Abstract
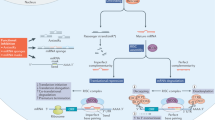
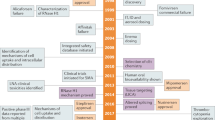
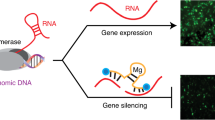
The availability of the human genome sequence has revolutionized the strategy of employing nucleic acids with sequences complementary to specific target genes to improve drug discovery and target validation. Development of sequence-specific DNA or RNA analogs that can block the activity of selected single-stranded genetic sequences offers the possibility of rational design with high specificity, lacking in many current drug treatments for various diseases including cancer, at relatively inexpensive costs. Antisense technology is one such example that has shown promising results and boasts of yielding the only approved drug to date in the genomics field. However, in vivo delivery issues have yet to be completely overcome for widespread clinical applications. In contrast to antisense oligonucleotides, the mechanism of silencing an endogenous gene by the introduction of a homologous double-stranded RNA (dsRNA), transgene or virus is called post-transcriptional gene silencing (PTGS) or RNA interference. PTGS is a natural mechanism whereby metazoan cells suppress expansion of genes when they come across dsRNA molecules with the same sequence. Short interfering RNA is currently the fastest growing sector of this antigene field for target validation and therapeutic applications. Although, in theory, the development of genomics-based agents to inhibit gene expression is simple and straightforward, the fundamental concern relies upon the capacity of the oligonucleotide to gain access to the target RNA. This paper summarizes the advances in the last decade in the field of PTGS using RNA interference approaches and provides relevant comparisons with other oligonucleotide-based approaches with a specific focus on oncology applications.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
Jemal A, Murray T, Ward E, Samuels A, Tiwari RC, Ghafoor A et al. Cancer statistics, 2005. CA Cancer J Clin 2005; 55: 10–30.
Scanlon KJ . Anti-genes: siRNA, ribozymes and antisense. Curr Pharm Biotechnol 2004; 5: 415–420.
Citti L, Rainaldi G . Synthetic hammerhead ribozymes as therapeutic tools to control disease genes. Curr Gene Ther 2005; 5: 11–24.
Frank-Kamenetskii MD, Mirkin SM . Triplex DNA structures. Annu Rev Biochem 1995; 64: 65–95.
Hoyne PR, Edwards LM, Viari A, Maher III LJ . Searching genomes for sequences with the potential to form intrastrand triple helices. J Mol Biol 2000; 302: 797–809.
Trojan LA, Kopinski P, Wei MX, Ly A, Glogowska A, Czarny J et al. IGF-I: from diagnostic to triple-helix gene therapy of solid tumors. Acta Biochim Pol 2002; 49: 979–990.
Dean DA . Peptide nucleic acids: versatile tools for gene therapy strategies. Adv Drug Deliv Rev 2000; 44: 81–95.
Nielsen PE . Targeting double stranded DNA with peptide nucleic acid (PNA). Curr Med Chem 2001; 8: 545–550.
Braasch DA, Corey DR . Novel antisense and peptide nucleic acid strategies for controlling gene expression. Biochemistry 2002; 41: 4503–4510.
Helene C, Toulme JJ . Specific regulation of gene expression by antisense, sense and antigene nucleic acids. Biochim Biophys Acta 1990; 1049: 99–125.
Dias N, Stein CA . Antisense oligonucleotides: basic concepts and mechanisms. Mol Cancer Ther 2002; 1: 347–355.
Paterson BM, Roberts BE, Kuff EL . Structural gene identification and mapping by DNA–mRNA hybrid-arrested cell-free translation. Proc Natl Acad Sci USA 1977; 74: 4370–4374.
Zamecnik PC, Stephenson ML . Inhibition of Rous sarcoma virus replication and cell transformation by a specific oligodeoxynucleotide. Proc Natl Acad Sci USA 1978; 75: 280–284.
Whitesell L, Rosolen A, Neckers LM . In vivo modulation of N-myc expression by continuous perfusion with an antisense oligonucleotide. Antisense Res Dev 1991; 1: 343–350 (Orr RM, 2001).
Lebedeva I, Stein CA . Antisense oligonucleotides: promise and reality. Annu Rev Pharmacol Toxicol 2001; 41: 403–419.
Crooke ST . Progress in antisense technology. Annu Rev Med 2004; 55: 61–95.
Orr RM . Technology evaluation: fomivirsen, Isis Pharmaceuticals Inc/CIBA vision. Curr Opin Mol Ther 2001; 3: 288–294.
Krieg AM . Mechanisms and applications of immune stimulatory CpG oligodeoxynucleotides. Biochim Biophys Acta 1999; 1489: 107–116.
Henry SP, Novotny W, Leeds J, Auletta C, Kornbrust DJ . Inhibition of coagulation by a phosphorothioate oligonucleotide. Antisense Nucleic Acid Drug Dev 1997; 7: 503–510.
Iversen PL, Cornish KG, Iversen LJ, Mata JE, Bylund DB . Bolus intravenous injection of phosphorothioate oligonucleotides causes hypotension by acting as alpha(1)-adrenergic receptor antagonists. Toxicol Appl Pharmacol 1999; 160: 289–296.
Burgess TL, Fisher EF, Ross SL, Bready JV, Qian YX, Bayewitch LA et al. The antiproliferative activity of c-myb and c-myc antisense oligonucleotides in smooth muscle cells is caused by a nonantisense mechanism. Proc Natl Acad Sci USA 1995; 92: 4051–4055.
Copple BL, Gmeiner WM, Iversen PL . Reaction between metabolically activated acetaminophen and phosphorothioate oligonucleotides. Toxicol Appl Pharmacol 1995; 133: 53–63.
Mata JE, Bishop MR, Tarantolo SR, Angel CR, Swanson SA, Iversen PL . Evidence of enhanced iron excretion during systemic phosphorothioate oligodeoxynucleotide treatment. J Toxicol Clin Toxicol 2000; 38: 383–387.
Henry SP, Geary RS, Yu R, Levin AA . Drug properties of second-generation antisense oligonucleotides: how do they measure up to their predecessors? Curr Opin Investig Drugs 2001; 2: 1444–1449.
Kurreck J . Antisense technologies. Improvement through novel chemical modifications. Eur J Biochem 2003; 270: 1628–1644 (review).
Summerton J, Weller D . Morpholino antisense oligomers: design, preparation and properties. Antisense Nucleic Acid Drug Dev 1997; 7: 187–195.
Arora V, Devi GR, Iversen PL . Neutrally charged phosphorodiamidate morpholino antisense oligomers: uptake, efficacy and pharmacokinetics. Curr Pharm Biotechnol 2004; 5: 431–439.
Hudziak RM, Barofsky E, Barofsky DF, Weller DL, Huang SB, Weller DD . Resistance of morpholino phosphorodiamidate oligomers to enzymatic degradation. Antisense Nucleic Acid Drug Dev 1996; 6: 267–272.
Taylor MF, Weller DD, Kobzik L . Effect of TNF-alpha antisense on cytokine production by primary murine alveolar macrophages. Antisense Nucleic Acid Drug Dev 1998; 8: 199–205.
Lacerra G, Sierakowska H, Carestia C, Fucharoen S, Summerton J, Weller D et al. Restoration of hemoglobin-A synthesis in erythroid cells from peripheral blood of thalassemic patients. Proc Natl Acad Sci USA 2000; 97: 9591–9596.
Arora V, Knapp DC, Smith BL, Statdfield ML, Stein DA, Reddy MT et al. c-myc antisense rat liver regeneration and indicates role for c-myc in regulating cytochrome P-450 3A activity. J Pharmacol Expt Ther 2000; 292: 921–928.
Nasevicius A, Ekker SC . Effective targeted gene knockout in zebrafish. Nat Genet 2000; 26: 216–220.
Qin G, Taylor M, Ning YY, Iversen P, Kobzik L . In vivo evaluation of a morpholine oligomer directed against TNF-α. Antisense Nucleic Acid Drug Dev 2000; 10: 11–16.
Arora V, Cate ML, Ghosh C, Iversen PL . Phosphorodiamidate Morpholino antisense oligomers inhibit expression of human cytochrome P450 3A4 and alter selected drug metabolism. Drug Metab Disposition 2002; 30: 757–762.
Arora V, Knapp DC, Reddy MT, Weller DD, Iversen PL . Bioavailability and efficacy of antisense morpholino oligomers targeted to c-Myc and cytochrome P-450 3A2 following oral administration in rats. J Pharm Sci 2002; 91: 1009–1018.
Kipshidze N, Keane E, Stein D, Chawla P, Skrinska V, Shankar LR et al. Local delivery of c-myc neutrally charged antisense oligonucleotides with transport catheter inhibits myointimal hyperplasia and positively affects vascular remodeling in the rabbit balloon injury model. Catheter Cardiovasc Interv 2001; 54: 247–256.
Ricker JL, Mata JE, Iversen PL, Gattone VH . c-myc antisense oligomer treatment meliorates murine infantile polycystic kidney disease. Kidney Int 2001; 61: 125–131.
Hudziak RM, Summerton J, Weller DD, Iversen PL . Antiproliferative effects of steric blocking phosphorodiamidate morpholino antisense agents directed against c-myc. Antisense Nucleic Acid Drug Dev 2000; 10: 163–176.
Devi GR, Oldenkamp JR, London CA, Iversen PL . Inhibition of human chorionic gonadotropin beta-subunit modulates the mitogenic effect of c-myc in human prostate cancer cells. Prostate 2002; 53: 200–210.
Knapp DC, Mata JE, Reddy MT, Devi GR, Iversen PL . Resistance to chemotherapeutic drugs overcome by c-Myc inhibition in a Lewis lung carcinoma murine model. Anticancer Drugs 2003; 14: 39–47.
London CA, Sekhon HS, Arora V, Stein DA, Iversen PL, Devi GR . A novel antisense inhibitor of MMP-9 attenuates angiogenesis, human prostate cancer cell invasion and tumorigenicity. Cancer Gene Ther 2003; 10: 823–832.
Devi GR . Prostate cancer: status of current treatments and emerging antisense-based therapies. Curr Opin Mol Ther 2002; 4: 138–148.
Ko YJ, Devi GR, London CA, Kayas A, Reddy MT, Iversen PL et al. Androgen receptor down-regulation in prostate cancer with phosphorodiamidate morpholino antisense oligomers. J Urol 2004; 172: 1140–1144.
Iversen PL, Arora V, Acker AJ, Mason DH, Devi GR . Efficacy of antisense morpholino oligomer targeted to c-myc in prostate cancer xenograft murine model and a Phase I safety study in humans. Clin Cancer Res 2003; 9: 2510–2519.
Devi GR, Beer TM, Corless CL, Arora V, Weller DL, Iversen PL . In vivo bioavailability and pharmacokinetics of a c-MYC antisense phosphorodiamidate morpholino oligomer, AVI-4126, in solid tumors. Clin Cancer Res 2005; 11: 3930–3938.
Gait MJ . Peptide-mediated cellular delivery of antisense oligonucleotides and their analogues. Cell Mol Life Sci 2003; 60: 844–853.
Moulton HM, Moulton JD . Arginine-rich cell-penetrating peptides with uncharged antisense oligomers. Drug Discov Today 2004; 9: 870.
Moulton HM, Nelson MH, Hatlevig SA, Reddy MT, Iversen PL . Cellular uptake of antisense morpholino oligomers conjugated to arginine-rich peptides. Bioconjug Chem 2004; 15: 290–299.
Cogoni C, Macino G . Post-transcriptional gene silencing across kingdoms. Curr Opin Genet Dev 2000; 10: 638–643.
Napoli C, Lemieux C, Jorgensen R . Introduction of a chimeric chalcone synthase gene into petunia results in reversible co-suppression of homologous genes in trans. Plant Cell 1990; 2: 279–289.
Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC . Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998; 391: 806–811.
Hammond SM, Bernstein E, Beach D, Hannon GJ . An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 2000; 404: 293–296.
Hannon GJ . RNA interference. Nature 2002; 418: 244–251.
Volpe TA, Kidner C, Hall IM, Teng G, Grewal SI, Martienssen RA . Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 2002; 297: 1833–1837.
Reinhart BJ, Bartel DP . Small RNAs correspond to centromere heterochromatic repeats. Science 2002; 297: 1831.
Matzke M, Matzke AJM . RNAi extends its reach. Science 2003; 301: 1060–1061.
Schramke V, Allshire R . Hairpin RNAs and retrotransposon LTRs effect RNAi and chromatin-based gene silencing. Science 2003; 301: 1069–1074.
Voinnet O . Induction and suppression of RNA silencing: insights from viral infections. Nat Rev Genet 2005; 6: 206–220.
Tomari Y, Zamore PD . Perspective: machines for RNAi. Genes Dev 2005; 19: 517–529.
Banan M, Puri N . The ins and outs of RNAi in mammalian cells. Curr Pharm Biotechnol 2004; 5: 441–450.
Lai EC . Micro RNAs are complementary to 3′ UTR sequence motifs that mediate negative post-transcriptional regulation. Nat Genet 2002; 30: 363–364.
Jackson AL, Bartz SR, Schelter J, Kobayashi SV, Burchard J, Mao M et al. Expression profiling reveals off-target gene regulation by RNAi. Nat Biotechnol 2003; 21: 635–637.
Scacheri PC, Rozenblatt-Rosen O, Caplen NJ, Wolfsberg TG, Umayam L, Lee JC et al. Short interfering RNAs can induce unexpected and divergent changes in the levels of untargeted proteins in mammalian cells. Proc Natl Acad Sci USA 2004; 101: 1892–1897.
Jackson AL, Linsley PS . Noise amidst the silence: off-target effects of siRNAs? Trends Genet 2004; 20: 521–524.
Kidner CA, Martienssen RA . The developmental role of microRNA in plants. Curr Opin Plant Biol 2005; 8: 38–44.
Tang G . siRNA and miRNA: an insight into RISCs. Trends Biochem Sci 2005; 30: 106–114.
Cullen BR . Transcription and processing of human microRNA precursors. Mol Cell 2004; 16: 861–865.
Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA 2002; 99: 15524–15529.
Michael MZ, O'Connor SM, van Holst Pellekaan NG, Young GP, James RJ . Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol Cancer Res 2003; 1: 882–891.
Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, Endoh H et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res 2004; 64: 3753–3756.
Metzler M, Wilda M, Busch K, Viehmann S, Borkhardt A . High expression of precursor microRNA-155/BIC RNA in children with Burkitt lymphoma. Genes Chromosomes Cancer 2004; 39: 167–169.
Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci USA 2004; 101: 2999–3004.
Hannon GJ, Rossi JJ . Unlocking the potential of the human genome with RNA interference. Nature 2004; 431: 371–378.
Song E, Lee SK, Dykxhoorn DM, Novina C, Zhang D, Crawford K et al. Sustained small interfering RNA-mediated human immunodeficiency virus type 1 inhibition in primary macrophages. J Virol 2003; 77: 7174–7181.
Song E, Lee SK, Wang J, Ince N, Ouyang N, Min J et al. RNA interference targeting Fas protects mice from fulminant hepatitis. Nat Med 2003; 9: 347–351.
Zentilin L, Giacca M . In vivo transfer and expression of genes coding for short interfering RNAs. Curr Pharm Biotechnol 2004; 5: 341–347.
Brummelkamp T, Bernards R, Agami R . Stable suppression of tumorigenicity by virus-mediated RNA interference. Cancer Cell 2002; 2: 243–247.
Lois C, Refaeli Y, Qin XF, Van Parijs L . Retroviruses as tools to study the immune system. Curr Opin Immunol 2001; 13: 496–504.
Abbas-Terki T, Blanco-Bose W, Deglon N, Pralong W, Aebischer P . Lentiviral-mediated RNA interference. Hum Gene Ther 2002; 13: 2197–2201.
Dirac AM, Bernards R . Reversal of senescence in mouse fibroblasts through lentiviral suppression of p53. J Biol Chem 2003; 278: 11731–11734.
Rubinson DA, Dillon CP, Kwiatkowski AV, Sievers C, Yang L, Kopinja J et al. A lentivirus-based system to functionally silence genes in primary mammalian cells, stem cells and transgenic mice by RNA interference. Nat Genet 2003; 33: 401–406.
Hommel JD, Sears RM, Georgescu D, Simmons DL, DiLeone RJ . Local gene knockdown in the brain using viral-mediated RNA interference. Nat Med 2003; 9: 1539–1544.
Lu S, Cullen BR . Adenovirus VA1 noncoding RNA can inhibit small interfering RNA and microRNA biogenesis. J Virol 2004; 78: 12868–12876.
Uchida H, Tanaka T, Sasaki K, Kato K, Dehari H, Ito Y et al. Adenovirus-mediated transfer of siRNA against survivin induced apoptosis and attenuated tumor cell growth in vitro and in vivo. Mol Ther 2004; 10: 162–171.
Arts GJ, Langemeijer E, Tissingh R, Ma L, Pavliska H, Dokic K et al. Adenoviral vectors expressing siRNAs for discovery and validation of gene function. Genome Res 2003; 13: 2325–2332.
Amalfitano A . Utilization of adenovirus vectors for multiple gene transfer applications. Methods 2004; 33: 173–178.
Amalfitano A . Use of multiply deleted adenovirus vectors to probe adenovirus vector performance and toxicities. Curr Opin Mol Ther 2003; 5: 362–366.
Shen C, Buck AK, Liu X, Winkler M, Reske SN . Gene silencing by adenovirus-delivered siRNA. FEBS Lett 2003; 539: 111–114.
Zhao LJ, Jian H, Zhu H . Specific gene inhibition by adenovirus-mediated expression of small interfering RNA. Gene 2003; 316: 137–141.
Grimm D, Kay MA, Kleinschmidt JA . Helper virus-free, optically controllable, and two-plasmid-based production of adeno-associated virus vectors of serotypes 1 to 6. Mol Ther 2003; 7: 839–850.
Thomas CE, Ehrhardt A, Kay MA . Progress and problems with the use of viral vectors for gene therapy. Nat Rev Genet 2003; 4: 346–358.
Kimchi-Sarfaty C, Gottesman MM . Transduction of multiple cell types using improved conditions for gene delivery and expression of SV40 pseudovirions packaged in vitro. Biotechniques 2004; 37: 270–275.
McCaffrey AP, Meuse L, Pham TT, Conklin DS, Hannon GJ, Kay MA . RNA interference in adult mice. Nature 2002; 418: 38–39.
Sørensen DR, Leirdal M, Sioud M . Gene silencing by systemic delivery of synthetic siRNAs in adult mice. J Mol Biol 2003; 327: 761–766.
Lewis DL, Hagstrom JE, Loomis AG, Wolff JA, Herweijer H . Efficient delivery of siRNA for inhibition of gene expression in postnatal mice. Nat Genet 2002; 32: 107–108.
Braasch DA, Jensen S, Liu Y, Kaur K, Arar K, White MA et al. RNA interference in mammalian cells by chemically-modified RNA. Biochemistry 2003; 42: 7967–7975.
Paddison PJ, Silva JM, Conklin DS, Schlabach M, Li M, Aruleba S et al. A resource for large-scale RNA-interference-based screens in mammals. Nature 2004; 428: 427–431.
Elbashir SM, Martinez J, Patkaniowska A, Lendeckel W, Tuschl T . Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J 2001; 20: 6877–6888.
Shang Y, Brown M . Molecular determinants for the tissue specificity of SERMs. Science 2002; 295: 2465–2468.
Vanhecke D, Janitz M . High-throughput gene silencing using cell arrays. Oncogene 2004; 23: 8353–8358.
Brummelkamp TR, Bernards R, Agami R . A system for stable expression of short interfering RNAs in mammalian cells. Science 2002; 296: 550–553.
Miyagishi M, Taira K . U6 promoter-driven siRNAs with four uridine 3′ overhangs efficiently suppress targeted gene expression in mammalian cells. Nat Biotechnol 2002; 20: 497–500.
Paul CP, Good PD, Winer I, Engelke DR . Effective expression of small interfering RNA in human cells. Nat Biotechnol 2002; 20: 505–508.
Sachse C, Echeverri CJ . Oncology studies using siRNA libraries: the dawn of RNAi-based genomics. Oncogene 2004; 23: 8384–8391.
Grzmil M, Thelen P, Hemmerlein B, Schweyer S, Voigt S, Mury D et al. Bax inhibitor-1 is overexpressed in prostate cancer and its specific down-regulation by RNA interference leads to cell death in human prostate carcinoma cells. Am J Pathol 2003; 163: 543–552.
Yin JQ, Gao J, Shao R, Tian WN, Wang J, Wan Y . siRNA agents inhibit oncogene expression and attenuate human tumor cell growth. J Exp Ther Oncol 2003; 3: 194–204.
Futami T, Miyagishi M, Seki M, Taira K . Induction of apoptosis in HeLa cells with siRNA expression vector targeted against bcl-2. Nucleic Acids Res Suppl 2002; 2: 251–252.
Duxbury MS, Ito H, Benoit E, Zinner MJ, Ashley SW, Whang EE . RNA interference targeting focal adhesion kinase enhances pancreatic adenocarcinoma gemcitabine chemosensitivity. Biochem Biophys Res Commun 2003; 311: 786–792.
Sanceau J, Truchet S, Bauvois B . Matrix metalloproteinase-9 silencing by RNA interference triggers the migratory-adhesive switch in Ewing's sarcoma cells. J Biol Chem 2003; 278: 36537–36546.
Zhang L, Yang N, Mohamed-Hadley A, Rubin SC, Coukos G . Vector-based RNAi, a novel tool for isoform-specific knock-down of VEGF and anti-angiogenesis gene therapy of cancer. Biochem Biophys Res Commun 2003; 303: 1169–1178.
De Schrijver E, Brusselmans K, Heyns W, Verhoeven G, Swinnen JV . RNA interference-mediated silencing of the fatty acid synthase gene attenuates growth and induces morphological changes and apoptosis of LNCaP prostate cancer cells. Cancer Res 2003; 63: 3799–3804.
Nieth C, Priebsch A, Stege A, Lage H . Modulation of the classical multidrug resistance (MDR) phenotype by RNA interference (RNAi). FEBS Lett 2003; 545: 144–150.
Spankuch-Schmitt B, Bereiter-Hahn J, Kaufmann M, Strebhardt K . Effect of RNA silencing of polo-like kinase-1 (PLK1) on apoptosis and spindle formation in human cancer cells. J Natl Cancer Inst 2002; 94: 1863–1877.
Konnikova L, Kotecki M, Kruger MM, Cochran BH . Knockdown of STAT3 expression by RNAi induces apoptosis in astrocytoma cells. BMC Cancer 2003; 3: 23.
Nagy P, Arndt-Jovin DJ, Jovin TM . Small interfering RNAs suppress the expression of endogenous and GFP-fused epidermal growth factor receptor (erbB1) and induce apoptosis in erbB1-overexpressing cells. Exp Cell Res 2003; 285: 39–49.
Zhang M, Zhang X, Bai CX, Chen J, Wei MQ . Inhibition of epidermal growth factor receptor expression by RNA interference in A549 cells. Acta Pharmacol Sin 2004; 25: 61–67.
Li XP, Li G, Peng Y, Kung HF, Lin MC . Suppression of Epstein–Barr virus-encoded latent membrane protein-1 by RNA interference inhibits the metastatic potential of nasopharyngeal carcinoma cells. Biochem Biophys Res Commun 2004; 315: 212–218.
Butz K, Ristriani T, Hengstermann A, Denk C, Scheffner M, Hoppe-Seyler F . siRNA targeting of the viral E6 oncogene efficiently kills human papillomavirus-positive cancer cells. Oncogene 2003; 22: 5938–5945.
Wohlbold L, van der Kuip H, Miething C, Vornlocher HP, Knabbe C, Duyster J et al. Inhibition of bcr-abl gene expression by small interfering RNA sensitizes for imatinib mesylate (STI571). Blood 2003; 102: 2236–2239.
Wilda M, Fuchs U, Wossmann W, Borkhardt A . Killing of leukemic cells with a BCR/ABL fusion gene by RNA interference (RNAi). Oncogene 2002; 21: 5716–5724.
Kosciolek BA, Kalantidis K, Tabler M, Rowley PT . Inhibition of telomerase activity in human cancer cells by RNA interference. Mol Cancer Ther 2003; 2: 209–216.
Persengiev SP, Zhu X, Green M . Nonspecific, concentration-dependant stimulation and repression of mammalian gene expression by small interfering RNAs (siRNAs). RNA 2004; 10: 12–18.
Scacheri PC, Rozenblatt-Rosen O, Caplen NJ, Wolfsberg TG, Umayam L, Lee JC et al. Short interfering RNAs can induce unexpected and divergent changes in the levels of untargeted proteins in mammalian cells. Proc Natl Acad Sci USA 2004; 101: 1892–1897.
Jackson AL, Linsley PS . Noise amidst the silence: off-target effects of siRNAs? Trends Genet 2004; 20: 521–524.
Jackson AL, Bartz SR, Schelter J, Kobayashi SV, Burchard J, Mao M et al. Expression profiling reveals off-target gene regulation by RNAi. Nat Biotechnol 2003; 21: 635–637.
Saxena S, Jonsson ZO, Dutta A . Small RNAs with imperfect match to endogenous mRNA repress translation: implications for off-target activity of small inhibitory RNA in mammalian cells. J Biol Chem 2003; 278: 44312–44319.
Agrawal S, Kandimalla ER . Antisense and siRNA as agonists of Toll-like receptors. Nat Biotechnol 2004; 22: 1533–1537.
Kariko K, Bhuyan P, Capodici J, Weissman D . Small interfering RNAs mediate sequence-independent gene suppression and induce immune activation by signaling through toll-like receptor 3. J Immunol 2004; 172: 6545–6549.
Sledz CA, Holko M, de Veer MJ, Silverman RH, Williams BR . Activation of the interferon system by short-interfering RNAs. Nat Cell Biol 2003; 5: 834–839.
Vester B, Wengel J . LNA (locked nucleic acid): high-affinity targeting of complementary RNA and DNA. Biochemistry 2004; 43: 13233–13241.
Acknowledgements
The field of genomics-based approaches including RNAi is rapidly growing and I apologize to all the authors whose work could not be cited owing to limitations in space and time. I thank Ms Dierdre Shipman for editorial assistance.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Devi, G. siRNA-based approaches in cancer therapy. Cancer Gene Ther 13, 819–829 (2006). https://doi.org/10.1038/sj.cgt.7700931
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.cgt.7700931
Keywords
This article is cited by
-
Clinical translation of gold nanoparticles
Drug Delivery and Translational Research (2023)
-
Investigation of the ionic conditions in SiRNA-mediated delivery through its carriers in the cell membrane: a molecular dynamic simulation
Scientific Reports (2022)
-
Immunomodulation therapy offers new molecular strategies to treat UTI
Nature Reviews Urology (2022)
-
Use of polymeric CXCR4 inhibitors as siRNA delivery vehicles for the treatment of acute myeloid leukemia
Cancer Gene Therapy (2020)
-
Enabling Combinatorial siRNA Delivery against Apoptosis-Related Proteins with Linoleic Acid and α-Linoleic Acid Substituted Low Molecular Weight Polyethylenimines
Pharmaceutical Research (2020)