Abstract
Cotton fibers elongate rapidly after initiation of elongation, eventually leading to the deposit of a large amount of cellulose. To reveal features of cotton fiber cells at the fast elongation and the secondary cell wall synthesis stages, we compared the respective transcriptomes and metabolite profiles. Comparative analysis of transcriptomes by cDNA array identified 633 genes that were differentially regulated during fiber development. Principal component analysis (PCA) using expressed genes as variables divided fiber samples into four groups, which are diagnostic of developmental stages. Similar grouping results are also found if we use non-polar or polar metabolites as variables for PCA of developing fibers. Auxin signaling, wall-loosening and lipid metabolism are highly active during fiber elongation, whereas cellulose biosynthesis is predominant and many other metabolic pathways are downregulated at the secondary cell wall synthesis stage. Transcript and metabolite profiles and enzyme activities are consistent in demonstrating a specialization process of cotton fiber development toward cellulose synthesis. These data demonstrate that cotton fiber cell at a certain stage has its own unique feature, and developmental stages of cotton fiber cells can be distinguished by their transcript and metabolite profiles. During the secondary cell wall synthesis stage, metabolic pathways are streamed into cellulose synthesis.
Similar content being viewed by others
Introduction
Flowering plants produce seeds that are composed of embryo, endosperm and seed coat. The seed coat is developed from integument, the outermost and sporophytic tissue of an ovule. Seed coats of different angiosperm species show variations in coloration and structure. Seeds of cotton (Gossypium spp.) are characterized by their long and single-celled epidermal trichomes, known as cotton fiber. Cotton fiber differentiates from the outer integument 2 to 3 days before anthesis and initiates the outgrowth from the flowering day to three days post-anthesis (DPA). After this initiation stage, fiber cells enter a fast elongation stage until about 20 DPA. At about 15 DPA, by beginning to synthesize a large amount of cellulose and by thickening their cell walls, cotton fibers enter the secondary cell wall synthesis stage. In the final stage (45-60 DPA), the fiber undergoes dehydration and maturation 1, 2.
Although the aforementioned four developmental stages are overlapping, each stage has its own features of physiological and cellular states. In the fast elongation stage, for example, length of a fiber cell may reach 2 cm or longer. Moreover, in the secondary cell wall formation stage, cellulose biosynthesis is so active that cellulose takes more than 95% of the dry weight 2, 3 in mature cotton fiber. Furthermore, growth and development of all fiber cells in a boll are highly synchronous and are not interfered by cell division, providing another advantage for using this system to investigate plant cell polar growth and cellulose biosynthesis.
During fiber elongation, many highly expressed genes are involved in osmosis regulation 4. Antisense suppression of a sucrose synthase (SuSy) gene disrupted fiber elongation, indicating the involvement of SuSy in osmosis regulation 5. Many members of the cell wall-loosening expansin family are highly expressed in elongating fiber cells; examination of the GUS report gene driven by a cotton expansin gene promoter further confirmed its fiber-specific expression pattern 6, 7. In addition, lipid transfer proteins also show higher levels of gene expression in fast elongating fiber cells 8. Actin is also reported to function in the fiber elongation 9.
Cellulose synthesis is a predominant event in fiber cells in the secondary cell wall synthesis stage. After identification of the first plant cellulose synthase (CESA) gene from cotton 10, dimerization of the cellulose synthase subunits via the zinc-binding domains was demonstrated by using cotton fiber. Peroxide, mainly as H2O2, promotes this dimerization and, subsequently, the cellulose synthesis 11, 12. A recent investigation of Arabidopsis thaliana using microarrays led to the identification of genes highly co-expressed with cellulose synthase genes and two mutants, irx8 and irx13, showing irregular xylem phenotypes 13.
Till date, there are more than 200 000 expressed sequence tags (ESTs) of Gossypium deposited in the NCBI database. A rich source of EST and cDNA sequences is invaluable not only for understanding mechanisms regulating cotton fiber development but also for cotton genomics studies and generating new molecular markers for breeding 14, 15, 16, 17, 18. More recently, approximately 180 000 cotton ESTs from more than 30 libraries have been integrated and analyzed through international collaborations. By sequence comparisons, 51 107 unique genes were identified and 33 665 represented partial or full-length non-repeated coding regions 19. This provides updated and comprehensive data of expressed genes of cotton.
Transcriptome analysis has been increasingly applied to reveal physiological states of cells and to identify gene functions. Microarray is a powerful and high throughput tool to detect differentially regulated genes. By analyzing gene expression profiles of rosette leaves of A. thaliana, Stitt and co-workers demonstrated that sugars play a profound role in diurnal gene regulation 20. Comparisons of gene expression patterns with that of IRREGULAR XYLEM1 (IRX1) and IRREGULAR XYLEM3 (IRX3) have led to the identification of five novel genes of Arabidopsis involved in secondary cell wall synthesis 21; this work provides evidence that it is possible to predict the function of a gene through co-expression analysis. For crops, a successful example is the use of TOM1 cDNA array to analyze tomato transcriptomes, which identified 869 differentially expressed genes in developing fruits 22. Microarray also has been applied to cotton research. An investigation of gene expression profiles was conducted by using a 70-mer oligo-nucloetide chip containing ∼12 000 ESTs, which identified 2 553 genes that are downregulated to a certain degree in the secondary cell wall synthesis stage. Only 81 genes, however, were found to have higher expression levels in this stage 23. A comparative analysis of transcript profiles of a naked seed mutant (NIN1) and its isogenic cotton line showed that many fiber-associated genes are suppressed by mutation 24. The transcriptomes of cotton fibers and that of the fuzzless-lintless (fl) mutant ovules were recently compared; in combination with the results obtained from in vitro ovule culture, the authors proposed that ethylene plays a major role in fiber elongation 25. Another investigation of 0 DPA ovule transcriptomes of the wide-type and six reduced fiber or fiberless mutants showed that 13 genes were downregulated in mutants, including an MYB transcription factor (GhMyb25) and a homeodomain protein, providing information for parallels between cotton fiber initiation and leaf trichome development 26.
Fiber length and strength are both key traits of fiber quality. Investigation of different cotton cultivars showed that fiber length is largely determined by the length of the elongation stage, and that fiber strength is tightly correlated with secondary cell wall synthesis and the array of crystal cellulose 1. To survey the events in developing cotton fiber cells at these two stages, we examined expression patterns of over 5 000 genes by a cDNA array, and analyzed metabolites by gas chromatography/mass spectrum (GC/MS). We show that developmental stages of cotton fibers can be separated by profiles of transcripts and metabolites, respectively. Many signaling and metabolic pathways that are highly active in fast elongating fibers are repressed in the secondary cell wall formation stage, when a vast amount of cellulose is synthesized. Our data point to a specialization process of cotton fiber development toward cellulose synthesis and deposition.
Materials and Methods
Plant material
Upland cotton, Gossypium hirsutum L. cv. Xuzhou-142, was grown in field. Bolls were tagged and the flowering day was taken as 0 DPA. Ovules from 3 days pre-anthesis (−3 DPA) to 24 DPA were collected in the morning, and the material was frozen in liquid nitrogen and stored at −70 °C before use.
RNA extraction and library construction
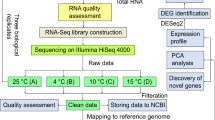
Fibers were deprived of the 3 to 18 DPA ovules in liquid nitrogen, followed by removing the ovule with forceps. The quality of fibers isolated from the 3 DPA ovules was also checked under a microscope. Total RNA was isolated from the −3 to 5 DPA ovules and the 6 to 24 DPA fibers by a cold-phenol method as described previously 27, 28. Poly (A)+RNA was purified by an Oligotex mRNA Midi kit (Qiagen, Valencia, CA, USA) according to the manufacturer's instruction, and cDNA libraries were constructed following the ZAP-cDNA Synthesis kit (Stratagene, La Jolla, CA, USA). The cDNA inserts were sequenced with T7 primer by the ABI3700 Automatic Sequencer. The clones having No Hits Found in BLAST searching were then sequenced by T3 primer.
For cDNA array hybridization, total RNAs were prepared by a hot borate method 29 from the 3 to 18 DPA fibers collected at a 3-day interval.
Microarray design and hybridization
Based on clustering and annotation, 5 122 unique EST clones were selected and their inserts were amplified by PCR, using T3 and T7 primers. House-keeping histones (H2A, H2B, H3 and H4) were included as internal controls. After purification and quantification, the PCR products were plotted onto the glass slides (Full Moon BioSystems, Sunnyvale, CA, USA) by GeneMachines OmniGrid Microarrayer (BST Group, Singapore).
RNAs from the 3-, 6-, 9-, 12-, 15- and 18 DPA fibers were compared to the 9 DPA fiber RNA for time course analysis. Probes preparation and slide hybridization were carried out by using the Micromax TSA Labeling and Detection kit (PerkinElmer, Wellesley, MA, USA), starting with 10 μg total RNA.
Image acquisition, data filtering and processing
Images were acquired by scanning the slides with a ScanArray 4000 Scanner (Packard BioChip Technologies, Meriden, CT, USA). Fluorescence signals were detected through ImaGene 5.6 (BioDiscovery, El Segundo, CA, USA). For each signal, mean value of the four repeats was taken. Signals lower than 2 000 were seen as low expression and omitted from further statistic analysis. All comparisons were carried out with four repeats (two biological and two technique repeats respectively). Log transformed normalized data were analyzed by fitting a mixed effects ANOVA model in MAANOVA package under R environment for multiple testing 30. In false discovery rate (FDR)-correction and Student's t-test analyses, the FDR-corrected p < 0.05, Student's t-test p < 0.05 and the average fold of change ≥ 2-fold were taken as standards for accessing differentially expressed genes. K-means cluster analysis was carried out on the stage-differential genes in the same software package. Principal component analysis (PCA) was performed using the software Matlab 7.0.4.
Identification of differentially regulated pathways was carried out using software KOBAS (for KEGG Orthology Based Annotation System) by setting cutoff values as e-values ≤1e-5, rank ≤10 and sequence identity ≥30% 31. The fatty acid metabolism pathway was enriched by adding lipase, fatty acid elongase, desaturase and very long-chain fatty acid condensing enzymes. After manually reviewing all identified plant metabolism pathways, p-value of a particular pathway was set following a hyper geometric distribution in the Matlab 7.0.4, and FDR correction was then applied to control the overall type I error rate of multiple hypotheses testing, using GeneTS (2.8.0) in the R (2.2.0) statistical software package 32, 33. Pathways with FDR-corrected q-values ≤0.05 were considered as differentially regulated.
Real-time RT-PCR
First-stand cDNA was synthesized from 1 μg total RNA. Histone H3 was used to normalize the template. Gene-specific primers were designed according to EST sequences using Primer Premier 5.0. Before analysis of gene expression, a standard curve was drawn for each pair of primers according to CT and concentration by Rotor-Gene 6.0 (Corbett Research, Australia) using 10× dilutions of modulates following a denaturing step from 60 to 95 °C, and primers without dimmers and with standard curve r2 > 0.95 were further applied to gene expression pattern analysis. Quantitative PCR was carried out by using the TaKaRa ExTaq R-PCR kit and SYBR green as the dye (TaKaRa, Dalian, China). The three-step method was used in the 35-cycle amplification following a denaturing process under Rotor-Gene 6.0. The expression level in the 9 DPA normal fiber was defined as 100. Each experiment was repeated at least three times. Gene-specific primers were available upon request.
Protein preparation and enzyme assay
Total proteins of cotton fiber cells for glucosidase assay were extracted as described 34. Fiber of 0.3 g was ground into fine powder in liquid nitrogen and then extracted with an ice-cold extraction buffer (HEPES-NaOH 0.05 M, MgCl2 0.01 M, Na2EDTA 0.001 M, DTT 0.0026 M, 10% ethylene glycol and 0.02% Triton X-100). After centrifugation, the liquid was desalted by Amicon ultra centrifugal filter 10 KD (Millipore Co., Cork, Ireland). The protein was quantified with the Protein Quatititative Analysis Kit (Shenergy Biocolor, Shanghai, China) and stored at −70 °C. The following substrates were purchased from Sigma (Sigma, St Louis, MO, USA): 4-nitrophenyl-β-L-arabinoside (N3512), 4-nitrophenyl-β-fucopyronoside (N3378), 4-nitrophenyl-β-galactopyronoside (N1252), 4-nitrophenyl-β-glucopyronoside (N7006) and 4-nitrophenyl-β-xylopyronoside (N2132). Proteins of 1 mg were added into a 390 μl PBS buffer containing 10 mM substrate and then incubated at 30 °C in a water bath. After reaction, the concentration of the product was determined spectrometrically at 402 nm.
Metabolite analysis
Metabolite extraction was carried out according to a published procedure 35. Ovule (3 DPA) or fiber of 0.3 g was ground in liquid nitrogen and rapidly added into 1.4 ml methanol to inactivate enzymes. Then water, ribitol (0.2 mg/ml in water) and nonadecanoic acid methyl ester (2 mg/ml in chloroform), 50 μl for each, were added, with the two solutes serving as internal standards. After preparation according to the protocol for plant leaf metabolite profiling (http://www.mpimp-golm.mpg.de/fiehn/forschung/blatt-protokoll-e.html), 2 μl samples were injected at a 1:25 split ratio into a GC/MS system (6890N Network GC System) and the MS data were obtained by the 5973 Mass Selective Detector (Agilent, Palo Alto, CA, USA). Peak identities were confirmed by National Institute of Standards and Technology (NIST) and Wiley libraries. Standards of fructose, glucose, galactosidase, sucrose and amino acids were also used for peak identification.
Results
Preparation of cotton fiber cDNA chip
We constructed a cDNA library of an upland cotton cultivar (G. hirsutum L. cv. Xuzhou-142) by using the fiber-containing ovules (-3 to 5 DPA) and the fibers (6-24 DPA), covering the developmental stages of fiber initiation/outgrowth, fast elongation and secondary cell wall synthesis. Through large-scale sequencing we generated ∼8 000 ESTs, of them 7 800 were sequenced from the 3′ end (by the T7 primer). These ESTs form 3 748 clusters with 2 496 singletons. In GenBank database, 4 188 ESTs have a BLASTX hit with an e-value less than 0.001. Based on Clusters of Orthologous Groups of proteins (COG) at NCBI (http://www.ncbi.nlm.nih.gov/COG) and KEGG Orthology (KO) at KEGG (http://www.genome.jp/dbget), these ESTs were classified according to their functional categories. They covered nearly all aspects of plant life ranging from genetic information maintenance to specific enzymes, with metabolism being the largest category of functions (1 210 genes). Of the plant hormone-related genes in the group of signal transduction and development, ethylene, auxin and GA-responsive genes are the major ones represented (Table 1).
Clustering assembly provided information to classify different ESTs in a statistical way. Some EST homologs in one cluster have different BLASTX hits in NCBI, which indicates that their differences occur in the coding region. To avoid loss of information, we selected 5 122 ESTs that differ in either cluster or BLASTX hits, and PCR products of these EST clones were arrayed to generate the cotton cDNA chip.
Separation of fiber developmental stages by transcript profiles
By using the cDNA chip, transcriptomes of cotton fiber cells at different developmental stages (3 to 18 DPA with a 3-day interval) were compared, with the 9 DPA fiber transcriptome as the reference. After hybridization, approximately 20% of the genes arrayed showed signal values <2 000, representing low (or no) expression. The remaining genes were considered expressed, among them 633 showed significant variations of expression levels in at least one combination, according to the analysis of variance (ANOVA, FDR < 0.05 and t-test < 0.05). These genes are hereafter called stage-differential genes (see Supplementary Table S1).
It is then of interest to see if fiber transcriptomes of different developmental stages are statistically different. The 2 030 genes expressed in all comparisons (stages) were then selected as variables for analysis. PCA divided the fiber samples of different harvesting times into four groups (Figure 1A). The first group is composed of 3 DPA only, representing fiber initiation and outgrowth; the second group includes three time points (6-, 9- and 12 DPA), representing the fast elongation stage; the third group contains one time point (15 DPA), spanning the overlapping stages of fast elongation and secondary cell wall biosynthesis; and the last group is formed by 18 DPA, when fiber elongation is completely ceased and secondary cell wall biosynthesis is highly active. These results demonstrate that developmental stages of cotton fiber cells can be recognized by their transcript profiles, and fiber cell at a certain stage has its own unique feature of transcriptome.
Statistical analysis of cDNA array data. (A) PCA of fiber samples. The expressed genes were used, and the 24 samples collected at different developmental stages were separated into four major groups. (B) K-means clustering of the stage-differential genes. X-axis indicates the DPA of sample collection, and Y-axis stands for gene expression levels relative to the 9-DPA fibers. (C) Distribution of functions of genes in different clusters. There are more hormone-responsive, signal transduction, transporter, peroxidase and fatty acid metabolism genes in clusters 1 and 2, whereas clusters 3 and 4 have more stress inducible, carbohydrate (particularly cellulose) metabolism and cell wall protein genes. Unknown and No-Hits-Found genes are not listed.
Based on K-means clustering of expression dynamics in developing fiber cells, the 633 stage-differential genes were classified into five groups, plus an unclassified group (Figure 1B). Genes in groups 1 and 2 have higher expression levels in the early stage (up to 9 DPA) than in the later stage (12 DPA and afterwards). Clearly, their expression has a close connection with fiber elongation. Genes of groups 3 and 4 show an opposite temporal expression pattern, as their expression levels in fibers are higher in the secondary cell wall synthesis stage (12-18 DPA) when compared to the elongation stage. The expression levels of group 5 genes are high at 3 DPA and decrease along with fiber development, and they are considered fiber initiation- and early elongation-related genes.
Classification by gene functions revealed that the fiber elongation-related groups (groups 1 and 2) have more information processing, hormone responsive, signal transduction, transporter, peroxidase and fatty acid metabolism genes, and the secondary cell wall-related groups (groups 3 and 4) are enriched in carbohydrate metabolism and cell wall protein genes (Figure 1C). This unbalanced distribution of functions reflects major physiological events of cell elongation and secondary cell wall formation of the fiber cells.
Changes of metabolites in developing fibers
Among the stage-differential genes, 372 have putative functions assigned, of which more than 1/3 are predicted to encode metabolic enzymes. Further investigation by KOBAS 27 mapped 1 116 out of the 5 122 cotton ESTs to 164 pathways, and 152 stage-differential genes to 85 pathways. After hyper geometric distribution and FDR corrections, seven metabolic pathways involving secondary metabolite, fatty acid and carbohydrate metabolisms show significant changes (q < 0.05) during cotton fiber development (Table 2).
We expect that this large portion of differential metabolism genes would be reflected by corresponding changes of metabolites in developing fiber cells. Metabolites from the 3 DPA fiber-containing ovules and 6 to 21 DPA fibers, again with a 3-day interval, were profiled by GC-MS, and those with a level higher than 1 mg/g fresh weight and an NIST fit higher than 700 in the MS data were selected for statistical analysis.
Among the lipid phase compounds, 37 metabolites (see Supplementary Table S2) were used for PCA, which then divided the samples into four groups. Four samples of 3 DPA form a group, which is most distantly related to others. The 6 and 9 DPA samples form the second group, representing fast elongating fibers. Samples from 12 to 18 DPA constitute a large group, suggesting a similar spectrum of lipophilic metabolites in a transitional period from the fast elongation to the secondary cell wall formation stages. The 21 DPA samples, representing the secondary cell wall formation stage, form another distinct group (Figure 2A). Hexadecanoic acid, octadecatrienoic acid and β-sitosterol are the most abundant lipids determined by weight in the 9 DPA fiber cells, representing 33%, 19% and 12% of the total lipid phase metabolites profiled, respectively (Figure 2B).
Metabolite profiles of cotton fibers of different developmental stages. (A) PCA of the samples based on metabolite profiles in the non-polar phase. The 3 DPA samples included ovule and fiber, and the others were of fiber cells only. Four major groups are formed. (B) Makeup of non-polar phase metabolites of the 9 DPA fiber. Hexadecanoic acid, octadecanoic acid, 9,12,15-octadecatrienoic acid and β-sitosterol are the major lipophilic metabolites according to their amounts by weight. (C) PCA of the samples based on metabolite profiles in the polar phase. (D) Makeup of polar phase metabolites of the 9 DPA fiber. Glucose, fructose, galactose and sucrose are the major polar components according to their amounts by weight, accounting for more than 90% of the total polar phase metabolites.
Based on similar parameters, 59 metabolites from the polar phase (see Supplementary Table S3) were employed for PCA, which also classified the samples into four groups, but with a slightly different grouping (Figure 2C). The 3 and 21 DPA samples again form two distinct groups well separated from others, those of elongating fibers (6-, 9-, 12-, and 15 DPA) are grouped together and those of 18 DPA are in an intermediate position between the group of fast elongation stage and the group of secondary cell wall formation stage (21 DPA). Carbohydrates are the major metabolites in the polar phase. In 9 DPA fibers, glucose, fructose, galactose and sucrose account for 50%, 33%, 5% and 7% of the total polar phase metabolites, respectively (Figure 2D).
Both polar and lipid phase metabolites identified the 3 and 21 DPA samples as distinct groups. Quantitative analysis showed that many components (e.g., sucrose) are present at a much higher level in 3 DPA samples; on the other hand, most metabolites are at very low levels in the 21 DPA fiber (see Supplementary Tables S2 and S3). In comparison with changes in gene expression, the metabolome dynamics exhibits a lag by approximately 3 days.
Genes upregulated during fiber elongation
The plant hormones auxin and gibberellin have been shown to promote fiber cell elongation in ovule culture systems 28. If the same occurs in planta, we would expect that genes induced by these two hormones are upregulated in the fast elongating fibers. Among the stage-differential genes, five putative auxin response genes are all highly expressed in the fast elongation stage (6- to 12 DPA), while their transcript levels are low both before and after the fast elongation stage (Figure 3A and 3B; see Supplementary Table S1). By contrast, expression of a putative auxin-repressed gene (homologous to AF336307) does not increase until the start of secondary cell wall synthesis. These data suggest that a high level of auxin signal is present in rapidly elongating fiber cells, and support the classical assumption that auxin plays a role in promoting cotton fiber elongation 36.
Expressions of selected stage-differential genes related to fiber elongation. (A, B) Auxin response genes. (A) Based on cDNA array, five genes of auxin responsive protein family members and two genes of auxin-related transcription factors (solid triangle) are upregulated in the fast elongation stage (6 to 12 DPA), and two auxin repressed genes (hollow diamond) show elevated expression in the secondary cell wall synthesis stage (from 12 DPA). (B) Expressions of three auxin responsive and one auxin repressed genes were also analyzed by real-time RT-PCR. 0: ovule (containing fiber initials) collected at 0 DPA; 3 to 18: DPA of fibers. (C) Expansin genes. Genes of four α-expansin proteins show a similar expression pattern with higher levels in the fiber outgrowth and fast elongation stages (solid triangle). The expression level of a β-expansin gene, however, increases at 18 DPA (hollow diamond). (D) Changes of fatty acid contents during fiber development. Levels of fatty acids decrease drastically at 21 DPA (t-test p < 0.0001). Data shown are the means of three biological repeats. Error bars indicate ± SD.
Aquaporins form protein complexes (water channels) across the membrane, and they facilitate water transport at a higher rate with less energy consumption than diffusion 37. Eight aquaporin-like genes are increasingly expressed in developing fiber cells from 3 to 15 DPA (see Supplementary information, Table S1). Metabolite analysis revealed a particular high level of sucrose in 3 DPA fiber cells, and this should result in high osmotic potential. The increased accumulation of aquaporins would allow water to enter fiber cells in an accelerated rate, leading to a high turgor pressure that in turn drives cell elongation. Two of the 10 aquaporin genes, however, do not show decreased expression at 18 DPA, and their role in fiber development needs further investigation.
For cell wall-loosening proteins, their overall expression dynamics shows an early-high and late-low pattern. Four genes that belong to the α-expansin family are highly expressed in the fiber outgrowth and fast elongation stages, and are generally downregulated when cells enter the secondary cell wall synthesis stage (Figure 3C). α-Expansin plays a role in cell wall-loosening and thus cell expansion 38, 39. This expression pattern shows that they play their role mainly in fast elongation stage. The expression of an At-Exp-Beta2.1 gene homolog, COT_CL_F12, increases at 18 DPA, indicating a different role from α-expansin (Figure 3C).
Lipid transfer proteins and lipid metabolism enzymes have been found to have a particularly high level of gene expression in fiber cells 8, 40, 41. Our array results revealed that these genes are upregulated mainly during fiber elongation. Expression levels of lipid biosynthesis genes rise from 6 DPA, and the high level is maintained throughout the fast elongation stage. Genes showing this expression pattern include those involved in the carbon chain increase cycle and later modification, such as the acyl-CoA-binding protein, fatty acid elongase, 3-ketoacyl-CoA synthase, β-ketoacyl-CoA synthase, ω-3 fatty acid desaturase and very-long-chain fatty acid condensing enzyme (see fast elongation stage 1). In accordance with the gene expression pattern, the total amount of fatty acids in fiber cells is greatly reduced in the 21 DPA fiber (Figure 3D and Supplementary Table S2).
Carbon fluxes in the secondary cell wall synthesis stage
Following the start of secondary cell wall synthesis, β-tubulin, cell wall protein and carbohydrate metabolism genes show increased expression levels in fiber cells. Here, we focus on the carbohydrate metabolism genes.
Mature cotton fiber is mainly composed of cellulose. Cellulose synthesis and degradation genes change their expression levels reciprocally from the fast elongation stage to the secondary cell wall synthesis stage. Five cellulose synthase genes show an increased level of expression in the secondary cell wall synthesis stage; by contrast, expression of a cellulase, which degrades (hydrolyzes) cellulose, is downregulated from 12 DPA and is further decreased at 18 DPA (Figure 4A and 4B).
Expressions of selected stage-differential genes related to secondary cell wall synthesis. (A, B) Cellulose metabolism. (A) Cellulose synthase genes (solid diamond) are markedly upregulated in the secondary cell wall synthesis stage; by contrast, a cellulase gene (hollow square) is downregulated. (B) Selected genes were also analyzed by real-time RT-PCR. (C, D) Sucrose metabolism. (C) Sucrose synthase genes (shown in hollow ring or triangles) show diverse changes in the secondary cell wall synthesis stage; by contrast, a vacuolar acid invertase gene (solid diamond) is downregulated. (D) The vacuolar acid invertase gene was also analyzed by real-time RT-PCR. (E) Changes of major carbohydrates during fiber development. Glucose and fructose maintain a high and stable level until 21 DPA when the level drops drastically (t-test p < 0.0001); sucrose amount is stable from 6 to 21 DPA. Data shown are the mean of three biological repeats. Error bars indicate ± SD.
Sucrose synthase and invertase are the major enzymes that catalyze sucrose conversion. Our array data show that genes of sucrose synthase family have a diverse expression pattern in developing fibers. Among the four genes arrayed, one is clearly downregulated at the secondary cell wall synthesis stage (18 DPA), whereas the other three are either slightly or dramatically upregulated at 18 DPA (Figure 4C), suggesting that these three sucrose synthases are involved in secondary cell wall synthesis. A putative vacuolar acid invertase gene (COT_CK_C03) shows high expression pattern related to the elongation stage according to our cDNA array and real-time RT-PCR results (Figure 4C and 4D); this invertase is likely active in fiber elongation stage.
A vast amount of cellulose synthesis requires an abundant supply of carbohydrate building blocks 42. Metabolite profiling showed that glucose accounts for ∼50% of the total amount of polar phase metabolites in rapidly elongating fiber cells, but its amount decreases from 9 to 21 DPA. Fructose level shows a less but similar change. In contrast to these two hexose moieties, sucrose maintains a relatively stable level in developing fibers; after a higher level at 3 DPA, there is no significant change from 6 to 21 DPA (Figure 4E). The dynamic changes of glucose content in developing fiber cells are more or less inversely correlated with the expression levels of cellulose synthase genes.
Pectin is a polysaccharide component of primary cell wall. Two genes of pectin esterase, which is involved in pectin hydrolysis into pectate, are drastically upregulated from 12 DPA and afterwards. On the other hand, two enzymes participating in pectin synthesis, UDP-glucose 6-dehydrogenase and UDP-D-glucuronate 4-epimerase, which turn UDP-glucose into UDP-D-glucuronate and then into UDP-galacturonate, respectively, are downregulated during secondary cell wall synthesis. UDP-D-galacturonate is the substrate for pectin biosynthesis (Figure 5A and 5B). These data are consistent with a previous report that pectin is decreased in both amount and molecular weight in cotton fiber cells of the late stage 43.
Metabolic pathways differentially regulated during fiber development. (A, B) Pectin and cellulose metabolism. (A) In the secondary cell wall synthesis stage (represented by 18 DPA), the pectin degradation genes are upregulated and the synthesis genes repressed, whereas the cellulose synthesis genes are upregulated. (B) Selected genes were also analyzed by real-time RT-PCR. (C, D) Glucose metabolism. (C) In the secondary cell wall synthesis stage, pathways converting carbohydrate polymers to smaller carbohydrates are enhanced, and pathways producing cellulose are promoted; those competing for glucose, including pectin and starch synthesis and TCA cycle, are repressed. (D) Selected genes were also analyzed by real-time RT-PCR. (E) Relative activities of glycosidases. The 18 DPA fiber proteins show significantly higher activities towards degrading p-nitrophenal conjugated arabinopyranoside, glucopyranoside and galactopyranoside than the 9 DPA fiber. The specific activities of the 9 DPA were taken as 100. (F) Schematic show of metabolite flux in fiber cells during secondary cell wall synthesis stage. Pathways of secondary metabolite, fatty acid and pectin biosynthesis are downregulated; carbon flux is directed to cellulose. Red: upregulation; blue: downregulation. Data shown are the means of three biological repeats. Error bars indicate ± SD. ** indicates very significant difference.
In addition to pectin esterase, other poly- or oligo-saccharide hydrolyzing enzymes are also upregulated at the transcriptional level in the secondary cell wall synthesis stage, represented by 18 DPA (Figure 5C and 5D). Having observed the dynamics of gene expression, we further investigated the changes of the enzyme activities. Enzyme assays of fiber cell extracts showed that specific activities of β-galactosidase, β-glucosidase and β-alabinosidase are increased by 16 to 30% from 9 to 18 DPA (Figure 5E). This provides further evidence for the accelerated degradation of poly- and oligo-saccharides. It is interesting to note that β-galactosidase, which participates in pectin degradation, has the highest activity among the enzymes assayed. Here, both gene expression patterns and enzyme activities point to active pectin degradation in fiber cells during secondary cell wall synthesis.
Besides repression of the pectin biosynthesis pathway, we found an inhibition of bypass pathway. The pectate generated from pectin degradation can be consumed in two directions. It can be degraded by galactosidase into galacturonate, and then converted into glucose derivatives through the pentose phosphate pathway. Alternatively, it is degraded by pectate lyase into 5-dehydro-4-deoxy-D-glucuronate, which is a termination end product. Our array data showed that genes controlling the two pathways have opposite regulations during fiber development. In contrast to pectin esterase and β-galactosidase that are upregulated in the secondary cell wall synthesis stage, pectate lyase genes are downregulated, thus the bypass pathway generating non-recycling monosaccharide is repressed (Figure 5A).
Discussion
Through transcriptome and metabolite analysis, we demonstrated that signaling and metabolic pathways are co-ordinated in cotton fiber cells to promote cell elongation in the early stage and to support cellulose synthesis in the later stage.
Transcriptome analysis through cDNA array showed that there are 633 genes that are differentially expressed in cotton fiber cells during development, accounting for more than 12.4% of the genes arrayed. It was reported that, in Arabidopsis, 49 genes (about 1% of the total arrayed) have at least a twofold change upon nitrate treatment 44. Comparisons of inflorescences among the five Arabidopsis floral organ mutants (apetala1, apetala2, apetala3, pistillata and agamous) and the wild type revealed 1 380 differential genes (≥2-fold change), which represent about 5% of the total genes (26 090) on the array 45. Using the Arabidopsis gene chip containing 8 100 genes, 2 781 genes were reported to be differentially regulated by at least two-fold between the single-cell typed guard cells and mesophyll cells 46. These ratios of differential genes seem to indicate that the simpler the material is, the more differential genes one may find, although there might be fewer expressed genes in total. Our cDNA array results further show that genes differentially regulated in cotton fiber during development are of great value in dissecting physiological events and developmental states of the cell.
Cotton fibers are single-celled, and their development in a boll is highly synchronous with four morphologically distinguishable, though overlapping, stages. These features make it feasible to follow the developmental course of cotton fiber in genomic and physiological studies. We showed that fibers of different physiological states can be separated by both gene expression and metabolite profiles, and their distribution patterns and groupings in statistics are diagnostic of developmental stages.
Mature cotton fiber, white in color, is composed of nearly pure cellulose; this requires not only the synthesis and deposition of a large amount of cellulose but also the clearance of other metabolites. Data gathered from transcript and metabolite profiles and from the enzyme assays all demonstrated the dynamic changes of metabolism network centering on cellulose synthesis during secondary cell wall synthesis. While cellulose synthesis is prevailing, many metabolism pathways that are active during fiber elongation are repressed, and secondary metabolism that is already low in fiber cells is further downregulated. In addition, hydrolysis of fatty acids and non-cellulose poly- and oligo-saccharides is upregulated, and the pathway producing carbohydrates that can be recycled into cellulose is favored. Thus, in the secondary cell wall synthesis stage, metabolic pathways are coordinated to direct carbon flux into cellulose (Figure 5F). Although the working hypothesis deduced from our transcriptome analysis needs to be experimentally tested, this type of metabolism regulation is likely common to many crop species whose seeds or fruits store a single or a few types of predominant metabolites, such as starch in cereal grains, fatty acids in oilseeds and sugars in fruits. A large amount of starch is stored in rice seed during seed maturation, and stage-specific genes involved in this process have been reported 47. In oilseed rape, oil bodies accumulate in large amounts later in developing embryos, when starch is degrading; at the same time, sucrose and hexoses are found to be mobilized for fatty acid synthesis via the oxidative pentose phosphate pathway 48, 49, 50. Dissection of the mechanism controlling carbon flux in developing seeds will provide valuable data for crop quality improvement.
Previous investigations of cotton fiber development by transcriptome analysis have isolated a large number of genes highly expressed at the elongation stage, but the number of upregulated genes in later stages is rather small 23, 25. For example, on the basis of a 12k G. arboreum array constructed from cDNAs of the 7-10 DPA fiber cells, 2 553 genes were found to have more or less a higher expression in the fast elongation stage, whereas only 81 showed an increased level of expression in the secondary cell wall synthesis stage 23. In our analysis, there are nearly equal numbers of genes (298 and 272) that show an upregulation in fiber cells of the fast elongation and the secondary cell wall synthesis stages, respectively. This even distribution of the two groups of genes is likely a result of the cDNA library used for our cDNA array, which covers the stages from early outgrowth to secondary cell wall formation. The strength of fiber is mainly determined by cellulose synthesis and deposition to the secondary cell wall, and transcriptome information at this stage provides new candidate genes for fiber improvement 51.
Accession numbers
Sequence data can be found in the GenBank libraries: DT046365 to DT054205 for ESTs sequenced with the T7 primer, DT054206 to DT054335 for those with the T3 primer. Microarray data were deposited into GEO database at NCBI according to MIAME guidelines under platform GPL3641 with series GSE4639. The samples have accession numbers from GSM104366 to GSM104379, GSM104383 to GSM 104392, GSM104394 and GSM104397 to GSM104398.
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
References
Applequist WL, Cronn R, Wendel JF . Comparative development of fiber in wild and cultivated cotton. Evol Dev 2001; 3:3–17.
Kim HJ, Triplett BA . Cotton fiber growth in planta and in vitro. Models for plant cell elongation and cell wall biogenesis. Plant Physiol 2001; 127:1361–1366.
Meinert MC, Delmer DP . Changes in biochemical composition of the cell wall in cotton fiber during development. Plant Physiol 1977; 59:1088–1097.
Smart LB, Vojdani F, Maeshima M, Wilkins Ta . Genes involved in osmoregulation during turgor-driven cell expansion of developing cotton fibers are differentially regulated. Plant Physiol 1998; 116:1539–1549.
Ruan YL, Llewellyn DJ, Furbank RT . Suppression of sucrose synthase gene expression represses cotton fiber cell initiation, elongation, and seed development. Plant Cell 2003; 15:952–964.
Orford SJ, Timmis JN . Specific expression of an expansin gene during elongation of cotton fibres. Biochim Biophys Acta 1998; 1398:342–346.
Harmer SE, Orford SJ, Timmis JN . Characterization of six alpha-expansin genes in Gossypium hirsutum (upland cotton). Mol Genet Genomics 2002; 268:1–9.
Orford SJ, Timmis JN . Expression of a lipid transfer protein gene family during cotton fibre development. Biochim Biophys Acta 2000; 1483:275–284.
Li XB, Fan XP, Wang XL, Cai L, Yang Wc . The cotton ACTIN1 gene is functionally expressed in fibers and participates in fiber elongation. Plant Cell 2005; 17:859–75.
Pear JR, Kawagoe Y, Schreckengost WE, Delmer DP, Stalker DM . Higher plants contain homologs of the bacterial celA genes encoding the catalytic subunit of cellulose synthase. Proc Natl Acad Sci USA 1996; 93:12637–12642.
Potikha TS, Collins CC, Johnson DI, Delmer DP, Levine A . The involvement of hydrogen peroxide in the differentiation of secondary walls in cotton fibers. Plant Physiol 1999; 119:849–858.
Kurek I, Kawagoe Y, Jacob-Wilk D, Doblin M, Delmer DP . Dimerization of cotton fiber cellulose synthase catalytic subunits occurs via oxidation of the zinc-binding domains. Proc Natl Acad Sci USA 2002; 99:11109–11114.
Persson S, Wei H, Milne J, Page GP, Somerville Cr . Identification of genes required for cellulose synthesis by regression analysis of public microarray data sets. Proc Natl Acad Sci USA 2005; 102:8633–8638.
Lacape JM, Nguyen TB, Thibivilliers S, et al. A combined RFLP-SSR-AFLP map of tetraploid cotton based on a Gossypium hirsutum ´ Gossypium barbadense backcross population. Genome 2003; 46:612–626.
Paterson AH, Saranga Y, Menz M, Jiang CX, Wright RJ . QTL analysis of genotype ´ environment interactions affecting cotton fiber quality. Theor Appl Genet 2003; 106:384–396.
Chee PW, Draye X, Jiang CX, et al. Molecular dissection of phenotypic variation between Gossypium hirsutum and Gossypium barbadense (cotton) by a backcross-self approach: III. Fiber length. Theor Appl Genet 2005; 111:772–781.
Han ZG, Wang CB, Song XL, et al. Characteristics, development and mapping of Gossypium hirsutum derived EST-SSRs in allotetraploid cotton. Theor Appl Genet 2006; 112:430–439.
Rong J, Pierce GJ, Waghmare VN, et al. Genetic mapping and comparative analysis of seven mutants related to seed fiber development in cotton. Theor Appl Genet 2005; 111:1137–46.19
Udall JA, Swanson JM, Haller K, et al. A global assembly of cotton ESTs. Genome Res 2006; 14:441–450.
Blasing OE, Gibon Y, Gunther M, et al. Sugars and circadian regulation make major contributions to the global regulation of diurnal gene expression in Arabidopsis. Plant Cell 2005; 17:3257–3281.
Brown DM, Zeef LA, Ellis J, Goodacre R, Turner Sr . Identification of novel genes in Arabidopsis involved in secondary cell wall formation using expression profiling and reverse genetics. Plant Cell 2005; 17:2281–2295.
Alba R, Payton P, Fei Z, et al. Transcriptome and selected metabolite analyses reveal multiple points of ethylene control during tomato fruit development. Plant Cell 2005; 17:2954–2965.
Arpat AB, Waugh M, Sullivan JP, et al. Functional genomics of cell elongation in developing cotton fibers. Plant Mol Biol 2004; 54:911–929.
Lee JJ, Hassan OS, Gao W, et al. Developmental and gene expression analyses of a cotton naked seed mutant. Planta 2006; 223:418–432.
Shi YH, Zhu SW, Mao XZ, et al. Transcriptome profiling, molecular biological and physiological studies reveal a major role for ethylene in cotton fiber cell elongation. Plant Cell 2006; 18:651–664.
Wu Y, Machado AC, White RG, Llewellyn DJ, Dennis ES . Expression profiling identifies genes expressed early during lint fibre initiation in cotton. Plant Cell Physiol 2006; 47:107–127.
Li CH, Zhu YQ, Meng YL, et al. Isolation of genes preferentially expressed in cotton fibers by cDNA filter arrays and RT-PCR. Plant Sci 2002; 163:1113–1120.
Wang S, Wang JW, Yu N, et al. Control of plant trichome development by a cotton fiber MYB gene. Plant Cell 2004; 16:2323–2334.
Wan CY, Wilkins TA . A modified hot borate method significantly enhances the yield of high-quality RNA from cotton (Gossypium hirsutum L.). Anal Biochem 1994; 223:7–12.
Wu H, Kerr MK, Cui XQ, Churchill GA . MAANOVA: a software package for the analysis of spotted cDNA microarray experiments in the analysis of gene expression data: an overview of methods and software. Parmigiani G, Garret ES, Irizarry RA, Zeger SL, eds. Springer, New York. 2003.
Mao X, Cai T, Olyarchuk JG, Wei L . Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 2005; 21:3787–3793.
Wichert S, Fokianos K, Strimmer K . Identifying periodically expressed transcripts in microarray time series data. Bioinformatics 2004; 20:5–20.
Storey JD . A direct approach to false discovery rates. J Roy Stat Soc B 2002; 64:479–498.
Pelleschi S, Rocher JP, Prioul JL . Effect of water restriction on carbohydrate metabolism and photosynthesis in mature maize leaves. Plant Cell Environ 1997; 20:493–503.
Fiehn O, Kopka J, Dormann P, Altmann T, Trethewey RN, Willmitzer L . Metabolite profiling for plant functional genomics. Nat Biotechnol 2000; 18:1157–1161.
Beasley CA, Ting IP . Effects of plant growth substances on in vitro fiber development from fertilized cotton ovules. Am J Bot 1973; 60:130–139.
Connolly DL, Shanahan CM, Weissberg PL . The aquaporins. A family of water channel proteins. Int J Biochem Cell Biol 1998; 30:169–172.
Cosgrove DJ . New genes and new biological roles for expansins. Curr Opin Plant Biol 2000; 3:73–78.
Cosgrove DJ, Li LC, Cho HT, Hoffmann-Benning S, Moore RC, Blecker D . The growing world of expansins. Plant Cell Physiol 2002; 43:1436–1444.
Song P, Allen RD . Identification of a cotton fiber-specific acyl carrier protein cDNA by differential display. Biochim Biophys Acta 1997; 1351:305–312.
Ji SJ, Lu YC, Feng JX, et al. Isolation and analyses of genes preferentially expressed during early cotton fiber development by subtractive PCR and cDNA array. Nucleic Acids Res 2003; 31:2534–2543.
Delmer DP, Haigler CH . The regulation of metabolic flux to cellulose, a major sink for carbon in plants. Metab Eng 2002; 4:22–28.
Tokumoto H, Wakabayashi K, Kamisaka S, Hoson T . Changes in the sugar composition and molecular mass distribution of matrix polysaccharides during cotton fiber development. Plant Cell Physiol 2002; 43:411–418.
Wang R, Guegler K, LaBrie ST, Crawford NM . Genomic analysis of a nutrient response in Arabidopsis reveals diverse expression patterns and novel metabolic and potential regulatory genes induced by nitrate. Plant Cell 2000; 12:1491–1509.
Wellmer F, Riechmann JL, Alves-Ferreira M, Meyerowitz EM . Genome-wide analysis of spatial gene expression in Arabidopsis flowers. Plant Cell 2004; 16:1314–1326.
Leonhardt N, Kwak JM, Robert N, Waner D, Leonhardt G, Schroeder JI . Microarray expression analyses of Arabidopsis guard cells and isolation of a recessive abscisic acid hypersensitive protein phosphatase 2C mutant. Plant Cell 2004; 16:596–615.
Ohdan T, Francisco PB, Sawada T, et al. Expression profiling of genes involved in starch synthesis in sink and source organs of rice. J Exp Bot 2005; 56:3229–3244.
Hill LM, Morley-Smith ER, Rawsthorne S . Metabolism of sugars in the endosperm of developing seeds of oilseed rape. Plant Physiol 2003; 131:228–236.
Hills MJ . Control of storage-product synthesis in seeds. Curr Opin Plant Biol 2004; 7:302–308.
Hutchings D, Rawsthorne S, Emes MJ . Fatty acid synthesis and the oxidative pentose phosphate pathway in developing embryos of oilseed rape (Brassica napus L.). J Exp Bot 2005; 56:577–585.
Haigler CH, Zhang DH, Wilkerson CG . Biotechnological improvement of cotton fibre maturity. Physiol Plantarum 2005; 124:285–294.
Acknowledgements
We thank Hong-You Qin (Shanghai Huaguan BioChip Co., Ltd, Shanghai, China) for help on cDNA array, and Bin Luo (Institute of Plant Physiology and Ecology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences) for comments on the manuscript preparation. This research was supported by Hi-Tech Research and Development Program of China (2006AA10Z102), National Basic Research Program of China (2002CB111303), The National Transgenic Plant Program (JY03-A-02) and Chinese Academy of Sciences (KSCX2-SW-329).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Gou, JY., Wang, LJ., Chen, SP. et al. Gene expression and metabolite profiles of cotton fiber during cell elongation and secondary cell wall synthesis. Cell Res 17, 422–434 (2007). https://doi.org/10.1038/sj.cr.7310150
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.cr.7310150
Keywords
This article is cited by
-
A comprehensive overview of cotton genomics, biotechnology and molecular biological studies
Science China Life Sciences (2023)
-
Coordination of floral and fiber development in cotton (Gossypium) by hormone- and flavonoid-signalling associated regulatory miRNAs
Plant Molecular Biology (2023)
-
Dynamic 3D genome architecture of cotton fiber reveals subgenome-coordinated chromatin topology for 4-staged single-cell differentiation
Genome Biology (2022)
-
AAQSP increases mapping resolution of stable QTLs through applying NGS-BSA in multiple genetic backgrounds
Theoretical and Applied Genetics (2022)
-
In silico analysis and expression profiling of Expansin A4, BURP domain protein RD22-like and E6-like genes associated with fiber quality in cotton
Molecular Biology Reports (2022)