Abstract
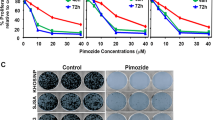
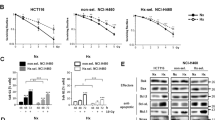
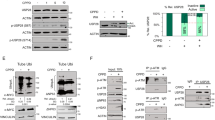
The inhibitor of apoptosis wild-type survivin is a multifunctional protein that suppresses apoptosis and regulates cell cycle progression. An association between wild-type survivin expression and radiosensitivity has been described in different tumor cells. The effects of siRNA-induced knockdown of wild-type survivin and survivin-splice variants survivin-2B and survivin-Δ3 were investigated under normoxic and hypoxic conditions in the human sarcoma cell line US 8–93 (mutant p53). Inhibition of the survivin isoforms by siRNA resulted in a decrease of target mRNA down to 14–70% compared to cells treated with control siRNA independent of the oxygen level. The mRNA expression of survivin isoforms was decreased by the factor of 1–12 when the cells were cultivated under hypoxic conditions. Moreover, the knockdown of wild-type survivin reduced colony formation independent of oxygen concentration down to 70% and induced formation of polyploid cells. Less reduction of plating efficiency was observed after specific knockdown of survivin-2B and survivin-Δ3 under hypoxic or normoxic conditions. A knockdown of wild-type survivin, survivin-Δ3 and survivin-2B isoforms in combination with irradiation caused no radiosensitization in cell line US 8–93, neither under hypoxic nor under normoxic conditions tested in the colony-forming assay. However, knockdown of wild-type survivin caused radiosensitization in the megacolony assay.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Abbreviations
- siRNA:
-
small interfering RNA
References
Altieri DC . Targeted therapy by disabling crossroad signaling networks: the survivin paradigm. Mol Cancer Ther 2006; 5: 478–482.
Altieri DC . Validating survivin as a cancer therapeutic target. Nat Rev Cancer 2003; 3: 46–54.
Kappler M, Kohler T, Kampf C, Diestelkotter P, Wurl P, Schmitz M et al. Increased survivin transcript levels: an independent negative predictor of survival in soft tissue sarcoma patients. Int J Cancer 2001; 95: 360–363.
Kappler M, Kotzsch M, Bartel F, Fussel S, Lautenschlager C, Schmidt U et al. Elevated expression level of survivin protein in soft-tissue sarcomas is a strong independent predictor of survival. Clin Cancer Res 2003; 9: 1098–1104.
Chen J, Wu W, Tahir SK, Kroeger PE, Rosenberg SH, Cowsert LM et al. Down-regulation of survivin by antisense oligonucleotides increases apoptosis, inhibits cytokinesis and anchorage-independent growth. Neoplasia 2000; 2: 235–241.
O'Connor DS, Schechner JS, Adida C, Mesri M, Rothermel AL, Li F et al. Control of apoptosis during angiogenesis by survivin expression in endothelial cells. Am J Pathol 2000; 156: 393–398.
Uren AG, Wong L, Pakusch M, Fowler KJ, Burrows FJ, Vaux DL et al. Survivin and the inner centromere protein INCENP show similar cell-cycle localization and gene knockout phenotype. Curr Biol 2000; 10: 1319–1328.
Reed JC . The Survivin saga goes in vivo. J Clin Invest 2001; 108: 965–969.
Marusawa H, Matsuzawa S, Welsh K, Zou H, Armstrong R, Tamm I et al. HBXIP functions as a cofactor of survivin in apoptosis suppression. EMBO J 2003; 22: 2729–2740.
Dohi T, Beltrami E, Wall NR, Plescia J, Altieri DC . Mitochondrial survivin inhibits apoptosis and promotes tumorigenesis. J Clin Invest 2004; 114: 1117–1127.
Hoffman WH, Biade S, Zilfou JT, Chen J, Murphy M . Transcriptional repression of the anti-apoptotic survivin gene by wild type p53. J Biol Chem 2002; 277: 3247–3257.
Mirza A, McGuirk M, Hockenberry TN, Wu Q, Ashar H, Black S et al. Human survivin is negatively regulated by wild-type p53 and participates in p53-dependent apoptotic pathway. Oncogene 2002; 21: 2613–2622.
Zhou M, Gu L, Li F, Zhu Y, Woods WG, Findley HW . DNA damage induces a novel p53-survivin signaling pathway regulating cell cycle and apoptosis in acute lymphoblastic leukemia cells. J Pharmacol Exp Ther 2002; 303: 124–131.
Zhu N, Gu L, Findley HW, Li F, Zhou M . An alternatively spliced survivin variant is positively regulated by p53 and sensitizes leukemia cells to chemotherapy. Oncogene 2004; 23: 7545–7551.
Kappler M, Bache M, Bartel F, Kotzsch M, Panian M, Wurl P et al. Knockdown of survivin expression by small interfering RNA reduces the clonogenic survival of human sarcoma cell lines independently of p53. Cancer Gene Ther 2004; 11: 186–193.
Li F, Ackermann EJ, Bennett CF, Rothermel AL, Plescia J, Tognin S et al. Pleiotropic cell-division defects and apoptosis induced by interference with survivin function. Nat Cell Biol 1999; 1: 461–466.
Kallio MJ, Nieminen M, Eriksson JE . Human inhibitor of apoptosis protein (IAP) survivin participates in regulation of chromosome segregation and mitotic exit. FASEB J 2001; 15: 2721–2723.
Carvalho A, Carmena M, Sambade C, Earnshaw WC, Wheatley SP . Survivin is required for stable checkpoint activation in taxol-treated HeLa cells. J Cell Sci 2003; 116: 2987–2998.
Lens SM, Wolthuis RM, Klompmaker R, Kauw J, Agami R, Brummelkamp T et al. Survivin is required for a sustained spindle checkpoint arrest in response to lack of tension. EMBO J 2003; 22: 2934–2947.
Williams NS, Gaynor RB, Scoggin S, Verma U, Gokaslan T, Simmang C et al. Identification and validation of genes involved in the pathogenesis of colorectal cancer using cDNA microarrays and RNA interference. Clin Cancer Res 2003; 9: 931–946.
Asanuma K, Moriai R, Yajima T, Yagihashi A, Yamada M, Kobayashi D et al. Survivin as a radioresistance factor in pancreatic cancer. Jpn J Cancer Res 2000; 91: 1204–1209.
Pennati M, Binda M, Colella G, Folini M, Citti L, Villa R et al. Radiosensitization of human melanoma cells by ribozyme-mediated inhibition of survivin expression. J Invest Dermatol 2003; 120: 648–654.
Rodel C, Haas J, Groth A, Grabenbauer GG, Sauer R, Rodel F . Spontaneous and radiation-induced apoptosis in colorectal carcinoma cells with different intrinsic radiosensitivities: survivin as a radioresistance factor. Int J Radiat Oncol Biol Phys 2003; 55: 1341–1347.
Rodel F, Hoffmann J, Distel L, Herrmann M, Noisternig T, Papadopoulos T et al. Survivin as a radioresistance factor, and prognostic and therapeutic target for radiotherapy in rectal cancer. Cancer Res 2005; 65: 4881–4887.
Chakravarti A, Zhai GG, Zhang M, Malhotra R, Latham DE, Delaney MA et al. Survivin enhances radiation resistance in primary human glioblastoma cells via caspase-independent mechanisms. Oncogene 2004; 23: 7494–7506.
Lu B, Mu Y, Cao C, Zeng F, Schneider S, Tan J et al. Survivin as a therapeutic target for radiation sensitization in lung cancer. Cancer Res 2004; 64: 2840–2845.
Asanuma K, Kobayashi D, Furuya D, Tsuji N, Yagihashi A, Watanabe N . A role for survivin in radioresistance of pancreatic cancer cells. Jpn J Cancer Res 2002; 93: 1057–1062.
Kappler M, Taubert H, Bartel F, Blumke K, Panian M, Schmidt H et al. Radiosensitization, after a combined treatment of survivin siRNA and irradiation, is correlated with the activation of caspases 3 and 7 in a wt-p53 sarcoma cell line, but not in a mt-p53 sarcoma cell line. Oncol Rep 2005; 13: 167–172.
Li F, Ling X . Survivin study: an update of ‘what is the next wave’? J Cell Physiol 2006; 208: 476–486.
Mahotka C, Wenzel M, Springer E, Gabbert HE, Gerharz CD . Survivin-deltaEx3 and survivin-2B: two novel splice variants of the apoptosis inhibitor survivin with different antiapoptotic properties. Cancer Res 1999; 59: 6097–6102.
Caldas H, Honsey LE, Altura RA . Survivin 2alpha: a novel Survivin splice variant expressed in human malignancies. Mol Cancer 2005; 4: 11.
Badran A, Yoshida A, Ishikawa K, Goi T, Yamaguchi A, Ueda T et al. Identification of a novel splice variant of the human anti-apoptopsis gene survivin. Biochem Biophys Res Commun 2004; 314: 902–907.
Li F . Role of survivin and its splice variants in tumorigenesis. Br J Cancer 2005; 92: 212–216.
Mahotka C, Liebmann J, Wenzel M, Suschek CV, Schmitt M, Gabbert HE et al. Differential subcellular localization of functionally divergent survivin splice variants. Cell Death Differ 2002; 9: 1334–1342.
Caldas H, Jiang Y, Holloway MP, Fangusaro J, Mahotka C, Conway EM et al. Survivin splice variants regulate the balance between proliferation and cell death. Oncogene 2005; 24: 1994–2007.
Noton EA, Colnaghi R, Tate S, Starck C, Carvalho A, Ko FP et al. Molecular analysis of survivin isoforms: evidence that alternatively spliced variants do not play a role in mitosis. J Biol Chem 2006; 281: 1286–1295.
Taubert H, Kappler M, Bache M, Bartel F, Kohler T, Lautenschlager C et al. Elevated expression of survivin-splice variants predicts a poor outcome for soft-tissue sarcomas patients. Oncogene 2005; 24: 5258–5261.
Yang L, Cao Z, Li F, Post DE, Van Meir EG, Zhong H et al. Tumor-specific gene expression using the survivin promoter is further increased by hypoxia. Gene Therapy 2004; 11: 1215–1223.
Mesri M, Morales-Ruiz M, Ackermann EJ, Bennett CF, Pober JS, Sessa WC et al. Suppression of vascular endothelial growth factor-mediated endothelial cell protection by survivin targeting. Am J Pathol 2001; 158: 1757–1765.
Bache M, Holzapfel D, Kappler M, Holzhausen HJ, Taubert H, Dunst J et al. Survivin protein expression and hypoxia in advanced cervical carcinoma of patients treated by radiotherapy. Gynecol Oncol 2007; 104: 139–144.
Taubert H, Schmidt H, Würl P, Hinze R, Meye A, Bache M et al. Morphological and molecular characterization of an undifferentiated soft tissue sarcoma cell line and derivative clones. Int J Oncol 1997; 25: 521–526.
Kummermehr J, Malinen E, Freykowski S, Sund M, Trott KR . The influence of autologous tumor fibroblasts on the radiosensitivity of squamous cell carcinoma megacolonies. Int J Radiat Oncol Biol Phys 2001; 50: 229–237.
Nordsmark M, Alsner J, Keller J, Nielsen OS, Jensen OM, Horsman MR et al. Hypoxia in human soft tissue sarcomas: adverse impact on survival and no association with p53 mutations. Br J Cancer 2001; 84: 1070–1075.
Shintani K, Matsumine A, Kusuzaki K, Matsubara T, Satonaka H, Wakabayashi T et al. Expression of hypoxia-inducible factor (HIF)-1alpha as a biomarker of outcome in soft-tissue sarcomas. Virchows Arch 2006; 449: 673–681.
Ueda Y, Nakagawa T, Kubota T, Ido K, Sato K . Glioma cells under hypoxic conditions block the brain microvascular endothelial cell death induced by serum starvation. J Neurochem 2005; 95: 99–110.
Peng XH, Karna P, Cao Z, Jiang BH, Zhou M, Yang L . Cross-talk between epidermal growth factor receptor and hypoxia-inducible factor-1alpha signal pathways increases resistance to apoptosis by up-regulating survivin gene expression. J Biol Chem 2006; 281: 25903–25914.
Chang Q, Qin R, Huang T, Gao J, Feng Y . Effect of antisense hypoxia-inducible factor 1alpha on progression, metastasis, and chemosensitivity of pancreatic cancer. Pancreas 2006; 32: 297–305.
Kwon DS, Kwon CH, Kim JH, Woo JS, Jung JS, Kim YK . Signal transduction of MEK/ERK and PI3K/Akt activation by hypoxia/reoxygenation in renal epithelial cells. Eur J Cell Biol 2006; 85: 1189–1199.
Li F . Survivin study: what is the next wave? J Cell Physiol 2003; 197: 8–29.
Acknowledgements
We thank our colleagues from the Department of Radiotherapy and the Institute of Pathology for contributing to this study and for their continuous support. We also thank Kathrin Spröte and Ute Rolle (Department of Radiotherapy, Martin-Luther-University Halle-Wittenberg) for their excellent technical assistance. We are very grateful to Uta Schramm und Dr Thomas Klapperstück (Department of Dermatology, Martin-Luther-University Halle-Wittenberg) for helping at the flow cytometric analyses. Furthermore, we thank Dr Axel Meye for helpful discussion. Major parts of the results were achieved during participation in the MSc course in Radiation Biology of University of College London (2005–2006) organized by Professor Klaus-R Trott and Dr Kevin M Prise. This work was supported by the State of Saxony-Anhalt (Grant number: 3584AB/1104 M) and by the Wilhelm Roux-Program of BMBF/NBL3 (FKZ: 16/18 and FKZ: 15/18).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kappler, M., Rot, S., Taubert, H. et al. The effects of knockdown of wild-type survivin, survivin-2B or survivin-Δ3 on the radiosensitization in a soft tissue sarcoma cells in vitro under different oxygen conditions. Cancer Gene Ther 14, 994–1001 (2007). https://doi.org/10.1038/sj.cgt.7701090
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.cgt.7701090
Keywords
This article is cited by
-
Normoxic accumulation of HIF1α is associated with glutaminolysis
Clinical Oral Investigations (2017)
-
Statistical and radiobiological analysis of the so-called thyroid stunning
EJNMMI Research (2015)
-
Clinical significance of hypoxia-inducible factor 1 and VEGF-A in osteosarcoma
International Journal of Clinical Oncology (2015)
-
Increased betulinic acid induced cytotoxicity and radiosensitivity in glioma cells under hypoxic conditions
Radiation Oncology (2011)
-
HIF-1α inhibition by siRNA or chetomin in human malignant glioma cells: effects on hypoxic radioresistance and monitoring via CA9 expression
BMC Cancer (2010)