is a major problem hampering the application of the gene-editing potential of CRISPR/Cas9. Herein, by screening a pool of more than 56 kinds of agents, we constructed a novel polyethylene glycol phospholipid-modified cationic lipid nanoparticle (PLNP)-based delivery system that can condense and encapsulate a Cas9/single-guide RNA (sgRNA) plasmid (DNA) to form a core–shell structure (PLNP/DNA) that mediated up to 47.4% successful transfection of Cas9/sgPLK-1 plasmids in A375 cells in vitro. An intratumor injection of Cas9/sgPLK-1 plasmids into melanoma tumor-bearing mice resulted in significant downregulation of Polo-like kinase 1 (PLK-1) protein and suppression of the tumor growth (>67%) in vivo. This approach provides a versatile method that could be used for delivering the CRISPR/Cas9 system with high efficiency and safety both in vitro and in vivo.
Similar content being viewed by others
Introduction
The RNA-guided CRISPR (clustered regularly interspaced short palindrome repeats)-associated nuclease Cas9 has emerged as an innovative and powerful means for gene editing and provides a platform for versatile genome editing.1 Mediated by a synthetic single-guide RNA (sgRNA), Cas9 can induce DNA double-strand breaks at targeted genomic loci and result in targeted loss-of function mutations at specific sites in the genome (Figure 1a).2, 3, 4 This system can be used to repress expression of targeted genes efficiently regardless of cell types in human cells.5, 6 These features endow the CRISPR/Cas9 system with promising potential in treating challenging diseases such as cancer.
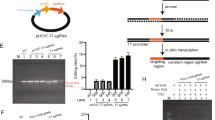
Schematic illustration of the strategy for delivering CRISPR/Cas9 system targeting tumors. (a) Schematic illustration of the CRISPR/Cas9 system. (b) The packaging and encapsulation processes of the Cas9-sgRNA plasmid by chondroitin sulfate, protamine, 1,2-dioleoyl-3-trimethylammoniumpropane (DOTAP), dioleoylphosphatidylethanolamine (DOPE) and DSPE-PEG. (c) The targeting sites of different single-guide RNAs (sgRNAs) on human Polo-like kinase 1 (PLK-1) locus indicated by blue lines with corresponding protospacer adjacent motifs (PAMs). (d) The map of the Cas9/sgPLK-1-fused plasmid. pCAG, chicken β-actin promoter with cytomegalovirus enhancer; T7, T7 RNA polymerase promoter; T7 promoter can drive transcription of the downstream coding of hCas9 in the presence of T7 RNA polymerase. EGFP, enhanced green fluorescent protein; gRNA, guide RNA; hCas9, human Cas9 endonuclease; hU6, human U6 promoter; pGK, mouse phosphoglycerate kinase 1 promoter; puro, Puro gene; 2A, ‘self-cleaving’ 2A sequence.
However, the expanding applications of the CRISPR/Cas9 system are limited by its large size, especially in the widely used Cas9 derived from Streptococcus pyogenes.7, 8 Although viral vectors show excellent gene transfection efficiency, they may result in off-target modification of the gene, mutagenesis and carcinogenesis.9 Furthermore, repeated administration of viral vectors may induce an immune response that may abolish the transgene expression. The development of nonviral vectors, especially nanomaterial-based vectors, is promising for delivering transfection agents.10
In this study, to achieve efficient and inheritable delivery of CRISPR/Cas9, we rationally designed various nonviral vectors (from a pool of more than 56 kinds of agents, Supplementary Table S1), including polymers (polyethyleneimine, polylysine and others, with various modifications) and nanoparticles (magnetic nanoparticles, gold nanoparticles and carbon nanoparticles with different surface modifications), to facilitate penetrating the cell membrane or localizing at specific areas of the cells especially their nuclei. We additionally screened their delivery efficiency in A375 cells (Supplementary Table S1). Most of these screened candidates showed lower transfection efficiency than those of the commercial lipid-based agents (Fugene 6 and Lipofectamines 2000 and 3000). Among all the agents, Lipofectamine 3000 yielded the highest transfection efficiency (13.1%) (Supplementary Table S1). Thus, the lipid systems may have great potential to carry large plasmids. Indeed, the lipid system is undoubtedly one of the most popular nonviral vehicles for gene delivery.11, 12, 13, 14, 15, 16, 17 The lipid bilayer can protect nucleic acids from degradation and enable efficient endosomal escape.18, 19 However, the present lipid systems, such as Lipofectamine 3000, are still not competent to deliver CRISPR/Cas9 because of their relatively low transfection efficiency. We speculated that there are two major reasons for this situation: (1) the volume of the Cas9/sgRNA-fused plasmid (>10 000 bp) is too large for ordinary vehicles; (2) the poorly encapsulated anionic plasmid would be repelled by the cell membrane that is also negatively charged.8 To surpass these barriers, we present a strategy to achieve exceptionally high transfection efficacy of Cas9/sgRNA both in vitro and in vivo by developing a novel cationic lipid-based delivery system.
Experimental procedures
Materials
PEG2000-DSPE (1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[methoxy(polyethylene glycol)-2000]) was from AVT (Shanghai, China). Cholesterol and protamine were from Sigma Aldrich (St Louis, MO, USA). Dioleoylphosphatidylethanolamine (DOPE) and 1,2-dioleoyl-3-trimethylammoniumpropane (DOTAP) were provided by Avanti Polar Lipids (Alabaster, AL, USA). Lipofectamine 2000 was from Invitrogen (New York, NY, USA); Cas9-sgPLK-1 plasmid DNA was from Viewsolid Biotech (Beijing, China) and small interfering RNA targeting for Polo-like kinase 1 (PLK-1) mRNA (siPLK-1) was from Takara (Dalian, China).
Cell culture
A375, MCF-7 and PC-3 cell lines were obtained from the Institute of Basic Medical Science, Chinese Academy of Medical Sciences (Beijing, China). The cells were maintained in complete Dulbecco’s modified Eagle’s medium (DMEM, Life Technology, Rockville, MD, USA) (containing 10% fetal bovine serum, 1% penicillin/streptomycin and 1% glutamine) and placed in 5% CO2 humidified incubator at 37 °C.
Preparation and characterization of the PLNP/DNA
The core–shell nanoparticles were prepared according to previous studies with minor modifications.20 Through a thin film hydration with an ultrasonic dispersion, we prepared the cationic lipid composed of DOTAP, DOPE and cholesterol (1.4:1:0.5, 0.8:1:0.5, 0.2:1:0.5, n/n/n). The negative core contained protamine Cas9-sgPLK-1 plasmid DNA and chondroitin sulfate (CS). In brief, we added the protamine solution to the mixture of Cas9-sgPLK-1 plasmid DNA and CS (Cas9-sgPLK-1 plasmid DNA/CS, 1/1, w/w) and obtained a negatively charged ternary complex by electrostatic interaction. The weight ratio of protamine/(Cas9-sgPLK-1 plasmid DNA+CS) in the optimized negative core was 1/1.2 (w/w). They were stored at room temperature for 15 min, followed by encapsulation by the cationic lipid shell (lipids/Cas9-sgPLK-1=2000/1, n/n) and incubated for another 15 min. Finally, the lipid nanoparticle (LNP)/DNA was further postmodified with 5% (molar ratio) polyethylene glycol phospholipid (DSPE-PEG) to yield PLNP/DNA. The PLNP/DNA was obtained after incubation at 55 °C for 15 min. The size and zeta potential of the nanoparticles above were examined in dynamic light scattering at room temperature (Malvern Zetasizer nano ZS, Malvern, UK).
High-resolution transmission electron microscope characterization
We suspended the PLNP/DNA-a in water and applied it to carbon-coated copper grids, negatively stained the PLNP/DNA-a with 2% uranyl acetate and dried it in the air. We observed the samples under high-resolution transmission electron microscope (Tecnai G2 F20 U-TWIN, operated at 200 kV, FEI, Hillsboro, OR, USA).
Loading capability calculation
To determine the encapsulation efficiency of the Cas9/sgRNA plasmid in the PLNP/DNA-a, the PLNP/DNA-a complexes were isolated by centrifugation at 4 °C and 15 000 r.p.m. for 30 min. The concentration of unbound Cas9-sgRNA plasmid in the supernatant was measured by a NanoDrop 2000 Spectrophotometer (Wilmington, DE, USA). The encapsulation efficiency can be calculated according to the equation:

where EE is encapsulation efficiency, C1 represents the original concentration of Cas9/sgRNA plasmid in the solution and C2 represents the unbound Cas9/sgRNA plasmid in the supernatant.
Cellular uptake study
DNA-a was labeled with N,N’-(dipropyl)-tetramethylindocarbocyanine (Cy3) Label IT Nucleic Acid Labeling Kit (Mirus Bio LLC, Madison, WI, USA) according to the supplier’s protocol. A375 cells were seeded onto 35 mm cover glass bottom culture dishes (NEST Science, Wuxi, China) with a cell number of 5 × 105 cells per well in complete DMEM (2 ml) for 24 h. The medium was replaced with Opti-MEM reduced serum medium containing DNA-a treatment, in which the DNA-a concentration was equivalent to 2 μg per well. After incubation with PLNP/DNA-a for 0, 1, 2 or 3 h, the cells were washed with PBS repeatedly and fixed with 4% paraformaldehyde for 15 min. The cells were washed with phosphate-buffered saline (PBS), stained with 4,6-diamidino-2-phenylindole and washed with PBS repeatedly. We observed the cells under confocal laser scanning microscope (CLSM 710; CarlZeiss, Oberlochen, Germany). The excitation for Cy3 is 543 nm, whereas that of 4,6-diamidino-2-phenylindole is 365 nm. The statistical analysis was carried out using ImageJ software (Bethesda, MD, USA).
In vitro transfection efficiency measurement
A375, MCF-7 and PC-3 cells were seeded into 35 mm cover glass bottom culture dishes (NEST Science, China) with a cell number of 5 × 105 cells per well in complete DMEM (2 ml) for 24 h. The medium was replaced with Opti-MEM reduced serum medium containing PBS, DNA-a, Lipo2000/DNA-a or PLNP/DNA-a treatment, in which the DNA-a concentration was equivalent to 2 μg per well. After transfection for 3 h, the medium was replaced with freshly prepared complete DMEM and incubated for another 48 h. We observed the cells under CLSM. The excitation for green fluorescent protein (GFP) is 488 nm. The statistical analysis was performed with CLSM 710 software.
Tumor suppression study
Female BALB/c nude mice weighing 18–20 g were from Vital River Laboratory Animal Center (Beijing, China). All care and handling of animals were performed with the approval of Institutional Authority for Laboratory Animal Care of Institute of Process Engineering, Chinese Academy of Sciences. The A375 cells with a number of 3 × 106 were inoculated subcutaneously. After the tumors grew to ∼100 mm3, the mice were randomly grouped. The tumor-bearing mice were administered in situ with saline, DNA-a, Lipo2000/DNA-a, LNP/DNA-a, PLNP/siPLK-1, PLNP/Cas9, PLNP/sgPLK-1a or PLNP/DNA-a treatment every other day. The plasmids in each injection were equivalent to 10 μg per mouse. The administration was finished on day 15, and the mice were killed on day 16. We evaluated the tumor volumes according to V (mm3)=0.5 × length × width2.
Western blot
The tumors were extracted from the mice, washed with PBS and lysed in 50 μl of lysis buffer. The protein was collected and the concentration was measured with a bicinchoninic acid protein assay kit (Pierce, Rockford, IL, USA). Cell lysates were run on an SDS gel and blotted with rabbit anti-human PLK-1 monoclonal (Cell Signaling Technology, Beverly, MA, USA) in a dilution of 1:1000. The lysates were processed with a precast gradient gel (Bio-Rad, Hercules, CA, USA). The images were obtained using Bio-Rad UNIVERSAL HOOD II (Bio-Rad Laboratory, Bossier City, LA, USA).
Gene sequencing
Deep sequencing of CRISPR-modified PLK-1 loci was carried as follows: the genomic region of PLK-1 was PCR amplified and purified with agarose gel. Deep sequencing libraries were made from 50 ng of the PCR products using the Nextera protocol and sequenced on Illumina MiSeq (150 bp paired-end) machines (San Diego, CA, USA). Data analysis was performed according to the procedures of standard Illumina sequencing analysis. Mutations were confirmed by comparison with the reference sequences using Burrows–Wheeler Aligner with custom scripts.
Primer:
Forward 5′-AGTGACTGCAGGGAAGCTG-3′.
Reverse 5′-GTCGTTGTCCTCGAAAAAGC-3′.
Results
Preparation of the PLNP/DNA
The basic principle of the strategy is to first tightly pack the cargo (Cas9/sgRNA) to form a core, which can greatly reduce the volume of the cargo, followed by the encapsulation of the core in a lipid shell, which can further stabilize the core and facilitate the entry of the resulting core–shell LNP into cells (Figure 1b). The core of the LNPs was formed by entangling the Cas9/sgRNA-fused plasmid DNA with CS that has a high density of anionic charges and is critical for endowing the Cas9/sgRNA-fused plasmid DNA with more capability to attach to protamine that was designed to serve as a ‘seed’ to initiate the condensation process of the Cas9/sgRNA-fused plasmid DNA/CS complex. The positive charges on the seed (protamine) and the negative charges on the Cas9/sgRNA-fused plasmid DNA/CS complex are the driving forces for the tight packing of the Cas9/sgRNA-fused plasmid DNA/CS complex on the protamine to form highly condensed, small and stable ternary core of the LNP. The core of the LNP was negatively charged because of the excessive amount of the anionic Cas9/sgRNA plasmid DNA/CS complex and its shielding effect on the cationic seed (protamine). As a core, the negatively charged ternary complexes (chondroitin/protamine/Cas9/sgRNA plasmid DNA) were encapsulated with cationic lipid composed of DOTAP, DOPE and cholesterol (improves the mechanical strength of the lipid layer) to yield positively charged lipid/chondroitin/protamine/Cas9-sgRNA plasmid DNA (LNP/DNA). The LNP/DNA was further postmodified with DSPE-PEG to yield lipid nanoparticle/DNA (PLNP/DNA) (Figure 1b) that increased stability and solubility, lowered toxicity, increased half-life and decreased clearance and immunogenicity.20, 21 We assume that the dense core structure of the PLNPs minimized the size of the CRISPR system, whereas the cationic shell of the PLNPs facilitated the PLNP–cell interactions and the internalization of the PLNPs22 and subsequent gene editing.
In this study, the sgRNA targeting the gene of PLK-1, a master regulator of mitosis, was selected for gene editing. Researchers found the overexpression of PLK-1 in tumor cells, such as melanoma, prostate and breast cancer cells. The downregulation of PLK-1 expression is confirmed to inhibit the tumor growth effectively.23, 24 We designed three sgRNA candidates (sgPLK-1a, b and c) (Figure 1c) targeting the PLK-1 gene and fused their gene sequences with the Cas9 gene in the plasmids (DNA-a, DNA-b and DNA-c) respectively with a fused GFP gene as a reporter (Figure 1d).
Characterization of the PLNP/DNA
We chose PLNP/DNA-a as a representative to demonstrate the characterization of the PLNP/DNAs. The high-resolution transmission electron microscopy showed the entangled and condensed Cas9-sgRNA plasmid DNA-a had an average size of ∼50 nm (Figure 2a and Supplementary Figure S1a). After lipid and PEG encapsulation, the product PLNP/DNA-a had a typical core–shell structure with an average size of ∼60 nm (Figure 2b and Supplementary Figure S1b). The DOTAP to DOPE ratio is an important factor affecting the size, distribution and zeta potential of the PLNP/DNA-a. By tuning the ratio from 1.4:1 to 0.2:1 using dynamic light scattering, we found the optimal ratio was 0.8:1 under which the PLNP/DNA-a had appropriate parameters, including size and zeta potential (Supplementary Figure S2). Under the optimized condition, the PLNP/DNA-a had a size of ∼156.5 nm and zeta potential of 23.2 mV as measured by dynamic light scattering (Supplementary Table S2). The other two kinds of plasmid DNAs (DNA-b and DNA-c) were encapsulated and modified with the same protocol and yielded PLNP/DNAs with similar sizes and zeta potentials (Supplementary Table S2).
High-resolution transmission electron microscopy (HR-TEM) and dynamic light scattering (DLS) analysis of the nanoparticles. HR-TEM image of chondroitin/protamine/Cas9-sgRNA plasmid DNA (a) and PLNP/DNA-a (b), DLS analysis of the PLNP/DNA-a and Lipo2000/DNA-a in serum-free Dulbecco’s modified Eagle’s medium (DMEM) (c), Opti-MEM reduced serum medium (d) and 10% fetal bovine serum (FBS) DMEM (e).
The stability of the transfection reagent in the serum is an important parameter for effective transfection. We chose PLNP/DNA-a as a representative and set a commercial reagent Lipofectamine 2000-encapsulated Cas9-sgPLK-1a DNA (Lipo2000/DNA-a) as a control and compared their stability in three kinds of culture media with different concentrations of serum: serum-free DMEM (Figure 2c), Opti-MEM reduced serum medium (Figure 2d) and DMEM with 10% fetal bovine serum (Figure 2e). Under the same conditions, the diameter of the PLNP/DNA-a was significantly smaller than that of Lipo2000/DNA-a. With the increase of the serum concentration, the size of the PLNP/DNA-a increased slightly. In contrast, the size of Lipo2000/DNA-a increased sharply with the increase of the serum concentration. This result indicates that the PLNP/DNA-a is more stable than Lipo2000/DNA-a in medium with a high concentration of serum. With the increase of incubation time, both the size and the size variation of PLNP/DNA-a were significantly smaller than those of Lipo2000/DNA-a. Thus, the PLNP/DNA-a is more stable than the commercial agents.
We evaluated the stability of the PLNP/DNA-a in various pH conditions in vitro. With the decrease of the pH value, the PLNP/DNA-a showed an increased diameter (Supplementary Figure S3), indicating the instability of the structure of the particles at a low pH. This means that the PLNP/DNA-a could break down and release the content under acidic conditions such as in lysosomes.
In vitro transfection
We first evaluated the cellular uptake of Cy3-labeled PLNP/DNA-a in human malignant melanoma cells (A375). The cellular uptake of Cy3-PLNP/DNA-a sharply increased within 2 h after the nanoparticle administration. At 3 h after administration, the cellular uptake of the nanoparticle reached the maximum level and the fluorescence intensity of Cy3-PLNP/DNA-a displayed an ∼25-fold increase compared with the initial stage (Supplementary Figure S4). To avoid unnecessary cytotoxicity, the cells were incubated with PLNP/DNA-a for 3 h in the following experiments. We tested the in vitro transfection efficiency of the PLNP/DNA-a with three representative cancer cell lines including A375 (human malignant melanoma), PC-3 (human prostate cancer cells) and MCF-7 (human breast cancer cells), setting Lipo2000/DNA-a as a control. To facilitate the characterization of positive transfection of the CRISPR system under CLSM and flow cytometry, GFP was also fused in the plasmids. The PLNP/DNA-a treatment induced a high intensity of green fluorescence in three cancer cell lines (Figure 3). In comparison, the Lipo2000/DNA-a-treated cells showed negligible fluorescence in the three cell types (Figure 3). As a negative control, the free plasmid (DNA-a) did not show any GFP fluorescence (not shown). Flow cytometry results indicated that PLNP/DNA-a yielded 47.4% positive transfection for A375 cell population, whereas Lipo2000/DNA-a only led to 3.09% successful transfection. High-throughput sequencing indicated that the PLNP/DNA-a induced various mutations ranging from 1 to 20 indels in the targeted site (Supplementary Figure S5), and the mutations reached 16.1%. However, Lipo2000/DNA-a induced only ∼1.2% mutations, showing much lower efficiency (Supplementary Figure S6) compared with the PLNP formulations. For the other two cell types, PC-3 and MCF-7, PLNP/DNA-a also showed much higher transfection ratio than Lipo2000/DNA-a. No GFP-positive cells were observed in the free plasmid (negative control)-treated cells. Similar results were also shown in PLNP/DNA-b and PLNP/DNA-c groups (Supplementary Figures S7 and S8). The in vitro Live/Dead assay indicated that compared with LNP/DNA, the PLNP/DNA was less cytotoxic (Supplementary Figure S9).
Moreover, the cells treated with PLNP/Cas9/sgLuc-GFP-fused plasmid, which contains sgRNA targeting to luciferase gene (sgLuc), showed minimal PE-Cy5 signal (Supplementary Figure S10). This means that the PLNP/Cas9/sgLuc-GFP-fused plasmid had a minimal effect on the downregulation of PLK-1. However, the cells displayed GFP fluorescence simultaneously, that is, the untargeted DNA can be transfected successfully but could not cause cell apoptosis, indicating that the mismatched sgRNA had little effect on the targeting gene (Supplementary Figure S10). These results indicated that the PLNP was an effective and safe tool to deliver the CRISPR/Cas9 system.
In vivo transfection
The excellent result of in vitro experiment of the PLNP/DNA-a drove us to verify whether it is also effective in tumor inhibition in vivo. The melanoma-bearing Balb/c nude mice were created by implanting A375 cells subcutaneously. After the tumors grew to ∼100 mm3, the transfection agents (Saline, Cas9-sgPLK-1a (DNA-a), Lipo2000/DNA-a, LNP/DNA-a, PLNP/siPLK-1, PLNP/Cas9, PLNP/sgPLK-1 and PLNP/DNA-a) were administered by intratumor injection to the tumor-bearing mice every other day. The PLNP/DNA-a inhibited the growth of the tumors most effectively compared with other groups.
Moreover, the average volume of the tumors treated by the PLNP/DNA-a was approximately one-third of the PBS (67% inhibition) and half of the PLNP/siPLK-1 group (50% inhibition) (Figure 4a). Under the same condition, the volumes of tumors in the mice treated with saline, free DNA-a, LNP/DNA-a, PLNP/Cas9 and PLNP/sgPLK-1 all increased significantly. For example, in the free DNA-a group, the average volume of the tumors on day 16 was >13-fold of those of the pretreated ones. As positive controls, the administration of PLNP/siPLK-1 or Lipo2000/DNA-a showed limited therapeutic efficacy in vivo (Figure 4a). The tumor weights administered with the PLNP/DNA-a were significantly lighter compared with those of other groups on day 16 (Figure 4b).
The drug administrations on A375 tumor-bearing model mice. (a) The volumes of the extracted tumors treated by different DNA-a formulations as a function of treatment time. The data were from the mean of each treatment group (mean±s.d., n=4). Black arrows indicate the day for drug administration. (b) The weights of the extracted tumors treated by different DNA-a formulations. The weights of the tumors administered with the PLNP/DNA-a are significantly lower than those of other groups on day 16 (*P<0.05; **P<0.01). (c) Western blot analysis of Polo-like kinase 1 (PLK-1) protein expression in tumors treated with different DNA-a formulations. The integrated optical density ratio of PLK-1 to GAPDH is shown below the western blot bands, and tumors were extracted on day 16. (d) Targeted indel mutations on PLK-1 induced by PLNP/DNA-a. Alleles shown were amplified from pooled genomic DNA isolated from the tumors in four mice. The wild-type sequence as a reference is shown in the way of reverse complement. The target sites were marked in green and protospacer adjacent motif (PAM) in gray. For the mutations, the red dashes represent deletions and blue lowercase letter represents insertions. The net change of bases for the mutation is listed to the right of each sequence (+, insertion; −, deletion).
We further carried out western blot to analyze expression level of the PLK-1 protein to verify whether the protein expression levels agree with the volume and weight changes of the treated tumors in vivo. The western blot demonstrated that the PLNP/DNA-a gave rise to the most efficient silencing of the target gene (Figure 4c) compared with other groups. The tendency of PLK-1 expression was well consistent with the tumor inhibition effect (Figure 4a).
We carried out deep sequencing on the PLK-1 locus of tumor genomic DNA. Sequencing analysis showed that the PLNP/DNA-a-treated tumors produced a wide variety of indels, ranging from 1 to 28 bp, and ∼3% of Cas9-mediated genome mutation in the tumors of live mice (Figure 4d and Supplementary Figure S11). For instance, we found 1-nucleotide or 2-nucleotide indels in frequent occurrence, leading to cleaving of the PLK-1 reading frame. The indels in targeted sites indicate the successful delivery of the Cas9-sgPLK-1 plasmid DNA to the tumors by the PLNP/DNAs.
The 3% mutation of the genomic locus of PLK-1 seems inconsistent with the effective inhibition of PLK-1 protein expression. The possible reasons for this are as follows. (1) The concepts of the two ratios are different: the mutation ratio indicates the ratio of the PLK-1 gene-mutated cells in all the extracted cells, whereas the protein expression ratio refers to the levels of PLK-1 protein compared with the reference protein GAPDH in the tumor tissues. (2) The protein level is affected by various factors such as transcription and translation processes, posttranslational modifications and regulatory factors from neighboring cells. Thus, the protein-level alteration cannot be strictly consistent with the gene-level modification. (3) The tumor tissue contains various cell types such as cancer fibroblast cells, immune cells, blood vessel cells and others. The PLK-1 gene may be mutated in all these cell types; however, the PLK-1 gene only shows a relatively high expression level in cancer cells, and thus the mutation ratio of the tumor tissues does not simply equal to the mutation ratio of the cancer cells, nor does it correlate with the PLK-1 protein level in the tumor tissue. (4) Drug administration and tissue resection are not always consistent in mutation detection and protein-level analysis.
Discussion
Efficient intracellular Cas9-sgRNA plasmid in vivo delivery is still challenged in its application. Herein, we investigate a general strategy for delivering Cas9-sgRNA plasmid by making use of the core–shell PLNPs that tightly packed the plasmid as a core and encapsulated the core with PEG-modified lipids. The Cas9-sgRNA plasmid with a size of up to 10 707 bp was packed into the PLNPs (Figure 1d), providing the prerequisites for effective transfection. In fact, to achieve effective transfection for the CRISPR/Cas9 system, we screened 56 different vehicle candidates, including various inorganic nanoparticles, polymers and some commercial reagents such as Lipofectamine 3000, TransIT-LT1 and Fugene (Supplementary Table S1), but most of them showed very limited transfection efficiency. We speculate that the major reason for this situation is that these vehicles failed to effectively pack the plasmid to a desirable state to facilitate the cellular internalization. Given the present knowledge about the CRISPR/Cas9 system, the reduction of the length of the plasmid may compromise its function. The best strategy is to minimize the volume of the plasmid without shortening the DNA sequence. In this study, we used chondroitin to endow the Cas9/sgRNA-fused plasmid DNA with more capability to attach to protamine that served as a ‘seed’ to condense the Cas9/sgRNA-fused plasmid DNA to form a compact core (DNA-a). The complex as a whole shows a negative charge that will hinder its internalization through the cell membrane with the same charge. The encapsulation of the cationic lipids (DOTAP, DOPE and cholesterol) on the core (DNA-a) dramatically promoted the transfection of the plasmid (Figure 3) and tumor therapy (Figure 4a). The cationic lipids are supposed to easily access the cell membrane through electrostatic interactions and merge with the cell membrane and facilitate the endocytosis of the Cas9-sgRNA plasmid DNA. However, the cationic lipids may be cytotoxic and may interact nonspecifically with serum or extracellular matrix, thus reducing cellular internalization. The further modification of DSPE-PEG on the surface of the cationic lipids significantly promoted the tumor therapy (Figure 4). Thus, our strategy rationally solved the series of problems in the delivery of CRISPR/Cas9 system: the volume of the plasmid, the penetration through the cell membrane, cytotoxicity and nonspecific interactions.
The in vitro experiments demonstrated that compared with the commercial reagent Lipofectamine 2000, our approach yielded a much higher transfection efficiency (Figure 3). The in vivo experiments showed that our approach was superior over Lipofectamine 2000 transduction (Supplementary Figure S11) or hydrodynamic injection.25, 26 Furthermore, the latter is not suitable for clinical therapy for solid tumors in the human body because hydrodynamic injection is performed by injecting the sample with a high speed in the blood vessel that has many potential risks.
In our experiment, the tumor inhibition induced by the CRISPR/Cas9 system was significantly better than that of siRNA (Figure 4). There may be three reasons for this result. (1) The PLK-1 gene could be completely silenced when a cell is successfully transfected by the CRISPR/Cas9 system and edited, whereas RNA interference could only cause transient gene silencing at the posttranscriptional level.27 (2) The off-target effect of the CRISPR/Cas9 system is justified to be lower than that of the RNA interference-based silencing.28 (3) Unmodified siRNA is more vulnerable to degradation, especially in the complicated in vivo environment, whereas the Cas9-sgRNA in our approach was highly condensed and encapsulated by the lipids that are resistant to enzymatic degradation. Oncogenes are usually overexpressed in various cancers. The CRISPR/Cas9 system, as a highly efficient gene editing toolbox, can completely knock out oncogenes and induce the cancer cell apoptosis when successfully transduced. Thus, CRISPR-based tumor therapy is promising.
Conclusions
We established a novel PLNP-based approach to deliver a CRISPR/Cas9 system and treated melanoma with a much higher efficiency than commercial transfection reagents (Lipofectamine 2000) both in vitro and in vivo. Compared with viral and/or hydrodynamic injection-based gene delivery methods, our approach is broadly applicable with high safety. Because of the powerful gene-editing ability of the CRISPR/Cas9 system and the excellent gene delivery capability of the PLNPs, we believe that our approach is promising in treating a wide range of gene-related diseases such as cancers, heritable diseases and infectious diseases.
References
Hsu, P. D., lander, E. S. & Zhang, F. Development and applications of CRISPR-Cas9 for genome engineering. Cell 157, 1262–1278 (2014).
Wang, T., Wei, J. J., Sabatini, D. M. & Lander, E. S. Genetic screens in human cells using the CRISPR-Cas9 system. Science 343, 80–84 (2014).
Chang, N., Sun, C., Gao, L., Zhu, D., Xu, X., Zhu, X., Xiong, J. W. & Xi, J. J. Genome editing with RNA-guided Cas9 nuclease in zebrafish embryos. Cell Res. 23, 465–472 (2013).
Xiao, A., Wang, Z., Hu, Y., Wu, Y., Luo, Z., Yang, Z., Zu, Y., Li, W., Huang, P. & Tong, X. Chromosomal deletions and inversions mediated by TALENs and CRISPR/Cas in zebrafish. Nucleic Acids Res. 41, 141 (2013).
Qi, L. S., Larson, M. H., Gilbert, L. A., Doudna, J. A., Weissman, J. S., Arkin, A. P. & Lim, W. A. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152, 1173–1183 (2013).
Mali, P., Yang, L., Esvelt, K. M., Aach, J., Guell, M., DiCarlo, J. E., Norville, J. E. & Church, G. M. RNA-guided human genome engineering via Cas9. Science 339, 823–826 (2013).
Ran, F. A., Cong, L., Yan, W. X., Scott, D. A., Gootenberg, J. S., Kriz, A. J., Zetsche, B., Shalem, O., Wu, X. & Makarova, K. S. In vivo genome editing using Staphylococcus aureus Cas9. Nature 520, 186–191 (2015).
Platt, R. J., Chen, S., Zhou, Y., Yim, M. J., Swiech, L., Kempton, H. R., Dahlman, J. E., Parnas, O., Eisenhaure, T. M. & Jovanovic, M. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell 159, 440–455 (2014).
Nault, J. C., Datta, S., Imbeaud, S., Franconi, A., Mallet, M., Couchy, G., Letouzé, E., Pilati, C., Verret, B., Blanc, J. F., Balabaud, C., Calderaro, J., Laurent, A., Letexier, M., Bioulac-Sage, P., Calvo, F. & Zucman-Rossi, J. Recurrent AAV2-related insertional mutagenesis in human hepatocellular carcinomas. Nat. Genet. 47, 1187–1193 (2015).
Sun, W., Ji, W., Hall, J. M., Hu, Q., Wang, C., Beisel, C. L. & Gu, Z. Self-assembled DNA nanoclews for the efficient delivery of CRISPR–Cas9 for genome editing. Angew. Chem. Int. Ed. 54, 12029–12033 (2015).
Gilleron, J., Querbes, W., Zeigerer, A., Borodovsky, A., Marsico, G., Schubert, U., Manygoats, K., Seifert, S., Andree, C. & Stöter, M. Image-based analysis of lipid nanoparticle-mediated siRNA delivery, intracellular trafficking and endosomal escape. Nat. Biotechnol. 31, 638–646 (2013).
Slingerland, M., Guchelaar, H. J. & Gelderblom, H. Liposomal drug formulations in cancer therapy: 15 years along the road. Drug Discov. Today 17, 160–166 (2012).
Zhang, L., Feng, Q., Wang, J., Sun, J., Shi, X. & Jiang, X. Microfluidic synthesis of rigid nanovesicles for hydrophilic reagents delivery. Angew. Chem. Int. Ed. 54, 3952–3956 (2015).
Sun, J. S., Zhang, L., Wang, J. D., Feng, Q., Liu, D. B., Yin, Q. F., Xu, D. Y., Wei, Y. J., Ding, B. Q., Shi, X. H. & Jiang, X. Y. Tunable rigidity of (polymeric core)-(lipid shell) nanoparticles for regulated cellular uptake. Adv. Mater. 27, 1402–1407 (2015).
Zhang, L., Feng, Q., Wang, J. L., Zhang, S., Ding, B. Q., Wei, Y. J., Dong, M. D., Ryu, J. Y., Yoon, T. Y., Shi, X. H., Sun, J. S. & Jiang, X. Y. Microfluidic synthesis of hybrid nanoparticles with controlled lipid layers: understanding flexibility-regulated cell-nanoparticle interaction. ACS Nano 9, 9912–9921 (2015).
Meng, H., Wang, M., Liu, H., Liu, X., Situ, A., Wu, B., Ji, Z., Chang, C. H. & Nel, A. E. Use of a lipid-coated mesoporous silica nanoparticle platform for synergistic gemcitabine and paclitaxel delivery to human pancreatic cancer in mice. ACS Nano 9, 3540–3557 (2015).
Gao, W., Vecchio, D., Li, J., Zhu, J., Zhang, Q., Fu, V., Li, J., Thamphiwatana, S., Lu, D. & Zhang, L. Hydrogel containing nanoparticle-stabilized liposomes for topical antimicrobial delivery. ACS Nano 8, 2900–2907 (2014).
Zuris, J. A., Thompson, D. B., Shu, Y., Guilinger, J. P., Bessen, J. L., Hu, J. H., Maeder, M. L., Joung, J. K., Chen, Z. Y. & Liu, D. R. Cationic lipid-mediated delivery of proteins enables efficient protein-based genome editing in vitro and in vivo. Nat. Biotechnol. 33, 73–80 (2014).
Tseng, Y. C., Mozumdar, S. & Huang, L. Lipid-based systemic delivery of siRNA. Adv. Drug. Deliv. Rev. 61, 721–731 (2009).
Immordino, M. L., Dosio, F. & Cattel, L. Stealth liposomes: review of the basic science, rationale, and clinical applications, existing and potential. Int. J. Nanomed. 1, 297–315 (2006).
Romberg, B., Hennink, W. E. & Storm, G. Sheddable coatings for long-circulating nanoparticles. Pharm. Res. 25, 55–71 (2008).
Feng, Q., Yu, M. Z., Wang, J. C., Hou, W. J., Gao, L. Y., Ma, X. F., Pei, X. W., Niu, Y. J., Liu, X. Y. & Qiu, C. Synergistic inhibition of breast cancer by co-delivery of VEGF siRNA and paclitaxel via vapreotide-modified core-shell nanoparticles. Biomaterials 35, 5028–5038 (2014).
Spänkuch-Schmitt, B., Bereiter-Hahn, J., Kaufmann, M. & Strebhardt, K. Effect of RNA silencing of polo-like kinase-1 (PLK1) on apoptosis and spindle formation in human cancer cells. J. Natl. Cancer Inst. 94, 1863–1877 (2002).
Zhang, L., Zheng, W., Tang, R., Wang, N., Zhang, W. & Jiang, X. Gene regulation with carbon-based siRNA conjugates for cancer therapy. Biomaterials 104, 269–278 (2016).
Xue, W., Chen, S., Yin, H., Tammela, T., Papagiannakopoulos, T., Joshi, N. S., Cai, W., Yang, G., Bronson, R. & Crowley, D. G. CRISPR-mediated direct mutation of cancer genes in the mouse liver. Nature 514, 380–384 (2014).
Yin, H., Xue, W., Chen, S., Bogorad, R. L., Benedetti, E., Grompe, M., Koteliansky, V., Sharp, P. A., Jacks, T. & Anderson, D. G. Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype. Nat. Biotechnol. 32, 551–553 (2014).
Shalem, O., Sanjana, N. E., Hartenian, E., Shi, X., Scott, D. A., Mikkelsen, T. S., Heckl, D., Ebert, B. L., Root, D. E. & Doench, J. G. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343, 84–87 (2014).
Heintze, J., Luft, C. & Ketteler, R. A CRISPR CASe for high-throughput silencing. Front. Genet. 4, 193 (2013).
Acknowledgements
We appreciate the support of National Nature Science Foundation of China (81361140345, 31470911, 81673039), Chinese Academy of Sciences (XDA09030305, XDA09030307) and the CAS/SAFEA International Partnership Program for Creative Research Teams.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information accompanies the paper on the NPG Asia Materials website
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Zhang, L., Wang, P., Feng, Q. et al. Lipid nanoparticle-mediated efficient delivery of CRISPR/Cas9 for tumor therapy. NPG Asia Mater 9, e441 (2017). https://doi.org/10.1038/am.2017.185
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/am.2017.185
This article is cited by
-
Nanomaterials for intelligent CRISPR-Cas tools: improving environment sustainability
Environmental Science and Pollution Research (2024)
-
Application of CRISPR/Cas system in optimizing nutrients and anti-nutrients content in fruits
Vegetos (2023)
-
Recent advances in the delivery and applications of nonviral CRISPR/Cas9 gene editing
Drug Delivery and Translational Research (2023)
-
Nanoparticles-mediated CRISPR-Cas9 gene therapy in inherited retinal diseases: applications, challenges, and emerging opportunities
Journal of Nanobiotechnology (2022)
-
BSA-PEI Nanoparticle Mediated Efficient Delivery of CRISPR/Cas9 into MDA-MB-231 Cells
Molecular Biotechnology (2022)